Current Concepts in Pharmacometabolomics, Biomarker Discovery, and Precision Medicine
Abstract
1. Introduction
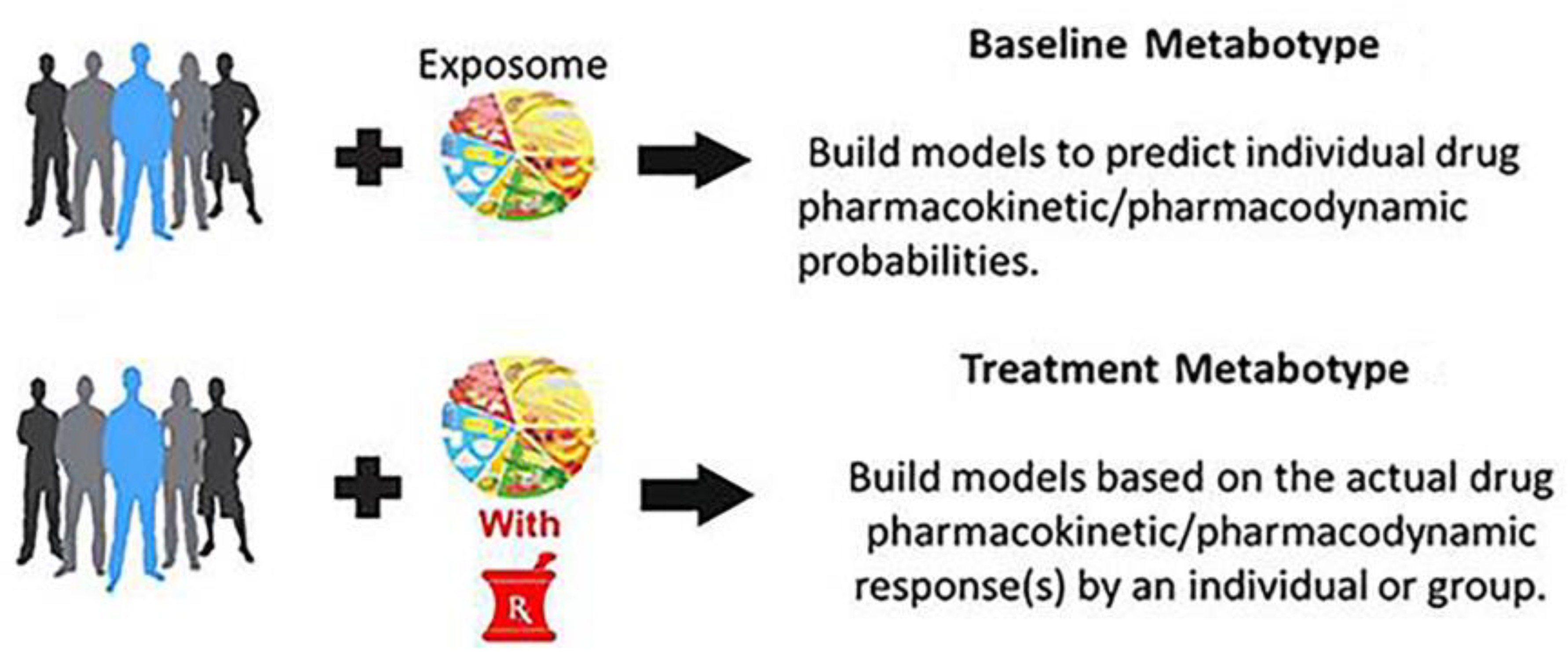
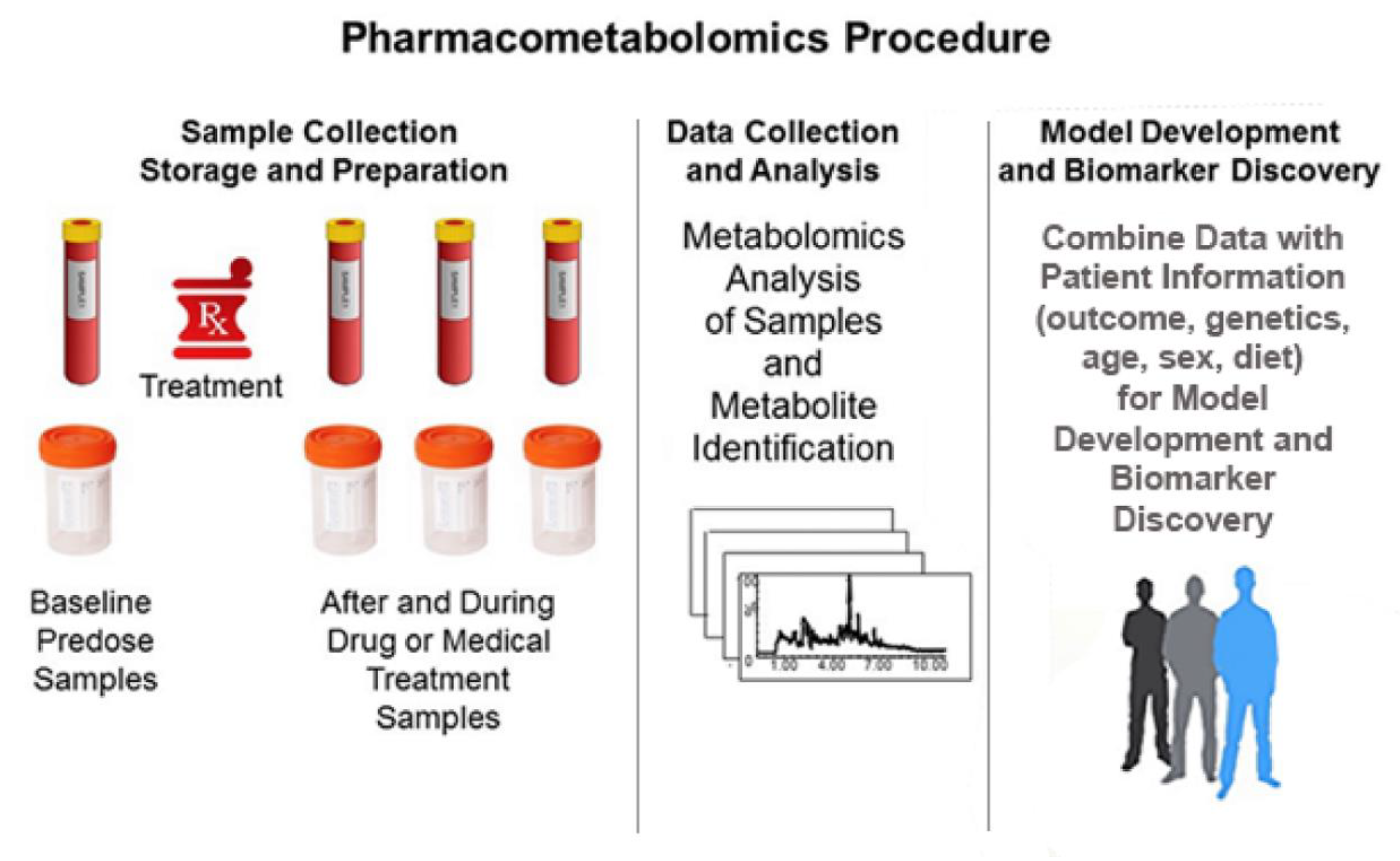
2. Pharmacometabolomics and “Metabotypes”
2.1. PMx Data Alone
2.2. PMx Data and PGx Data
2.3. PMx Data and Gut Flora Metagenomics Data
2.4. PMx Data and Multi-Scale Omics Data
3. Gut Microflora Metagenome and Drug Metabolism
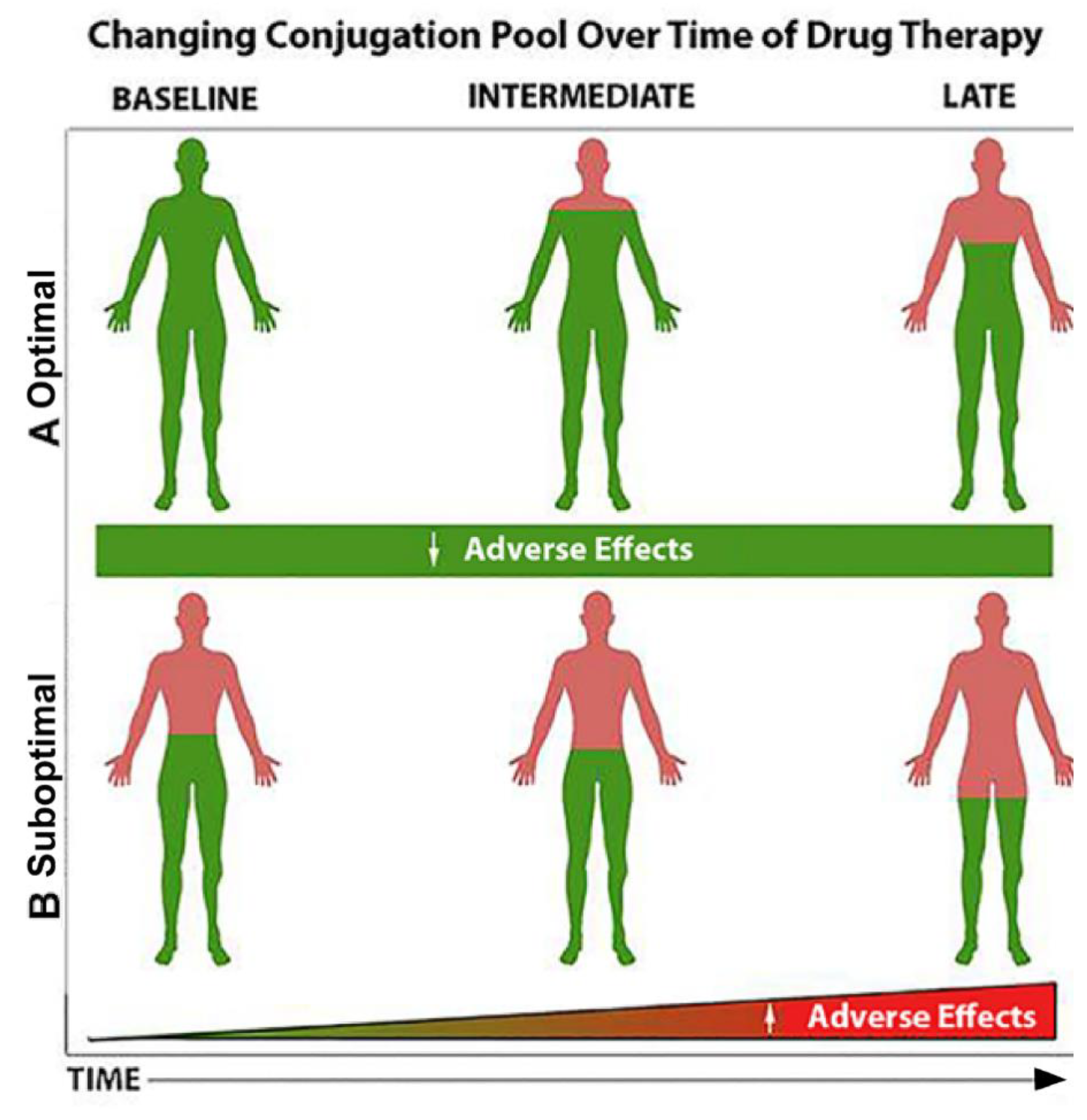
4. Where We are Today and the Future of PMx
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wilkinson, G.R. Drug metabolism and variability among patients in drug response. N. Engl. J. Med. 2005, 352, 2211–2221. [Google Scholar] [CrossRef] [PubMed]
- Lazarou, J.; Pomeranz, B.H.; Corey, P.N. Incidence of adverse drug reactions in hospitalized patientsa meta-analysis of prospective studies. JAMA 1998, 279, 1200–1205. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, G.; Mohorn, P.; Yacoub, K.; May, D.W. Adverse drug reaction deaths reported in United States vital statistics, 1999–2006. Ann. Pharmacother. 2012, 46, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Mancinelli, L.; Cronin, M.; Sadée, W. Pharmacogenomics: The promise of personalized medicine. AAPS Pharmsci. 2000, 2, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Pirmohamed, M. Pharmacogenetics and pharmacogenomics. Br. J. Clin. Pharmacol. 2001, 52, 345–347. [Google Scholar] [CrossRef] [PubMed]
- Daly, A.K. Genome-wide association studies in pharmacogenomics. Nat. Rev. Genet. 2010, 11, 241–246. [Google Scholar] [CrossRef]
- Roden, D.M.; Wilke, R.A.; Kroemer, H.K.; Stein, C.M. Pharmacogenomics. Circulation 2011, 123, 1661–1670. [Google Scholar] [CrossRef]
- Table of Pharmacogenomic Biomarkers in Drug Labelin. Available online: https://www.fda.gov/drugs/science-research-drugs/table-pharmacogenomic-biomarkers-drug-labeling (accessed on 1 March 2020).
- Ventola, C.L. Role of pharmacogenomic biomarkers in predicting and improving drug response: Part 1: The clinical significance of pharmacogenetic variants. P T A Peer-Rev. J. Formul. Manag. 2013, 38, 545–560. [Google Scholar]
- Clayton, T.A.; Lindon, J.C.; Cloarec, O.; Antti, H.; Charuel, C.; Hanton, G.; Provost, J.P.; Le Net, J.L.; Baker, D.; Walley, R.J.; et al. Pharmaco-metabonomic phenotyping and personalized drug treatment. Nature 2006, 440, 1073–1077. [Google Scholar] [CrossRef]
- Lindon, J.C.; Nicholson, J.K.; Holmes, E.; Antti, H.; Bollard, M.E.; Keun, H.; Beckonert, O.; Ebbels, T.M.; Reily, M.D.; Robertson, D.; et al. Contemporary issues in toxicology the role of metabonomics in toxicology and its evaluation by the COMET project. Toxicol. Appl. Pharmacol. 2003, 187, 137–146. [Google Scholar] [CrossRef]
- Ebbels, T.M.; Keun, H.C.; Beckonert, O.P.; Bollard, M.E.; Lindon, J.C.; Holmes, E.; Nicholson, J.K. Prediction and classification of drug toxicity using probabilistic modeling of temporal metabolic data: The consortium on metabonomic toxicology screening approach. J. Proteome Res. 2007, 6, 4407–4422. [Google Scholar] [CrossRef] [PubMed]
- Kaddurah-Daouk, R.; Kristal, B.S.; Weinshilboum, R.M. Metabolomics: A global biochemical approach to drug response and disease. Annu. Rev. Pharmacol. Toxicol. 2008, 48, 653–683. [Google Scholar] [CrossRef] [PubMed]
- Kaddurah-Daouk, R.; McEvoy, J.; Baillie, R.A.; Lee, D.; Yao, J.K.; Doraiswamy, P.M.; Krishnan, K.R. Metabolomic mapping of atypical antipsychotic effects in schizophrenia. Mol. Psychiatry 2007, 12, 934–945. [Google Scholar] [CrossRef] [PubMed]
- Pharmacometabolomics Research Network. Available online: http://pharmacometabolomics.duhs.duke.edu/ (accessed on 1 March 2020).
- Pharmacogenomics Research Network. Available online: https://www.pgrn.org (accessed on 1 March 2020).
- Kaddurah-Daouk, R.; Weinshilboum, R.M.; Network, P.R. Pharmacometabolomics: Implications for clinical pharmacology and systems pharmacology. Clin. Pharmacol. Ther. 2014, 95, 154–167. [Google Scholar] [CrossRef] [PubMed]
- Kaddurah-Daouk, R.; Weinshilboum, R. Pharmacometabolomics research network. metabolomic signatures for drug response phenotypes: Pharmacometabolomics enables precision medicine. Clin. Pharmacol. Ther. 2015, 98, 71–75. [Google Scholar]
- Kaddurah-Daouk, R.; Baillie, R.A.; Zhu, H.; Zeng, Z.B.; Wiest, M.M.; Nguyen, U.T.; Wojnoonski, K.; Watkins, S.M.; Trupp, M.; Krauss, R.M. Enteric microbiome metabolites correlate with response to simvastatin treatment. PLoS ONE 2011, 6, e25482. [Google Scholar] [CrossRef]
- Kaddurah-Daouk, R.; Hankemeier, T.; Scholl, E.H.; Baillie, R.; Harms, A.; Stage, C.; Dalhoff, K.P.; Jűrgens, G.; Taboureau, O.; Nzabonimpa, G.S.; et al. Pharmacometabolomics Informs About Pharmacokinetic Profile of Methylphenidate. CPT Pharmacomet. Syst. Pharm. 2018, 7, 525–533. [Google Scholar] [CrossRef]
- Wikoff, W.R.; Frye, R.F.; Zhu, H.; Gong, Y.; Boyle, S.; Churchill, E.; Cooper-Dehoff, R.M.; Beitelshees, A.L.; Chapman, A.B.; Fiehn, O.; et al. Pharmacometabolomics reveals racial differences in response to atenolol treatment. PLoS ONE 2013, 8, e57639. [Google Scholar] [CrossRef]
- Elbadawi-Sidhu, M.; Baillie, R.A.; Zhu, H.; Chen, Y.D.; Goodarzi, M.O.; Rotter, J.I.; Krauss, R.M.; Fiehn, O.; Kaddurah-Daouk, R. Pharmacometabolomic signature links simvastatin therapy and insulin resistance. Metab. Off. J. Metab. Soc. 2017, 13, 11. [Google Scholar] [CrossRef]
- Wilson, I.D. Drugs, bugs, and personalized medicine: Pharmacometabonomics enters the ring. Proc. Natl. Acad. Sci. USA 2009, 106, 14187–14188. [Google Scholar] [CrossRef]
- Everett, J.R. Pharmacometabonomics in humans: A new tool for personalized medicine. Pharmacogenomics 2015, 16, 737–754. [Google Scholar] [CrossRef] [PubMed]
- Beger, R.D.; Dunn, W.; Schmidt, M.A.; Gross, S.S.; Kirwan, J.A.; Cascante, M.; Brennan, L.; Wishart, D.S.; Oresic, M.; Hankemeier, T.; et al. Metabolomics enables precision medicine: “A White Paper, Community Perspective”. Metab. Off. J. Metab. Soc. 2016, 12, 149. [Google Scholar] [CrossRef] [PubMed]
- Rattray, N.J.W.; Daouk, R.K. Pharmacometabolomics and precision medicine special issue editorial. Metabolomics 2017, 13, 59. [Google Scholar] [CrossRef][Green Version]
- Jain, D.; Ahmad, T.; Cairo, M.; Aronow, W. Cardiotoxicity of cancer chemotherapy: Identification, prevention and treatment. Ann. Transl. Med. 2017, 5, 348. [Google Scholar] [CrossRef]
- Mosedale, M.; Watkins, P.B. Drug-induced liver injury: Advances in mechanistic understanding that will inform risk management. Clin. Pharmacol. Ther. 2017, 101, 469–480. [Google Scholar] [CrossRef] [PubMed]
- Rasool, M.; Malik, A.; Naseer, M.I.; Manan, A.; Ansari, S.A.; Begum, I.; Qazi, M.H.; Pushparaj, P.N.; Abuzenadah, A.M.; Al-Qahtani, M.H.; et al. The role of epigenetics in personalized medicine: Challenges and opportunities. BMC Med Genom. 2015, 8, S5. [Google Scholar] [CrossRef]
- Nicholson, J.K. Global systems biology, personalized medicine and molecular epidemiology. Mol. Syst. Biol. 2006, 2, 52. [Google Scholar] [CrossRef]
- Drug Development Tool Qualification Process. Available online: https://www.fda.gov/drugs/drug-development-tool-ddt-qualification-programs/drug-development-tool-qualification-process-transparency-provisions (accessed on 1 March 2020).
- Leptak, C.; Menetski, J.P.; Wagner, J.A.; Aubrecht, J.; Brady, L.; Brumfield, M.; Chin, W.W.; Hoffmann, S.; Kelloff, G.; Lavezzari, G.; et al. What evidence do we need for biomarker qualification? Sci. Transl. Med. 2017, 9, eaal4599. [Google Scholar] [CrossRef]
- Resources for Biomarker Requestors. Available online: https://www.fda.gov/drugs/cder-biomarker-qualification-program/resources-biomarker-requestors (accessed on 1 March 2020).
- Context. Available online: https://www.fda.gov/Drugs/DevelopmentApprovalProcess/DrugDevelopmentToolsQualificationProgram/BiomarkerQualificationProgram/ucm535395.htm (accessed on 1 March 2020).
- BEST (Biomarkers, EndpointS, and other Tools) Resource. Available online: https://www.ncbi.nlm.nih.gov/books/NBK338448/?report=reader (accessed on 1 March 2020).
- Wishart, D.S.; Xia, J. MetPA: A web-based metabolomics tool for pathway analysis and visualization. Bioinformatics 2010, 26, 2342–2344. [Google Scholar]
- Pazos, F.; Chagoyen, M. Tools for the functional interpretation of metabolomic experiments. Brief. Bioinform. 2012, 14, 737–744. [Google Scholar]
- Everett, J.R.; Loo, R.L.; Pullen, F.S. Pharmacometabonomics and personalized medicine. Ann. Clin. Biochem. 2013, 50, 523–545. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.P.; Yerges-Armstrong, L.M.; Ellero-Simatos, S.; Georgiades, A.; Kaddurah-Daouk, R.; Hankemeier, T. Integration of pharmacometabolomic and pharmacogenomic approaches reveals novel insights into antiplatelet therapy. Clin. Pharmacol. Ther. 2013, 94, 570–573. [Google Scholar] [CrossRef] [PubMed]
- Rotroff, D.M.; Corum, D.G.; Motsinger-Reif, A.; Fiehn, O.; Bottrel, N.; Drevets, W.C.; Singh, J.; Salvadore, G.; Kaddurah-Daouk, R. Metabolomic signatures of drug response phenotypes for ketamine and esketamine in subjects with refractory major depressive disorder: New mechanistic insights for rapid acting antidepressants. Transl. Psychiatry 2016, 6, e894. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, S.; Ahmed, A.T.; Arnold, M.; Liu, D.; Luo, C.; Zhu, H.; Mahmoudiandehkordi, S.; Neavin, D.; Louie, G.; Dunlop, B.W.; et al. Metabolomic signature of exposure and response to citalopram/escitalopram in depressed outpatients. Transl. Psychiatry 2019, 9, 173. [Google Scholar] [CrossRef]
- Beger, R.D.; Flynn, T.J. Pharmacometabolomics in drug safety and drug-exposome interactions. Metabolomics 2016, 12, 123. [Google Scholar] [CrossRef]
- Palleria, C.; Di Paolo, A.; Giofrè, C.; Caglioti, C.; Leuzzi, G.; Siniscalchi, A.; De Sarro, G.; Gallelli, L. Pharmacokinetic drug-drug interaction and their implication in clinical management. J. Res. Med Sci. Off. J. Isfahan Univ. Med Sci. 2013, 18, 601–610. [Google Scholar]
- Bushra, R.; Aslam, N.; Khan, A.Y. Food-drug interactions. Oman Med. J. 2011, 26, 77–83. [Google Scholar] [CrossRef]
- Swanson, H.I. Drug Metabolism by the host and gut microbiota: A partnership or rivalry? Drug Metab. Dispos. Biol. Fate Chem. 2015, 43, 1499–1504. [Google Scholar] [CrossRef]
- Holbrook, A.M.; Pereira, J.A.; Labiris, R.; McDonald, H.; Douketis, J.D.; Crowther, M.; Wells, P.S. Systematic overview of warfarin and its drug and food interactions. JAMA Intern. Med. 2005, 165, 1095–1106. [Google Scholar] [CrossRef]
- Cao, Z.; Kamlage, B.; Wagner-Golbs, A.; Maisha, M.; Sun, J.; Schnackenberg, L.K.; Pence, L.; Schmitt, T.C.; Daniels, J.R.; Rogstad, S.; et al. An integrated analysis of metabolites, peptides, and inflammation biomarkers for assessment of preanalytical variability of human plasma. J. Proteome Res. 2019, 18, 2411–2421. [Google Scholar] [CrossRef]
- Szymańska, E.; Saccenti, E.; Smilde, A.K.; Westerhuis, J.A. Double-check: Validation of diagnostic statistics for PLS-DA models in metabolomics studies. Metabolomics 2012, 8, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Stone, N.J.; Robinson, J.G.; Lichtenstein, A.H.; Merz, C.N.; Blum, C.B.; Eckel, R.H.; Goldberg, A.C.; Gordon, D.; Levy, D.; Lloyd-Jones, D.M.; et al. 2013 ACC/AHA guideline on the treatment of blood cholesterol to reduce atherosclerotic cardiovascular risk in adults. Circulation 2014, 129, S1–S45. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.; Ewy, W.; Corrigan, B.W.; Ouellet, D.; Hermann, D.; Kowalski, K.G.; Lockwood, P.; Koup, J.R.; Donevan, S.; El-Kattan, A.; et al. How modeling and simulation have enhanced decision making in new drug development. J. Pharmacokinet. Pharmacodyn. 2005, 32, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Zhou, Z.; Zhou, J.; Chen, S.Q. Pharmacogenomics of drug metabolizing enzymes and transporters: Relevance to precision medicine. Genom. Proteom. Bioinform. 2016, 14, 298–313. [Google Scholar] [CrossRef]
- Turner, R.M.; Park, B.K.; Pirmohamed, M. Parsing interindividual drug variability: An emerging role for systems pharmacology. Wiley Interdiscip. Rev. Syst. Biol. Med. 2015, 7, 221–241. [Google Scholar] [CrossRef]
- Kantae, V.; Krekels, E.H.; Van Esdonk, M.J.; Lindenburg, P.; Harms, A.C.; Knibbe, C.A.; Van der Graaf, P.H.; Hankemeier, T. Integration of pharmacometabolomics with pharmacokinetics and pharmacodynamics: Towards personalized drug therapy. Metab. Off. J. Metab. Soc. 2017, 13, 9. [Google Scholar] [CrossRef]
- Shuker, N.; Shuker, L.; van Rosmalen, J.; Roodnat, J.I.; Borra, L.C.; Weimar, W.; Hesselink, D.A.; van Gelder, T. A high intrapatient variability in tacrolimus exposure is associated with poor long-term outcome of kidney transplantation. Transpl. Int. 2016, 29, 1158–1167. [Google Scholar] [CrossRef]
- Goldsmith, P.M.; Bottomley, M.J.; Okechukwu, O.; Ross, V.C.; Ghita, R.; Wandless, D.; Falconer, S.J.; Papachristos, S.; Nash, P.; Androshchuk, V.; et al. Impact of intrapatient variability (IPV) in tacrolimus trough levels on long-term renal transplant function: Multicentre collaborative retrospective cohort study protocol. BMJ Open 2017, 7, e016144. [Google Scholar] [CrossRef]
- Cattaneo, D.; Gervasoni, C.; Meraviglia, P.; Landonio, S.; Fucile, S.; Cozzi, V.; Baldelli, S.; Pellegrini, M.; Galli, M.; Clementi, E. Inter- and intra-patient variability of raltegravir pharmacokinetics in HIV-1-infected subjects. J. Antimicrob. Chemother. 2011, 67, 460–464. [Google Scholar] [CrossRef]
- Siccardi, M.; D’Avolio, A.; Rodriguez-Novoa, S.; Cuenca, L.; Simiele, M.; Baietto, L.; Calcagno, A.; Moss, D.; Bonora, S.; Soriano, V.; et al. Intrapatient and interpatient pharmacokinetic variability of raltegravir in the clinical setting. Ther. Drug Monit. 2012, 34, 232–235. [Google Scholar] [CrossRef]
- MetaboLights. Available online: https://www.ebi.ac.uk/metabolights (accessed on 1 March 2020).
- Metabolomics Workbench. Available online: http://www.metabolomicsworkbench.org/ (accessed on 1 March 2020).
- COnsortium of METabolomics Studies. Available online: https://epi.grants.cancer.gov/comets/ (accessed on 1 March 2020).
- Clinical Trials. Available online: https://clinicaltrials.gov/ (accessed on 3 September 2019).
- van Roekel, E.H.; Loftfield, E.; Kelly, R.S.; Zeleznik, O.A.; Zanetti, K.A. Metabolomics in epidemiologic research: Challenges and opportunities for early-career epidemiologists. Metabolomics 2019, 15, 9. [Google Scholar] [CrossRef] [PubMed]
- Fearnley, L.G.; Inouye, M. Metabolomics in epidemiology: From metabolite concentrations to integrative reaction networks. Int. J. Epidemiol. 2016, 45, 1319–1328. [Google Scholar] [CrossRef] [PubMed]
- Clayton, T.A.; Baker, D.; Lindon, J.C.; Everett, J.R.; Nicholson, J.K. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc. Natl. Acad. Sci. USA 2009, 106, 14728–14733. [Google Scholar] [CrossRef] [PubMed]
- Muhrez, K.; Benz-de Bretagne, I.; Nadal-Desbarats, L.; Blasco, H.; Gyan, E.; Choquet, S.; Montigny, F.; Emond, P.; Barin-Le Guellec, C. Endogenous metabolites that are substrates of organic anion transporter’s (OATs) predict methotrexate clearance. Pharmacol. Res. 2017, 118, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Krausz, K.W.; Idle, J.R.; Gonzalez, F.J. Identification of novel toxicity-associated metabolites by metabolomics and mass isotopomer analysis of acetaminophen metabolism in wild-type and Cyp2e1-null mice. J. Biol. Chem. 2008, 283, 4543–4559. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, S.; Yan, K.; Pence, L.; Simpson, P.M.; Gill, P.; Letzig, L.G.; Beger, R.D.; Sullivan, J.E.; Kearns, G.L.; Reed, M.D.; et al. Targeted liquid chromatography–mass spectrometry analysis of serum acylcarnitines in acetaminophen toxicity in children. Biomark. Med. 2014, 8, 147–159. [Google Scholar] [CrossRef]
- McEvoy, J.; Baillie, R.A.; Zhu, H.; Buckley, P.; Keshavan, M.S.; Nasrallah, H.A.; Dougherty, G.G.; Yao, J.K.; Kaddurah-Daouk, R. Lipidomics reveals early metabolic changes in subjects with schizophrenia: Effects of atypical antipsychotics. PLoS ONE 2013, 8, e68717. [Google Scholar] [CrossRef]
- Kaddurah-Daouk, R.; Baillie, R.A.; Zhu, H.; Zeng, Z.B.; Wiest, M.M.; Nguyen, U.T.; Watkins, S.M.; Krauss, R.M. Lipidomic analysis of variation in response to simvastatin in the Cholesterol and Pharmacogenetics Study. Metab. Off. J. Metab. Soc. 2010, 6, 191–201. [Google Scholar] [CrossRef]
- Trupp, M.; Zhu, H.; Wikoff, W.R.; Baillie, R.A.; Zeng, Z.B.; Karp, P.D.; Fiehn, O.; Krauss, R.M.; Kaddurah-Daouk, R. Metabolomics reveals amino acids contribute to variation in response to simvastatin treatment. PLoS ONE 2012, 7, e38386. [Google Scholar] [CrossRef]
- Cooper-DeHoff, R.M.; Hou, W.; Weng, L.; Baillie, R.A.; Beitelshees, A.L.; Gong, Y.; Shahin, M.H.; Turner, S.T.; Chapman, A.; Gums, J.G.; et al. Is diabetes mellitus-linked amino acid signature associated with β-blocker-induced impaired fasting glucose? Circ. Cardiovasc. Genet. 2014, 7, 199–205. [Google Scholar] [CrossRef]
- Rotroff, D.M.; Shahin, M.H.; Gurley, S.B.; Zhu, H.; Motsinger-Reif, A.; Meisner, M.; Beitelshees, A.L.; Fiehn, O.; Johnson, J.A.; Elbadawi-Sidhu, M.; et al. Pharmacometabolomic assessments of atenolol and hydrochlorothiazide treatment reveal novel drug response phenotypes. CPT Pharmacomet. Syst. Pharmacol. 2015, 4, 669–679. [Google Scholar] [CrossRef] [PubMed]
- Backshall, A.; Sharma, R.; Clarke, S.J.; Keun, H.C. Pharmacometabonomic profiling as a predictor of toxicity in patients with inoperable colorectal cancer treated with capecitabine. Clin. Cancer Res. 2011, 17, 3019–3028. [Google Scholar] [CrossRef] [PubMed]
- Miolo, G.; Muraro, E.; Caruso, D.; Crivellari, D.; Ash, A.; Scalone, S.; Lombardi, D.; Rizzolio, F.; Giordano, A.; Corona, G. Pharmacometabolomics study identifies circulating spermidine and tryptophan as potential biomarkers associated with the complete pathological response to trastuzumab-paclitaxel neoadjuvant therapy in HER-2 positive breast cancer. Oncotarget 2016, 7, 39809–39822. [Google Scholar] [CrossRef] [PubMed]
- Yerges-Armstrong, L.M.; Ellero-Simatos, S.; Georgiades, A.; Zhu, H.; Lewis, J.P.; Horenstein, R.B.; Beitelshees, A.L.; Dane, A.; Reijmers, T.; Hankemeier, T.; et al. Purine pathway implicated in mechanism of resistance to aspirin therapy: Pharmacometabolomics-informed pharmacogenomics. Clin. Pharmacol. Ther. 2013, 94, 525–532. [Google Scholar] [CrossRef]
- Ellero-Simatos, S.; Lewis, J.P.; Georgiades, A.; Yerges-Armstrong, L.M.; Beitelshees, A.L.; Horenstein, R.B.; Dane, A.; Harms, A.C.; Ramaker, R.; Vreeken, R.J.; et al. Pharmacometabolomics reveals that serotonin is implicated in aspirin response variability. CPT Pharmacomet. Syst. Pharmacol. 2014, 3, 125. [Google Scholar] [CrossRef]
- Lanznaster, D.; de Assis, D.R.; Corcia, P.; Pradat, P.F.; Blasco, H. Metabolomics biomarkers: A strategy toward therapeutics improvement in ALS. Front. Neurol. 2018, 9, 1126. [Google Scholar] [CrossRef]
- de Jager, J.; Kooy, A.; Lehert, P.; Wulffelé, M.G.; Van der Kolk, J.; Bets, D.; Verburg, J.; Donker, A.J.; Stehouwer, C.D. Long term treatment with metformin in patients with type 2 diabetes and risk of vitamin B-12 deficiency: Randomised placebo controlled trial. BMJ (Clin. Res. Ed.), 2010; 340, c2181. [Google Scholar]
- Reinstatler, L.; Qi, Y.P.; Williamson, R.S.; Garn, J.V.; Oakley, G.P., Jr. Association of biochemical B₁₂ deficiency with metformin therapy and vitamin B₁₂ supplements: The national health and nutrition examination survey, 1999–2006. Diabetes Care 2012, 35, 327–333. [Google Scholar] [CrossRef]
- Ko, S.H.; Ko, S.H.; Ahn, Y.B.; Song, K.H.; Han, K.D.; Park, Y.M.; Ko, S.H.; Kim, H.S. Association of vitamin B12 deficiency and metformin use in patients with type 2 diabetes. J. Korean Med Sci. 2014, 29, 965–972. [Google Scholar] [CrossRef]
- Aroda, V.R.; Edelstein, S.L.; Goldberg, R.B.; Knowler, W.C.; Marcovina, S.M.; Orchard, T.J.; Bray, G.A.; Schade, D.S.; Temprosa, M.G.; White, N.H.; et al. Long-term metformin use and vitamin B12 deficiency in the diabetes prevention program outcomes study. J. Clin. Endocrinol. Metab. 2016, 101, 1754–1761. [Google Scholar] [CrossRef]
- Ahmed, M.A.; Muntingh, G.; Rheeder, P. Vitamin B12 deficiency in metformin-treated type-2 diabetes patients, prevalence and association with peripheral neuropathy. BMC Pharmacol. Toxicol. 2016, 17, 44. [Google Scholar] [CrossRef]
- Alvarez, M.; Rincon, O.; Saavedra, G.; Moreno, S.M. Vitamin B12 deficiency and diabetic neuropathy in patients taking metformin: A cross-sectional study. Endocr. Connect. 2019, 8, 1324–1329. [Google Scholar] [CrossRef] [PubMed]
- Alharbi, T.J.; Tourkmani, A.M.; Abdelhay, O.; Alkhashan, H.I.; Al-Asmari, A.K.; Rsheed, A.M.; Abuhaimed, S.N.; Mohammed, N.; AlRasheed, A.N.; AlHarbi, N.G. The association of metformin use with vitamin B12 deficiency and peripheral neuropathy in Saudi individuals with type 2 diabetes mellitus. PLoS ONE 2018, 13, e0204420. [Google Scholar] [CrossRef] [PubMed]
- Chapman, L.E.; Darling, A.L.; Brown, J.E. Association between metformin dose and vitamin B12 deficiency in patients with type 2 diabetes. Medicine 2019, 98, e17918. [Google Scholar]
- Orlenko, A.; Moore, J.H.; Orzechowski, P.; Olson, R.S.; Cairns, J.; Caraballo, P.J.; Weinshilboum, R.M.; Wang, L.; Breitenstein, M.K. Considerations for automated machine learning in clinical metabolic profiling: Altered homocysteine plasma concentration associated with metformin exposure. Pac. Symp. Biocomput. Pac. Symp. Biocomput. 2018, 23, 460–471. [Google Scholar] [PubMed]
- Out, M.; Kooy, A.; Lehert, P.; Schalkwijk, C.A.; Stehouwer, C.D. Long-term treatment with metformin in type 2 diabetes and methylmalonic acid: Post hoc analysis of a randomized controlled 4.3year trial. J. Diabetes Its Complicat. 2018, 32, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Abo, R.; Hebbring, S.; Ji, Y.; Zhu, H.; Zeng, Z.B.; Batzler, A.; Jenkins, G.D.; Biernacka, J.; Snyder, K.; Drews, M.; et al. Merging pharmacometabolomics with pharmacogenomics using ‘1000 Genomes’ single-nucleotide polymorphism imputation: Selective serotonin reuptake inhibitor response pharmacogenomics. Pharm. Genom. 2012, 22, 247–253. [Google Scholar] [CrossRef]
- Ji, Y.; Hebbring, S.; Zhu, H.; Jenkins, G.D.; Biernacka, J.; Snyder, K.; Drews, M.; Fiehn, O.; Zeng, Z.; Schaid, D.; et al. Glycine and a glycine dehydrogenase (GLDC) SNP as citalopram/escitalopram response biomarkers in depression: Pharmacometabolomics-informed pharmacogenomics. Clin. Pharmacol. Ther. 2011, 89, 97–104. [Google Scholar] [CrossRef]
- Shahin, M.H.; Gong, Y.; Frye, R.F.; Rotroff, D.M.; Beitelshees, A.L.; Baillie, R.A.; Chapman, A.B.; Gums, J.G.; Turner, S.T.; Boerwinkle, E.; et al. Sphingolipid metabolic pathway impacts thiazide diuretics blood pressure response: Insights from genomics, metabolomics, and lipidomics. J. Am. Heart Assoc. 2017, 7, e006656. [Google Scholar] [CrossRef]
- Neavin, D.; Kaddurah-Daouk, R.; Weinshilboum, R. Pharmacometabolomics informs pharmacogenomics. Metab. Off. J. Metab. Soc. 2016, 12, 121. [Google Scholar] [CrossRef]
- de Oliveira, F.A.; Shahin, M.H.; Gong, Y.; McDonough, C.W.; Beitelshees, A.L.; Gums, J.G.; Chapman, A.B.; Boerwinkle, E.; Turner, S.T.; Frye, R.F.; et al. Novel plasma biomarker of atenolol-induced hyperglycemia identified through a metabolomics-genomics integrative approach. Metab. Off. J. Metab. Soc. 2016, 12, 129. [Google Scholar] [CrossRef]
- Frankel, A.E.; Coughlin, L.A.; Kim, J.; Froehlich, T.W.; Xie, Y.; Frenkel, E.P.; Koh, A.Y. Metagenomic shotgun sequencing and unbiased metabolomic profiling identify specific human gut microbiota and metabolites associated with immune checkpoint therapy efficacy in melanoma patients. Neoplasia (N. Y.) 2017, 19, 848–855. [Google Scholar] [CrossRef] [PubMed]
- Morais, S.M.; Silva, K.A.; Araujo, H.; Vieira, I.G.; Alves, D.R.; Fontenelle, R.O.; Silva, A. Anacardic acid constituents from cashew nut shell liquid: NMR characterization and the effect of unsaturation on its biological activities. Pharmaceuticals 2017, 10, 31. [Google Scholar] [CrossRef] [PubMed]
- Hollands, A.; Corriden, R.; Gysler, G.; Dahesh, S.; Olson, J.; Ali, S.R.; Kunkel, M.T.; Lin, A.E.; Forli, S.; Newton, A.C.; et al. Natural product anacardic acid from cashew nut shells stimulates neutrophil extracellular trap production and bactericidal activity. J. Biol. Chem. 2016, 291, 13964–13973. [Google Scholar] [CrossRef]
- Dethlefsen, L.; Relman, D.A. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4554–4561. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2013, 505, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.A.; Schmidt, C.M.; Goodwin, T.J. Pharmacogenomics in Spaceflight. In Handbook of Space Pharmaceuticals; Pathak, Y., Araújo dos Santos, M., Zea, L., Eds.; Springer International Publishing: Cham/Basel, Switzerland, 2018; pp. 1–39. [Google Scholar]
- Garrett-Bakelman, F.E.; Darshi, M.; Green, S.J.; Gur, R.C.; Lin, L.; Macias, B.R.; McKenna, M.J.; Meydan, C.; Mishra, T.; Nasrini, J.; et al. The NASA twins study: A multidimensional analysis of a year-long human spaceflight. Science 2019, 364, eaau8650. [Google Scholar] [PubMed]
- Koppel, N.; Rekdal, V.M.; Balskus, E.P. Chemical transformation of xenobiotics by the human gut microbiota. Science 2017, 356, eaag2770. [Google Scholar] [CrossRef]
- Sender, R.; Fuchs, S.; Milo, R. Revised estimates for the number of human and bacteria cells in the body. Plos Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef]
- Bi, Y.; Qin, N.; Yang, R. Human microbiota: A neglected “organ” in precision medicine. Infect. Dis. Transl. Med. 2015, 1, 63–72. [Google Scholar]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207. [Google Scholar]
- Heintz-Buschart, A.; Wilmes, P. Human gut microbiome: Function matters. Trends Microbiol. 2018, 26, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Clarke, G.; Sandhu, K.V.; Griffin, B.T.; Dinan, T.G.; Cryan, J.F.; Hyland, N.P. Gut reactions: Breaking down xenobiotic-microbiome interactions. Pharmacol. Rev. 2019, 71, 198. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Southall, N.; Wang, Y.; Yasgar, A.; Shinn, P.; Jadhav, A.; Nguyen, D.T.; Austin, C.P. The NCGC pharmaceutical collection: A comprehensive resource of clinically approved drugs enabling repurposing and chemical genomics. Sci. Transl. Med. 2011, 3, 80ps16. [Google Scholar] [CrossRef]
- Haiser, H.J.; Turnbaugh, P.J. Is it time for a metagenomic basis of therapeutics? Science 2012, 336, 1253–1255. [Google Scholar] [CrossRef] [PubMed]
- Yip, L.Y.; Chan, E.C.Y. Investigation of host-gut microbiota modulation of therapeutic outcome. Drug Metab. Dispos. 2015, 43, 1619–1631. [Google Scholar] [CrossRef] [PubMed]
- Wilson, I.D.; Nicholson, J.K. Gut microbiome interactions with drug metabolism, efficacy, and toxicity. Transl. Res. 2017, 179, 204–222. [Google Scholar] [CrossRef]
- Saito, Y.; Sato, T.; Nomoto, K.; Tsuji, H. Identification of phenol- and p-cresol-producing intestinal bacteria by using media supplemented with tyrosine and its metabolites. Fems Microbiol. Ecol. 2018, 94, fiy125. [Google Scholar] [CrossRef]
- Mitchell, J.R.; Jollow, D.J.; Potter, W.Z.; Davis, D.C.; Gillette, J.R.; Brodie, B.B. Acetaminophen-induced hepatic necrosis. I. Role of drug metabolism. J. Pharmacol. Exp. Ther. 1973, 187, 185. [Google Scholar]
- Ben-Shachar, R.; Chen, Y.; Luo, S.; Hartman, C.; Reed, M.; Nijhout, H.F. The biochemistry of acetaminophen hepatotoxicity and rescue: A mathematical model. Theor. Biol. Med Model. 2012, 9, 55. [Google Scholar] [CrossRef]
- Heruth, D.P.; Shortt, K.; Zhang, N.; Li, D.Y.; Zhang, L.Q.; Ye, S.Q. Genetic association of single nucleotide polymorphisms with acetaminophen-induced hepatotoxicity. J. Pharmacol. Exp. Ther. 2018, 367, 95–100. [Google Scholar] [CrossRef]
- Moyer, A.M.; Fridley, B.L.; Jenkins, G.D.; Batzler, A.J.; Pelleymounter, L.L.; Kalari, K.R.; Ji, Y.; Chai, Y.; Nordgren, K.K.; Weinshilboum, R.M. Acetaminophen-NAPQI hepatotoxicity: A cell line model system genome-wide association study. Toxicol. Sci. Off. J. Soc. Toxicol. 2011, 120, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Mohn, E.S.; Kern, H.J.; Saltzman, E.; Mitmesser, S.H.; McKay, D.L. Evidence of drug-nutrient interactions with chronic use of commonly prescribed medications: An update. Pharmaceutics 2018, 10, 36. [Google Scholar] [CrossRef]
- Sanderson, S.M.; Gao, X.; Dai, Z.; Locasale, J.W. Methionine metabolism in health and cancer: A nexus of diet and precision medicine. Nat. Rev. Cancer 2019, 19, 625–637. [Google Scholar] [CrossRef] [PubMed]
- Flores, R.; Shi, J.; Fuhrman, B.; Xu, X.; Veenstra, T.D.; Gail, M.H.; Gajer, P.; Ravel, J.; Goedert, J.J. Fecal microbial determinants of fecal and systemic estrogens and estrogen metabolites: A cross-sectional study. J. Transl. Med. 2012, 10, 253. [Google Scholar] [CrossRef] [PubMed]
- Drozda, K.; Pacanowski, M.A.; Grimstein, C.; Zineh, I. Pharmacogenetic labeling of FDA-approved drugs: A regulatory retrospective. Jacc. Basic Transl. Sci. 2018, 3, 545–549. [Google Scholar] [CrossRef] [PubMed]
- Burt, T.; Dhillon, S. Pharmacogenomics in early-phase clinical development. Pharmacogenomics 2013, 14, 1085–1097. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Yi, S.; Gu, N.; Shin, D.; Yu, K.S.; Yoon, S.H.; Cho, J.Y.; Jang, I.J. Utility of integrated analysis of pharmacogenomics and pharmacometabolomics in early phase clinical trial: A case study of a new molecular entity. Genom. Inform. 2018, 16, 52–58. [Google Scholar] [CrossRef]
| BEST Biomarker Category | Relationship between Metabolites and Biomarker Category | Potential Context of Use (COU) in a Clinical Study |
|---|---|---|
| Prognosis Biomarker | Metabolites that indicate a likelihood of a future clinical event | Stratify Patients Enrichment: Inclusion/Exclusion Data |
| Diagnostic Biomarker | Metabolites that detect the presence of a disease or identify individuals with a subtype of the disease | Patient Selection |
| Monitoring Biomarker | Metabolites that are measured continually over time to assess status of a disease or medical condition or for evidence of exposure to (or effect of) a medical product or an environmental agent | Indicate Toxicity or assess safety Provide evidence of exposure |
| Predictive Biomarker | Metabolites that predict outcome | Identify individuals based on effect from a specific intervention or exposure |
| Safety Biomarker | Metabolites that are related to adverse and safety events | Indicate the presence or extent of toxicity related to an intervention or exposure |
| Pharmacodynamic Response Biomarker | Metabolites that are related to response in an individual or group of individuals who have been exposed to a medical product or an environmental agent | Efficacy biomarkers/surrogate endpoint Show biological response related to an intervention or exposure |
| Susceptibility/Risk Biomarker | Metabolites related to developing a disease or medical condition in an patient that does not currently have clinically apparent disease or medical condition | Indicate the potential for developing a disease or sensitivity to an exposure |
| Provisional Biomarker | Metabolites that are in discovery and show potential as biomarkers, although they have not been validated as true biomarkers | Discovery-associated analytes that assist in identification of signals with potential biological meaning. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beger, R.D.; Schmidt, M.A.; Kaddurah-Daouk, R. Current Concepts in Pharmacometabolomics, Biomarker Discovery, and Precision Medicine. Metabolites 2020, 10, 129. https://doi.org/10.3390/metabo10040129
Beger RD, Schmidt MA, Kaddurah-Daouk R. Current Concepts in Pharmacometabolomics, Biomarker Discovery, and Precision Medicine. Metabolites. 2020; 10(4):129. https://doi.org/10.3390/metabo10040129
Chicago/Turabian StyleBeger, Richard D., Michael A Schmidt, and Rima Kaddurah-Daouk. 2020. "Current Concepts in Pharmacometabolomics, Biomarker Discovery, and Precision Medicine" Metabolites 10, no. 4: 129. https://doi.org/10.3390/metabo10040129
APA StyleBeger, R. D., Schmidt, M. A., & Kaddurah-Daouk, R. (2020). Current Concepts in Pharmacometabolomics, Biomarker Discovery, and Precision Medicine. Metabolites, 10(4), 129. https://doi.org/10.3390/metabo10040129






