Targeted Quantification of Carbon Metabolites Identifies Metabolic Progression Markers and an Undiagnosed Case of SDH-Deficient Clear Cell Renal Cell Carcinoma in a German Cohort
Abstract
:1. Introduction
2. Results
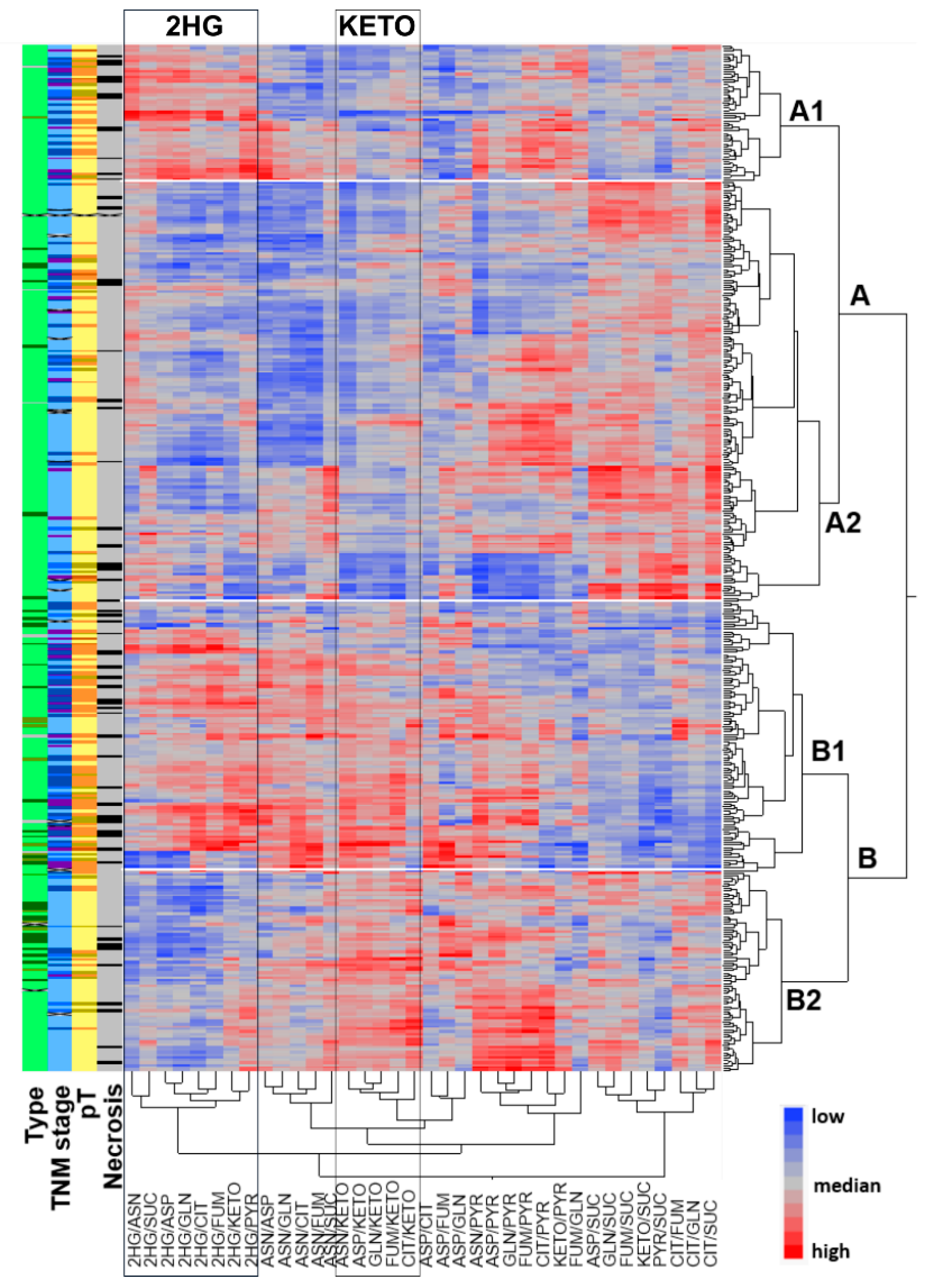
2.1. Metabolic Profiling of 420 RCCs Identified Two Main Clusters Separated by α-KG Levels and Two Sub-Clusters with Differential Concentrations of 2HG
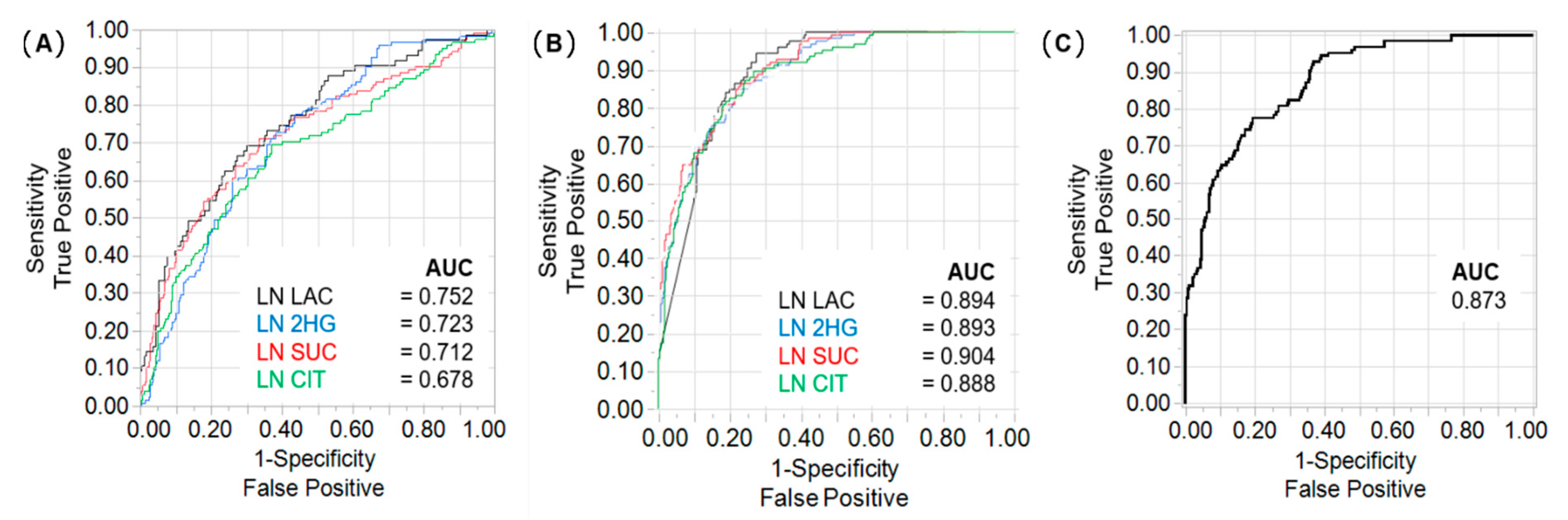
2.2. Lactate, 2HG, Succinate, and Citrate Are Independent Predictors of Tumour Stage and CSS
2.3. Differences in Prognostic Metabolites between Patients with ccRCC and papRCC
2.4. Metabologenomics Identified a Previously Undiagnosed Case of SDH-Deficient RCC
3. Discussion
4. Materials and Methods
4.1. Patient Cohort and Sample Collection
4.2. Metabolite Profile
4.3. Genetic Testing and Variant Classification
4.4. Western Blot
4.5. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Escudier, B.; Porta, C.; Schmidinger, M.; Rioux-Leclercq, N.; Bex, A.; Khoo, V.; Grunwald, V.; Gillessen, S.; Horwich, A. Renal cell carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-updagger. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol./ESMO 2019, 30, 706–720. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.; Swanson, A.A.; Chen, Y.B.; Lopez, T.; Milosevic, D.; Kipp, B.R.; Leibovich, B.C.; Thompson, R.H.; Herrera-Hernandez, L.; Cheville, J.C.; et al. Incidence of succinate dehydrogenase and fumarate hydratase-deficient renal cell carcinoma based on immunohistochemical screening with SDHA/SDHB and FH/2SC. Hum. Pathol. 2019, 91, 114–122. [Google Scholar] [CrossRef]
- Gill, A.J.; Hes, O.; Papathomas, T.; Sedivcova, M.; Tan, P.H.; Agaimy, A.; Andresen, P.A.; Kedziora, A.; Clarkson, A.; Toon, C.W.; et al. Succinate dehydrogenase (SDH)-deficient renal carcinoma: A morphologically distinct entity: A clinicopathologic series of 36 tumors from 27 patients. Am. J. Surg. Pathol. 2014, 38, 1588–1602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Launonen, V.; Vierimaa, O.; Kiuru, M.; Isola, J.; Roth, S.; Pukkala, E.; Sistonen, P.; Herva, R.; Aaltonen, L.A. Inherited susceptibility to uterine leiomyomas and renal cell cancer. Proc. Natl. Acad. Sci. USA 2001, 98, 3387–3392. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricketts, C.J.; De Cubas, A.A.; Fan, H.; Smith, C.C.; Lang, M.; Reznik, E.; Bowlby, R.; Gibb, E.A.; Akbani, R.; Beroukhim, R.; et al. The Cancer Genome Atlas Comprehensive Molecular Characterization of Renal Cell Carcinoma. Cell Rep. 2018, 23, 313–326.e315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, L.; Huang, Z.; Gao, Y.; Yan, X.; Xing, J.; Hang, W. LC-MS based serum metabonomic analysis for renal cell carcinoma diagnosis, staging, and biomarker discovery. J. Proteome Res. 2011, 10, 1396–1405. [Google Scholar] [CrossRef]
- Hakimi, A.A.; Reznik, E.; Lee, C.H.; Creighton, C.J.; Brannon, A.R.; Luna, A.; Aksoy, B.A.; Liu, E.M.; Shen, R.; Lee, W.; et al. An Integrated Metabolic Atlas of Clear Cell Renal Cell Carcinoma. Cancer Cell 2016, 29, 104–116. [Google Scholar] [CrossRef] [Green Version]
- Falegan, O.S.; Ball, M.W.; Shaykhutdinov, R.A.; Pieroraio, P.M.; Farshidfar, F.; Vogel, H.J.; Allaf, M.E.; Hyndman, M.E. Urine and Serum Metabolomics Analyses May Distinguish between Stages of Renal Cell Carcinoma. Metabolites 2017, 7, 6. [Google Scholar] [CrossRef] [Green Version]
- Poplawski, P.; Tohge, T.; Boguslawska, J.; Rybicka, B.; Tanski, Z.; Trevino, V.; Fernie, A.R.; Piekielko-Witkowska, A. Integrated transcriptomic and metabolomic analysis shows that disturbances in metabolism of tumor cells contribute to poor survival of RCC patients. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 744–752. [Google Scholar] [CrossRef] [PubMed]
- Jing, L.; Guigonis, J.M.; Borchiellini, D.; Durand, M.; Pourcher, T.; Ambrosetti, D. LC-MS based metabolomic profiling for renal cell carcinoma histologic subtypes. Sci. Rep. 2019, 9, 15635. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Kawasaki, Y.; Maekawa, M.; Takasaki, S.; Saigusa, D.; Ota, H.; Shimada, S.; Yamashita, S.; Mitsuzuka, K.; Yamaguchi, H.; et al. Value of global metabolomics in association with diagnosis and clinicopathological factors of renal cell carcinoma. Int. J. Cancer 2019, 145, 484–493. [Google Scholar] [CrossRef]
- Richter, S.; Gieldon, L.; Pang, Y.; Peitzsch, M.; Huynh, T.; Leton, R.; Viana, B.; Ercolino, T.; Mangelis, A.; Rapizzi, E.; et al. Metabolome-guided genomics to identify pathogenic variants in isocitrate dehydrogenase, fumarate hydratase, and succinate dehydrogenase genes in pheochromocytoma and paraganglioma. Genet. Med. Off. J. Am. Coll. Med. Genet. 2019, 21, 705–717. [Google Scholar] [CrossRef] [PubMed]
- Pignot, G.; Elie, C.; Conquy, S.; Vieillefond, A.; Flam, T.; Zerbib, M.; Debre, B.; Amsellem-Ouazana, D. Survival analysis of 130 patients with papillary renal cell carcinoma: Prognostic utility of type 1 and type 2 subclassification. Urology 2007, 69, 230–235. [Google Scholar] [CrossRef]
- Gieldon, L.; William, D.; Hackmann, K.; Jahn, W.; Jahn, A.; Wagner, J.; Rump, A.; Bechmann, N.; Nolting, S.; Knosel, T.; et al. Optimizing Genetic Workup in Pheochromocytoma and Paraganglioma by Integrating Diagnostic and Research Approaches. Cancers 2019, 11, 809. [Google Scholar] [CrossRef] [Green Version]
- Briston, T.; Stephen, J.M.; Thomas, L.W.; Esposito, C.; Chung, Y.L.; Syafruddin, S.E.; Turmaine, M.; Maddalena, L.A.; Greef, B.; Szabadkai, G.; et al. VHL-Mediated Regulation of CHCHD4 and Mitochondrial Function. Front. Oncol. 2018, 8, 388. [Google Scholar] [CrossRef] [Green Version]
- Cancer Genome Atlas Research, N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013, 499, 43–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shim, E.H.; Livi, C.B.; Rakheja, D.; Tan, J.; Benson, D.; Parekh, V.; Kho, E.Y.; Ghosh, A.P.; Kirkman, R.; Velu, S.; et al. L-2-Hydroxyglutarate: An epigenetic modifier and putative oncometabolite in renal cancer. Cancer Discov. 2014, 4, 1290–1298. [Google Scholar] [CrossRef] [Green Version]
- Lucarelli, G.; Galleggiante, V.; Rutigliano, M.; Sanguedolce, F.; Cagiano, S.; Bufo, P.; Lastilla, G.; Maiorano, E.; Ribatti, D.; Giglio, A.; et al. Metabolomic profile of glycolysis and the pentose phosphate pathway identifies the central role of glucose-6-phosphate dehydrogenase in clear cell-renal cell carcinoma. Oncotarget 2015, 6, 13371–13386. [Google Scholar] [CrossRef] [Green Version]
- Courtney, K.D.; Bezwada, D.; Mashimo, T.; Pichumani, K.; Vemireddy, V.; Funk, A.M.; Wimberly, J.; McNeil, S.S.; Kapur, P.; Lotan, Y.; et al. Isotope Tracing of Human Clear Cell Renal Cell Carcinomas Demonstrates Suppressed Glucose Oxidation In Vivo. Cell Metab. 2018, 28, 793–800.e792. [Google Scholar] [CrossRef] [Green Version]
- Semenza, G.L. HIF-1 mediates the Warburg effect in clear cell renal carcinoma. J. Bioenerg. Biomembr. 2007, 39, 231–234. [Google Scholar] [CrossRef]
- Girgis, H.; Masui, O.; White, N.M.; Scorilas, A.; Rotondo, F.; Seivwright, A.; Gabril, M.; Filter, E.R.; Girgis, A.H.; Bjarnason, G.A.; et al. Lactate dehydrogenase A is a potential prognostic marker in clear cell renal cell carcinoma. Mol. Cancer 2014, 13, 101. [Google Scholar] [CrossRef] [Green Version]
- Shen, J.; Chen, Z.; Zhuang, Q.; Fan, M.; Ding, T.; Lu, H.; He, X. Prognostic Value of Serum Lactate Dehydrogenase in Renal Cell Carcinoma: A Systematic Review and Meta-Analysis. PLoS ONE 2016, 11, e0166482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlo, M.I.; Mukherjee, S.; Mandelker, D.; Vijai, J.; Kemel, Y.; Zhang, L.; Knezevic, A.; Patil, S.; Ceyhan-Birsoy, O.; Huang, K.C.; et al. Prevalence of Germline Mutations in Cancer Susceptibility Genes in Patients With Advanced Renal Cell Carcinoma. JAMA Oncol. 2018, 4, 1228–1235. [Google Scholar] [CrossRef] [Green Version]
- Wallace, P.W.; Conrad, C.; Bruckmann, S.; Pang, Y.; Caleiras, E.; Murakami, M.; Korpershoek, E.; Zhuang, Z.; Rapizzi, E.; Kroiss, M.; et al. Metabolomics, machine learning and immunohistochemistry to predict succinate dehydrogenase mutational status in phaeochromocytomas and paragangliomas. J. Pathol. 2020, 251, 378–387. [Google Scholar] [CrossRef]
- Williamson, S.R.; Gill, A.J.; Argani, P.; Chen, Y.B.; Egevad, L.; Kristiansen, G.; Grignon, D.J.; Hes, O. Report From the International Society of Urological Pathology (ISUP) Consultation Conference on Molecular Pathology of Urogenital Cancers: III: Molecular Pathology of Kidney Cancer. Am. J. Surg. Pathol. 2020, 44, e47–e65. [Google Scholar] [CrossRef] [PubMed]
- Srigley, J.R.; Delahunt, B.; Eble, J.N.; Egevad, L.; Epstein, J.I.; Grignon, D.; Hes, O.; Moch, H.; Montironi, R.; Tickoo, S.K.; et al. The International Society of Urological Pathology (ISUP) Vancouver Classification of Renal Neoplasia. Am. J. Surg. Pathol. 2013, 37, 1469–1489. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. Off. J. Am. Coll. Med. Genet. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Povysil, G.; Tzika, A.; Vogt, J.; Haunschmid, V.; Messiaen, L.; Zschocke, J.; Klambauer, G.; Hochreiter, S.; Wimmer, K. panelcn.MOPS: Copy-number detection in targeted NGS panel data for clinical diagnostics. Hum. Mutat. 2017, 38, 889–897. [Google Scholar] [CrossRef] [PubMed]



| Feature | Details |
|---|---|
| Age at resection | 68 (27–90) years 1 |
| Sex | 36% females |
| Tumour type | 360 clear cell; 38 papillary (type 1 and 2); 6 mixed clear cell/papillary; 13 chromophobe; 1 mixed chromophobe/clear cell; 1 mucinous tubular/spindle cell; 1 unknown |
| Primary tumour stage | 266 pT1; 40 pT2; 105 pT3; 8 pT4; 1 unknown |
| TNM stage | 247 stage I; 33 stage II; 81 stage III; 44 stage IV; 15 unknown |
| Necrosis | 16.0% (67/419, 1 unknown) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
William, D.; Erdmann, K.; Ottemöller, J.; Mangelis, A.; Conrad, C.; Peitzsch, M.; Schröck, E.; Eisenhofer, G.; Zacharis, A.; Füssel, S.; et al. Targeted Quantification of Carbon Metabolites Identifies Metabolic Progression Markers and an Undiagnosed Case of SDH-Deficient Clear Cell Renal Cell Carcinoma in a German Cohort. Metabolites 2021, 11, 764. https://doi.org/10.3390/metabo11110764
William D, Erdmann K, Ottemöller J, Mangelis A, Conrad C, Peitzsch M, Schröck E, Eisenhofer G, Zacharis A, Füssel S, et al. Targeted Quantification of Carbon Metabolites Identifies Metabolic Progression Markers and an Undiagnosed Case of SDH-Deficient Clear Cell Renal Cell Carcinoma in a German Cohort. Metabolites. 2021; 11(11):764. https://doi.org/10.3390/metabo11110764
Chicago/Turabian StyleWilliam, Doreen, Kati Erdmann, Jonas Ottemöller, Anastasios Mangelis, Catleen Conrad, Mirko Peitzsch, Evelin Schröck, Graeme Eisenhofer, Aristeidis Zacharis, Susanne Füssel, and et al. 2021. "Targeted Quantification of Carbon Metabolites Identifies Metabolic Progression Markers and an Undiagnosed Case of SDH-Deficient Clear Cell Renal Cell Carcinoma in a German Cohort" Metabolites 11, no. 11: 764. https://doi.org/10.3390/metabo11110764
APA StyleWilliam, D., Erdmann, K., Ottemöller, J., Mangelis, A., Conrad, C., Peitzsch, M., Schröck, E., Eisenhofer, G., Zacharis, A., Füssel, S., Aust, D., Klink, B., & Richter, S. (2021). Targeted Quantification of Carbon Metabolites Identifies Metabolic Progression Markers and an Undiagnosed Case of SDH-Deficient Clear Cell Renal Cell Carcinoma in a German Cohort. Metabolites, 11(11), 764. https://doi.org/10.3390/metabo11110764