Improved Dried Blood Spot-Based Metabolomics: A Targeted, Broad-Spectrum, Single-Injection Method
Abstract
1. Introduction
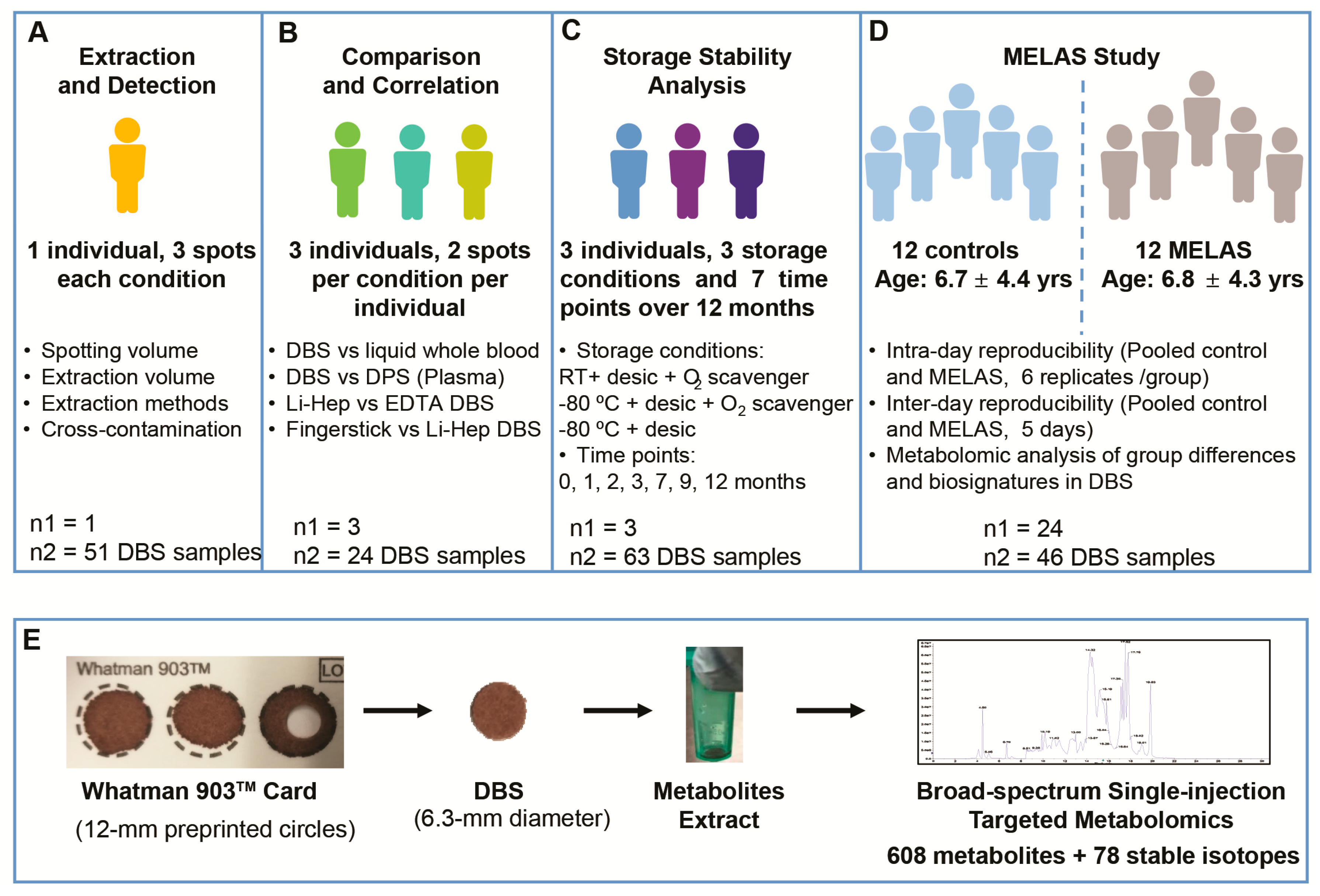
2. Materials and Methods
2.1. Informed Consent
2.2. DBS and DPS (Dried Plasma Spot) Preparation
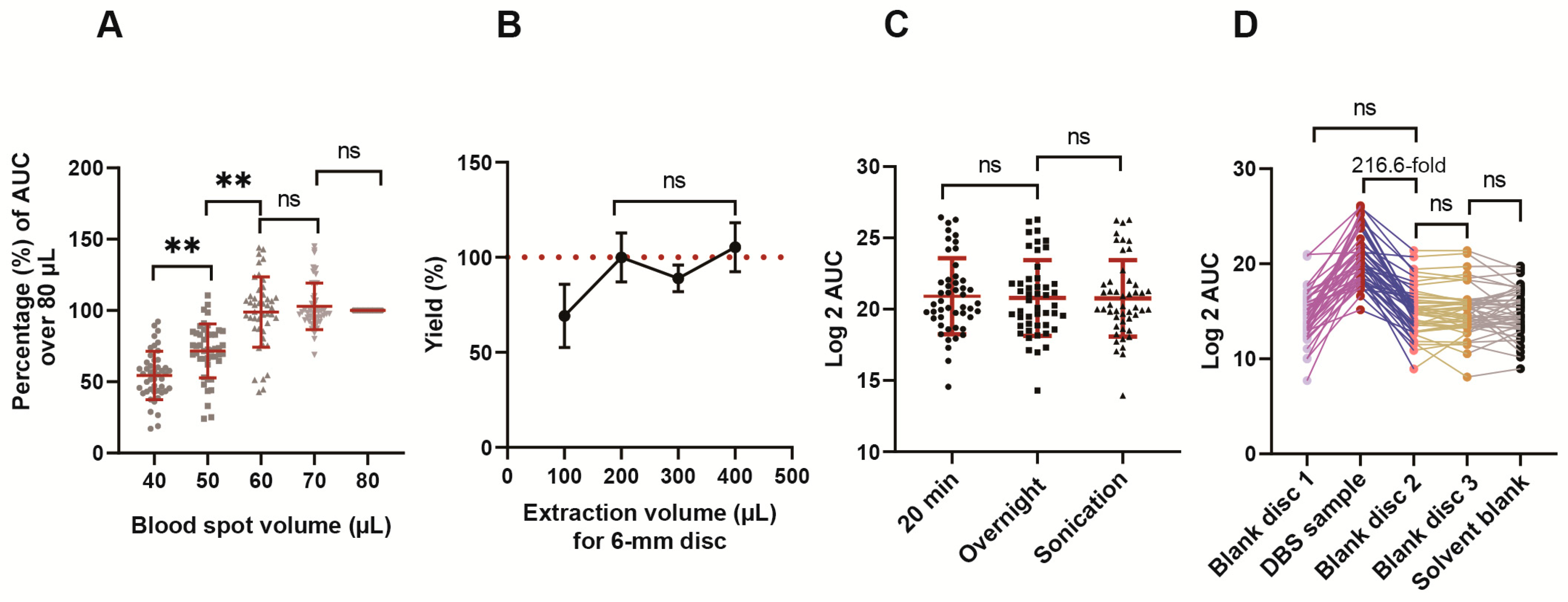
2.3. Method Optimization
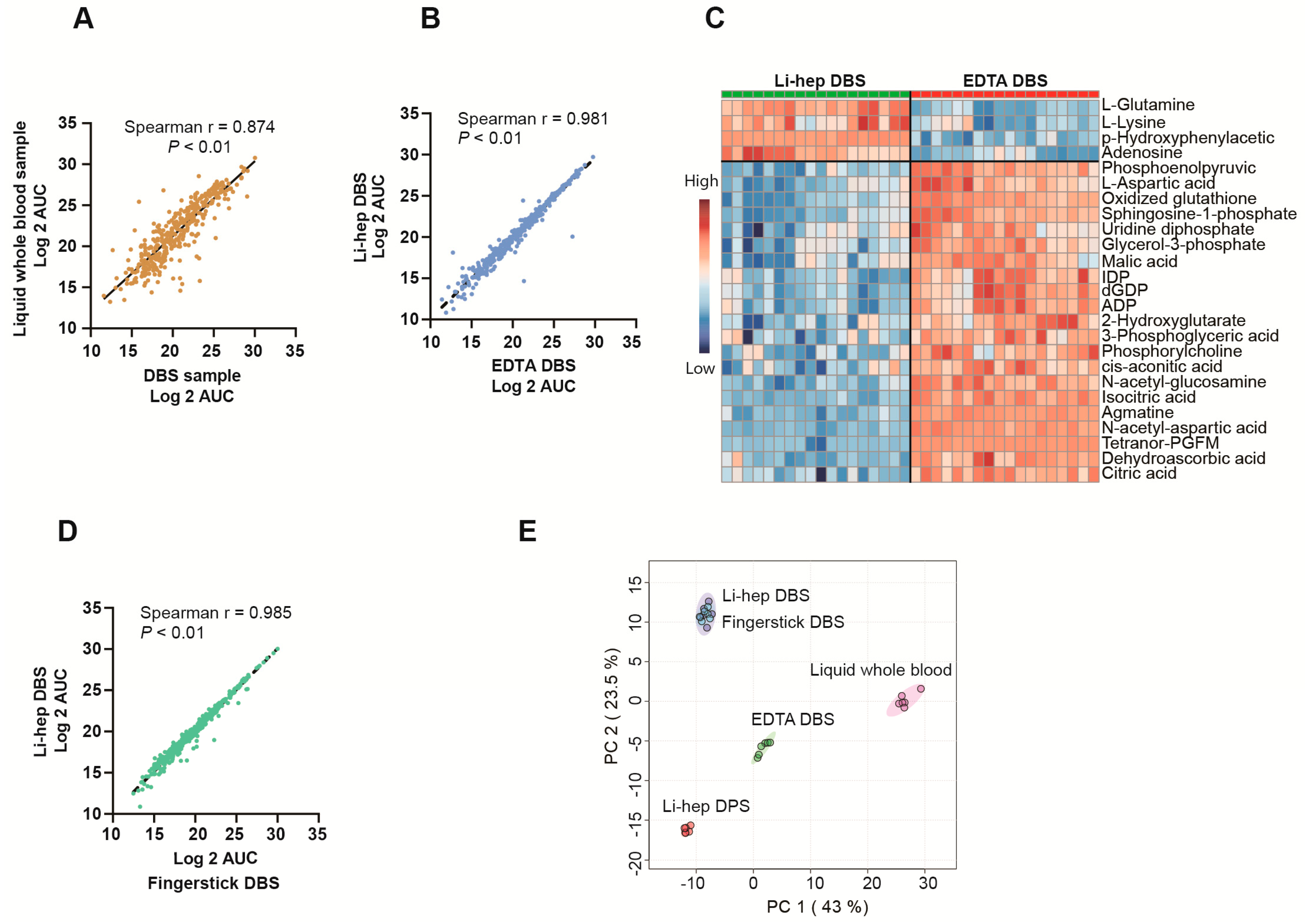
2.4. Sample Type Comparisons
2.5. Stability Analysis
2.6. Hematocrit Effects
2.7. Metabolite Extraction
2.8. Metabolomic Analysis
2.9. Clinical Application in MELAS
2.10. Bioinformatic and Statistical Analysis
3. Results
3.1. Method Optimization
3.2. Sample Type Comparisons
3.3. One-Year Stability Analysis
3.4. Clinical Application in MELAS
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Naviaux, R.K.; Naviaux, J.C.; Li, K.; Bright, A.T.; Alaynick, W.A.; Wang, L.; Baxter, A.; Nathan, N.; Anderson, W.; Gordon, E. Metabolic features of chronic fatigue syndrome. Proc. Natl. Acad. Sci. USA 2016, 113, E5472–E5480. [Google Scholar] [CrossRef] [PubMed]
- Naviaux, R.K.; Naviaux, J.C.; Li, K.; Wang, L.; Monk, J.M.; Bright, A.T.; Koslik, H.J.; Ritchie, J.B.; Golomb, B.A. Metabolic features of Gulf War illness. PLoS ONE 2019, 14, e0219531. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Kirwan, J.A.; Brennan, L.; Broadhurst, D.; Fiehn, O.; Cascante, M.; Dunn, W.B.; Schmidt, M.A.; Velagapudi, V. Preanalytical Processing and Biobanking Procedures of Biological Samples for Metabolomics Research: A White Paper, Community Perspective (for “Precision Medicine and Pharmacometabolomics Task Group”—The Metabolomics Society Initiative). Clin. Chem. 2018, 64, 1158–1182. [Google Scholar] [CrossRef]
- Guthrie, R.; Susi, A. A Simple Phenylalanine Method for Detecting Phenylketonuria in Large Populations of Newborn Infants. Pediatrics 1963, 32, 338–343. [Google Scholar]
- Zytkovicz, T.H.; Fitzgerald, E.F.; Marsden, D.; Larson, C.A.; Shih, V.E.; Johnson, D.M.; Strauss, A.W.; Comeau, A.M.; Eaton, R.B.; Grady, G.F. Tandem mass spectrometric analysis for amino, organic, and fatty acid disorders in newborn dried blood spots: A two-year summary from the New England Newborn Screening Program. Clin. Chem. 2001, 47, 1945–1955. [Google Scholar] [CrossRef]
- Kvaskoff, D.; Ko, P.; Simila, H.A.; Eyles, D.W. Distribution of 25-hydroxyvitamin D3 in dried blood spots and implications for its quantitation by tandem mass spectrometry. J. Chromatogr. B 2012, 901, 47–52. [Google Scholar] [CrossRef]
- Martial, L.C.; Aarnoutse, R.E.; Schreuder, M.F.; Henriet, S.S.; Bruggemann, R.J.; Joore, M.A. Cost Evaluation of Dried Blood Spot Home Sampling as Compared to Conventional Sampling for Therapeutic Drug Monitoring in Children. PLoS ONE 2016, 11, e0167433. [Google Scholar] [CrossRef]
- Albani, V.; Celis-Morales, C.; Marsaux, C.F.; Forster, H.; O’Donovan, C.B.; Woolhead, C.; Macready, A.L.; Fallaize, R.; Navas-Carretero, S.; San-Cristobal, R.; et al. Exploring the association of dairy product intake with the fatty acids C15:0 and C17:0 measured from dried blood spots in a multipopulation cohort: Findings from the Food4Me study. Mol. Nutr. Food Res. 2016, 60, 834–845. [Google Scholar] [CrossRef]
- Mercolini, L.; Mandrioli, R.; Sorella, V.; Somaini, L.; Giocondi, D.; Serpelloni, G.; Raggi, M.A. Dried blood spots: Liquid chromatography-mass spectrometry analysis of Delta9-tetrahydrocannabinol and its main metabolites. J. Chromatogr. A 2013, 1271, 33–40. [Google Scholar] [CrossRef]
- Martial, L.C.; Kerkhoff, J.; Martinez, N.; Rodriguez, M.; Coronel, R.; Molinas, G.; Roman, M.; Gomez, R.; Aguirre, S.; Jongedijk, E.; et al. Evaluation of dried blood spot sampling for pharmacokinetic research and therapeutic drug monitoring of anti-tuberculosis drugs in children. Int. J. Antimicrob. Agents 2018, 52, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Bjorkesten, J.; Enroth, S.; Shen, Q.; Wik, L.; Hougaard, D.M.; Cohen, A.S.; Sorensen, L.; Giedraitis, V.; Ingelsson, M.; Larsson, A.; et al. Stability of Proteins in Dried Blood Spot Biobanks. Mol. Cell. Proteom. 2017, 16, 1286–1296. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; McDaniel, J.; Chen, E.Y.; Rockwell, H.E.; Drolet, J.; Vishnudas, V.K.; Tolstikov, V.; Sarangarajan, R.; Narain, N.R.; Kiebish, M.A. Dynamic and temporal assessment of human dried blood spot MS/MS(ALL) shotgun lipidomics analysis. Nutr. Metab. Lond 2017, 14, 28. [Google Scholar] [CrossRef] [PubMed]
- Koulman, A.; Prentice, P.; Wong, M.C.Y.; Matthews, L.; Bond, N.J.; Eiden, M.; Griffin, J.L.; Dunger, D.B. The development and validation of a fast and robust dried blood spot based lipid profiling method to study infant metabolism. Metabolomics 2014, 10, 1018–1025. [Google Scholar] [CrossRef] [PubMed]
- Haijes, H.A.; Willemsen, M.; Van der Ham, M.; Gerrits, J.; Pras-Raves, M.L.; Prinsen, H.; Van Hasselt, P.M.; De Sain-van der Velden, M.G.M.; Verhoeven-Duif, N.M.; Jans, J.J.M. Direct Infusion Based Metabolomics Identifies Metabolic Disease in Patients’ Dried Blood Spots and Plasma. Metabolites 2019, 9, 12. [Google Scholar] [CrossRef]
- Petrick, L.; Edmands, W.; Schiffman, C.; Grigoryan, H.; Perttula, K.; Yano, Y.; Dudoit, S.; Whitehead, T.; Metayer, C.; Rappaport, S. An untargeted metabolomics method for archived newborn dried blood spots in epidemiologic studies. Metabolomics 2017, 13, 27. [Google Scholar] [CrossRef]
- Wang, Q.; Sun, T.; Cao, Y.; Gao, P.; Dong, J.; Fang, Y.; Fang, Z.; Sun, X.; Zhu, Z. A dried blood spot mass spectrometry metabolomic approach for rapid breast cancer detection. Onco Targets Ther. 2016, 9, 1389–1398. [Google Scholar] [CrossRef]
- Jing, Y.; Wu, X.; Gao, P.; Fang, Z.Z.; Wu, J.J.; Wang, Q.J.; Li, C.; Zhu, Z.T.; Cao, Y.F. Rapid Differentiating Colorectal Cancer and Colorectal Polyp Using Dried Blood Spot Mass Spectrometry Metabolomic Approach. IUBMB Life 2017, 69, 347–354. [Google Scholar] [CrossRef]
- Gorman, G.S.; Chinnery, P.F.; DiMauro, S.; Hirano, M.; Koga, Y.; McFarland, R.; Suomalainen, A.; Thorburn, D.R.; Zeviani, M.; Turnbull, D.M. Mitochondrial diseases. Nat. Rev. Dis. Primers 2016, 2, 16080. [Google Scholar] [CrossRef]
- Koga, Y.; Povalko, N.; Nishioka, J.; Katayama, K.; Yatsuga, S.; Matsuishi, T. Molecular pathology of MELAS and L-arginine effects. Biochim. Biophys. Acta 2012, 1820, 608–614. [Google Scholar] [CrossRef]
- Li, K.; Naviaux, J.C.; Bright, A.T.; Wang, L.; Naviaux, R.K. A robust, single-injection method for targeted, broad-spectrum plasma metabolomics. Metabolomics 2017, 13, 122. [Google Scholar] [CrossRef] [PubMed]
- Grant, R.P.; Hoofnagle, A.N. From lost in translation to paradise found: Enabling protein biomarker method transfer by mass spectrometry. Clin. Chem. 2014, 60, 941–944. [Google Scholar] [CrossRef] [PubMed]
- Kaza, M.; Karazniewicz-Lada, M.; Kosicka, K.; Siemiatkowska, A.; Rudzki, P.J. Bioanalytical method validation: New FDA guidance vs. EMA guideline. Better or worse? J. Pharm. Biomed. Anal. 2019, 165, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.W.; Lin, S.W.; Chen, G.Y.; Kuo, C.H. Estimation and Correction of the Blood Volume Variations of Dried Blood Spots Using a Postcolumn Infused-Internal Standard Strategy with LC-Electrospray Ionization-MS. Anal. Chem. 2016, 88, 6457–6464. [Google Scholar] [CrossRef]
- Zheng, N.; Yuan, L.; Ji, Q.C.; Mangus, H.; Song, Y.; Frost, C.; Zeng, J.; Aubry, A.F.; Arnold, M.E. “Center punch” and “whole spot” bioanalysis of apixaban in human dried blood spot samples by UHPLC-MS/MS. J. Chromatogr. B 2015, 988, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Capiau, S.; Wilk, L.S.; Aalders, M.C.; Stove, C.P. A Novel, Nondestructive, Dried Blood Spot-Based Hematocrit Prediction Method Using Noncontact Diffuse Reflectance Spectroscopy. Anal. Chem. 2016, 88, 6538–6546. [Google Scholar] [CrossRef]
- Newman, M.S.; Brandon, T.R.; Groves, M.N.; Gregory, W.L.; Kapur, S.; Zava, D.T. A liquid chromatography/tandem mass spectrometry method for determination of 25-hydroxy vitamin D2 and 25-hydroxy vitamin D3 in dried blood spots: A potential adjunct to diabetes and cardiometabolic risk screening. J. Diabetes Sci. Technol. 2009, 3, 156–162. [Google Scholar] [CrossRef]
- Wilhelm, A.J.; den Burger, J.C.; Vos, R.M.; Chahbouni, A.; Sinjewel, A. Analysis of cyclosporin A in dried blood spots using liquid chromatography tandem mass spectrometry. J. Chromatogr. B 2009, 877, 1595–1598. [Google Scholar] [CrossRef]
- Kayiran, S.M.; Ozbek, N.; Turan, M.; Gurakan, B. Significant differences between capillary and venous complete blood counts in the neonatal period. Clin. Lab. Haematol. 2003, 25, 9–16. [Google Scholar] [CrossRef]
- Han, J.; Higgins, R.; Lim, M.D.; Lin, K.; Yang, J.; Borchers, C.H. Short-Term Stabilities of 21 Amino Acids in Dried Blood Spots. Clin. Chem. 2018, 64, 400–402. [Google Scholar] [CrossRef]
- Rola, R.; Kowalski, K.; Bienkowski, T.; Kolodynska-Goworek, A.; Studzinska, S. Development of a method for multiple vitamin D metabolite measurements by liquid chromatography coupled with tandem mass spectrometry in dried blood spots. Analyst 2018, 144, 299–309. [Google Scholar] [CrossRef] [PubMed]
- El-Gharbawy, A.; Vockley, J. Inborn Errors of Metabolism with Myopathy: Defects of Fatty Acid Oxidation and the Carnitine Shuttle System. Pediatr. Clin. N. Am. 2018, 65, 317–335. [Google Scholar] [CrossRef] [PubMed]
- Vissing, C.R.; Duno, M.; Wibrand, F.; Christensen, M.; Vissing, J. Hydroxylated Long-Chain Acylcarnitines are Biomarkers of Mitochondrial Myopathy. J. Clin. Endocrinol. Metab. 2019, 104, 5968–5976. [Google Scholar] [CrossRef] [PubMed]
- Braverman, N.E.; Moser, A.B. Functions of plasmalogen lipids in health and disease. Biochim. Biophys. Acta 2012, 1822, 1442–1452. [Google Scholar] [CrossRef] [PubMed]
- Nagan, N.; Zoeller, R.A. Plasmalogens: Biosynthesis and functions. Prog. Lipid Res. 2001, 40, 199–229. [Google Scholar] [CrossRef]
- Su, X.Q.; Wang, J.; Sinclair, A.J. Plasmalogens and Alzheimer’s disease: A review. Lipids Health Dis. 2019, 18, 100. [Google Scholar] [CrossRef]
- Calvano, C.D.; Ventura, G.; Sardanelli, A.M.M.; Savino, L.; Losito, I.; Michele, G.; Palmisano, F.; Cataldi, T.R.I. Searching for Potential Lipid Biomarkers of Parkinson’s Disease in Parkin-Mutant Human Skin Fibroblasts by HILIC-ESI-MS/MS: Preliminary Findings. Int. J. Mol. Sci. 2019, 20, 3341. [Google Scholar] [CrossRef]
- Schedin, S.; Sindelar, P.J.; Pentchev, P.; Brunk, U.; Dallner, G. Peroxisomal impairment in Niemann-Pick type C disease. J. Biol. Chem. 1997, 272, 6245–6251. [Google Scholar] [CrossRef]
- Messias, M.C.F.; Mecatti, G.C.; Priolli, D.G.; de Oliveira Carvalho, P. Plasmalogen lipids: Functional mechanism and their involvement in gastrointestinal cancer. Lipids Health Dis. 2018, 17, 41. [Google Scholar] [CrossRef]
- Enns, G.M.; Moore, T.; Le, A.; Atkuri, K.; Shah, M.K.; Cusmano-Ozog, K.; Niemi, A.K.; Cowan, T.M. Degree of glutathione deficiency and redox imbalance depend on subtype of mitochondrial disease and clinical status. PLoS ONE 2014, 9, e100001. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, K.; Naviaux, J.C.; Monk, J.M.; Wang, L.; Naviaux, R.K. Improved Dried Blood Spot-Based Metabolomics: A Targeted, Broad-Spectrum, Single-Injection Method. Metabolites 2020, 10, 82. https://doi.org/10.3390/metabo10030082
Li K, Naviaux JC, Monk JM, Wang L, Naviaux RK. Improved Dried Blood Spot-Based Metabolomics: A Targeted, Broad-Spectrum, Single-Injection Method. Metabolites. 2020; 10(3):82. https://doi.org/10.3390/metabo10030082
Chicago/Turabian StyleLi, Kefeng, Jane C. Naviaux, Jonathan M. Monk, Lin Wang, and Robert K. Naviaux. 2020. "Improved Dried Blood Spot-Based Metabolomics: A Targeted, Broad-Spectrum, Single-Injection Method" Metabolites 10, no. 3: 82. https://doi.org/10.3390/metabo10030082
APA StyleLi, K., Naviaux, J. C., Monk, J. M., Wang, L., & Naviaux, R. K. (2020). Improved Dried Blood Spot-Based Metabolomics: A Targeted, Broad-Spectrum, Single-Injection Method. Metabolites, 10(3), 82. https://doi.org/10.3390/metabo10030082



