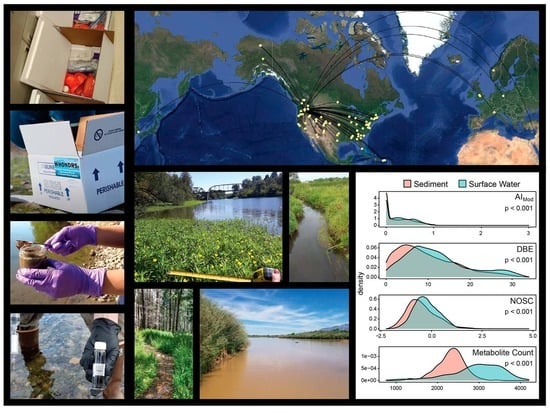
Using Community Science to Reveal the Global Chemogeography of River Metabolomes
Abstract
1. Introduction
2. Results and Discussion
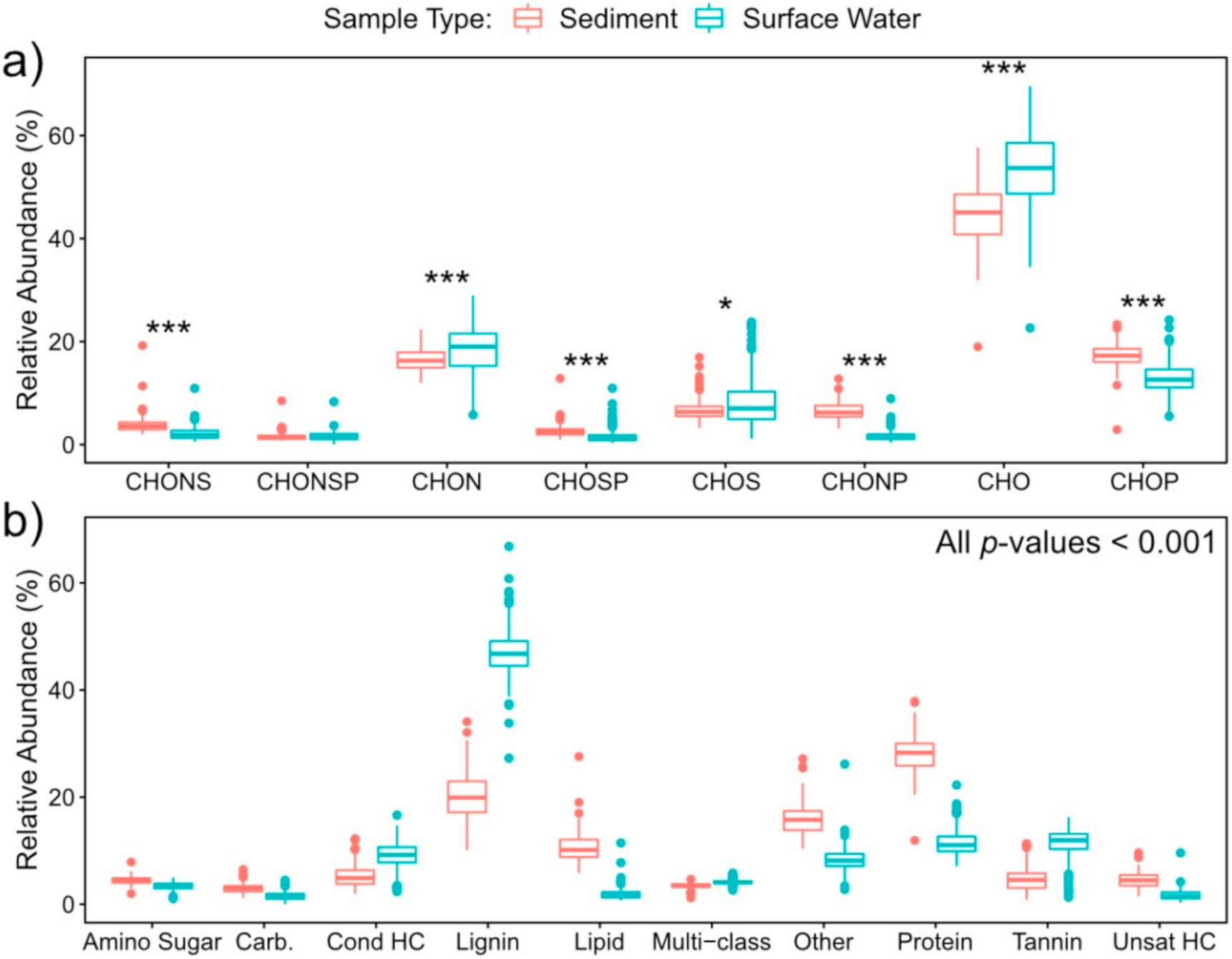
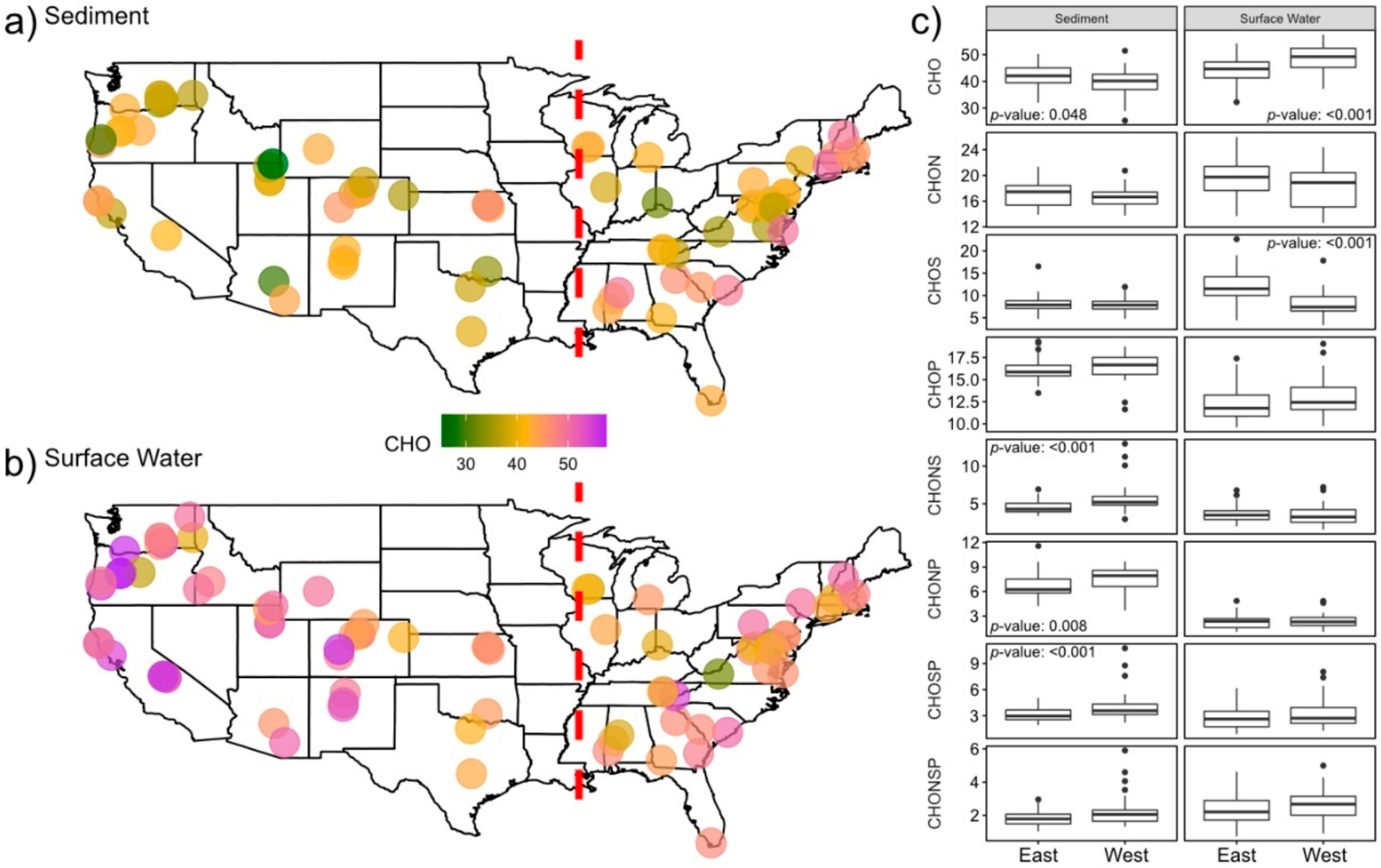
2.1. Surface Water Metabolome Is More Unsaturated, Aromatic, Oxidized, Rich, and Variable Than Sediment Metabolome
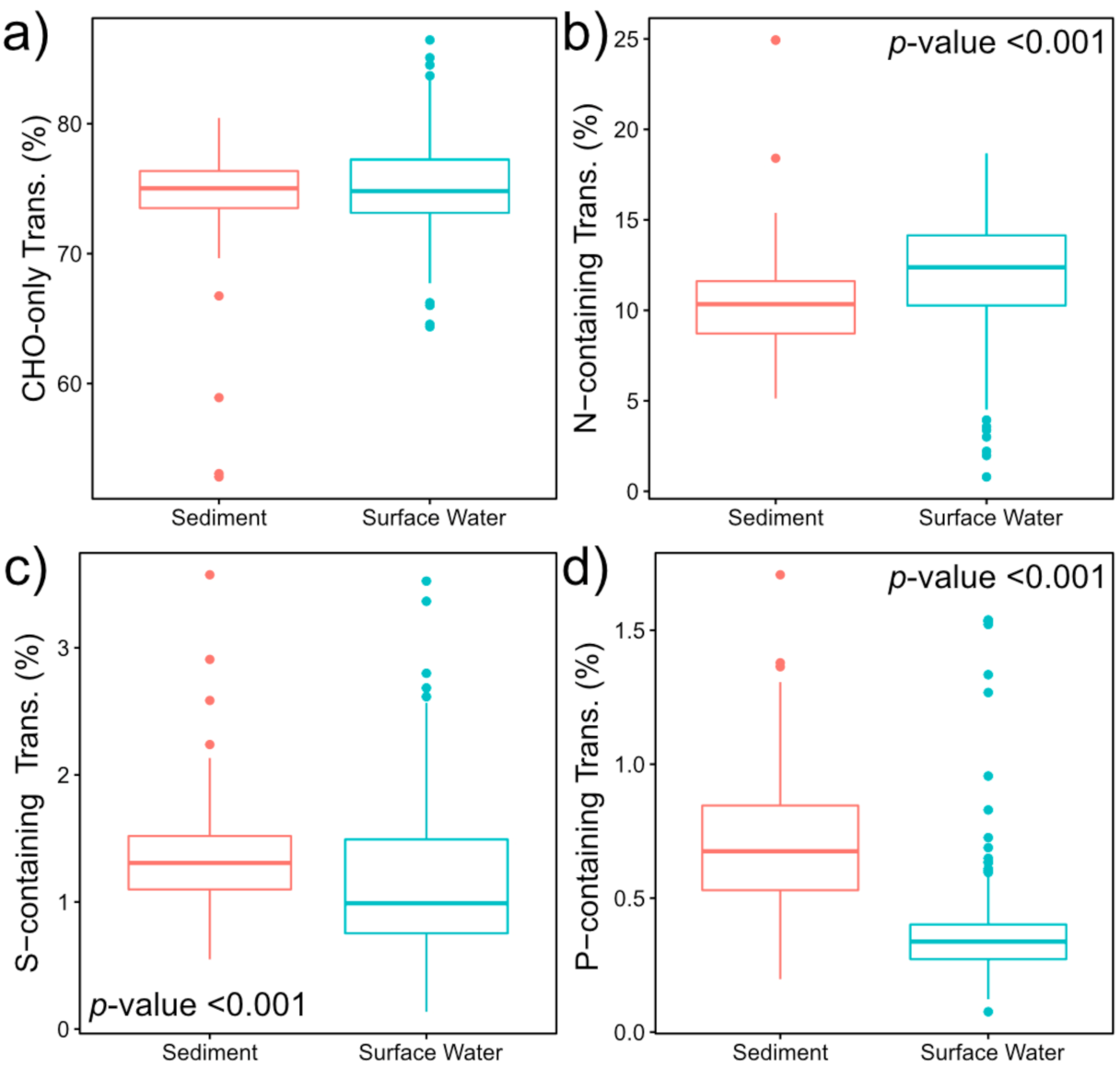
2.2. Nitrogen-, Sulfur- and Phosphorous-Containing Transformations Vary Across Surface Water and Sediment Metabolomes
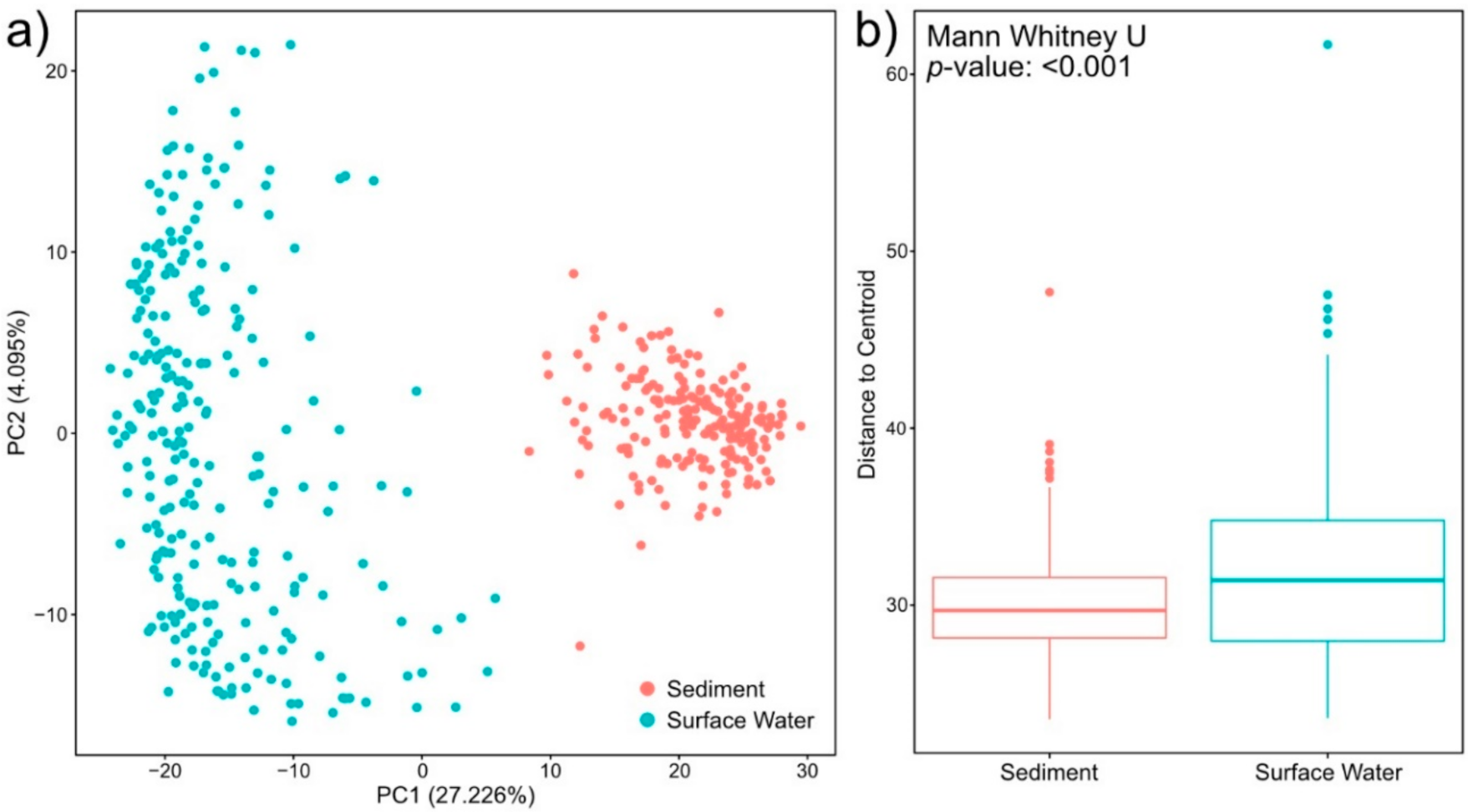
2.3. Sediment Metabolomes Are More Spatially Variable Than Surface Water Metabolomes
3. Materials and Methods
3.1. WHONDRS Summer 2019 Sampling Campaign
3.2. Sample Collection and Laboratory Pre-Processing
3.3. Fourier Transform Ion Cyclotron Resonance Mass Spectrometry (FTICR-MS)
3.4. FTICR-MS Data Analysis
3.5. Biochemical Transformation Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability
Acknowledgments
Conflicts of Interest
References
- Battin, T.J.; Luyssaert, S.; Kaplan, L.A.; Aufdenkampe, A.K.; Richter, A.; Tranvik, L.J. The boundless carbon cycle. Nat. Geosci. 2009, 2, 598–600. [Google Scholar] [CrossRef]
- Cole, J.J.; Prairie, Y.T.; Caraco, N.F.; McDowell, W.H.; Tranvik, L.J.; Striegl, R.G.; Duarte, C.M.; Kortelainen, P.; Downing, J.A.; Middelburg, J.J.; et al. Plumbing the Global Carbon Cycle: Integrating Inland Waters into the Terrestrial Carbon Budget. Ecosystems 2007, 10, 172–185. [Google Scholar] [CrossRef]
- Marín-Spiotta, E.; Gruley, K.E.; Crawford, J.; Atkinson, E.E.; Miesel, J.R.; Greene, S.; Cardona-Correa, C.; Spencer, R.G.M. Paradigm shifts in soil organic matter research affect interpretations of aquatic carbon cycling: Transcending disciplinary and ecosystem boundaries. Biogeochemistry 2014, 117, 279–297. [Google Scholar] [CrossRef]
- Regnier, P.; Friedlingstein, P.; Ciais, P.; Mackenzie, F.T.; Gruber, N.; Janssens, I.A.; Laruelle, G.G.; Lauerwald, R.; Luyssaert, S.; Andersson, A.J.; et al. Anthropogenic perturbation of the carbon fluxes from land to ocean. Nat. Geosci. 2013, 6, 597–607. [Google Scholar] [CrossRef]
- Hotchkiss, E.R.; Hall, R.O., Jr.; Sponseller, R.A.; Butman, D.; Klaminder, J.; Laudon, H.; Rosvall, M.; Karlsson, J. Sources of and processes controlling CO2 emissions change with the size of streams and rivers. Nat. Geosci. 2015, 8, 696–699. [Google Scholar] [CrossRef]
- Catalán, N.; Casas-Ruiz, J.P.; Arce, M.I.; Abril, M.; Bravo, A.G.; Campo, R.D.; Estévez, E.; Freixa, A.; Giménez-Grau, P.; González-Ferreras, A.M.; et al. Behind the Scenes: Mechanisms Regulating Climatic Patterns of Dissolved Organic Carbon Uptake in Headwater Streams. Glob. Biogeochem. Cycles 2018, 32, 1528–1541. [Google Scholar] [CrossRef]
- Aufdenkampe, A.K.; Mayorga, E.; Raymond, P.A.; Melack, J.M.; Doney, S.C.; Alin, S.R.; Aalto, R.E.; Yoo, K. Riverine coupling of biogeochemical cycles between land, oceans, and atmosphere. Front. Ecol. Environ. 2011, 9, 53–60. [Google Scholar] [CrossRef]
- Raymond, P.A.; Hartmann, J.; Lauerwald, R.; Sobek, S.; McDonald, C.; Hoover, M.; Butman, D.; Striegl, R.; Mayorga, E.; Humborg, C.; et al. Global carbon dioxide emissions from inland waters. Nature 2013, 503, 355–359. [Google Scholar] [CrossRef] [PubMed]
- Moody, C.S.; Worrall, F.; Evans, C.D.; Jones, T.G. The rate of loss of dissolved organic carbon (DOC) through a catchment. J. Hydrol. 2013, 492, 139–150. [Google Scholar] [CrossRef]
- Cory, R.M.; Ward, C.P.; Crump, B.C.; Kling, G.W. Sunlight controls water column processing of carbon in arctic fresh waters. Science 2014, 345, 925. [Google Scholar] [CrossRef]
- Newbold, J.D.; Bott, T.L.; Kaplan, L.A.; Dow, C.L.; Jackson, J.K.; Aufdenkampe, A.K.; Martin, L.A.; Horn, D.J.V.; Long, A.A. Uptake of nutrients and organic C in streams in New York City drinking-water-supply watersheds. Freshw. Sci. 2006, 25, 998–1017. [Google Scholar] [CrossRef]
- Bernhardt, E.S.; McDowell, W.H. Twenty years apart: Comparisons of DOM uptake during leaf leachate releases to Hubbard Brook Valley streams in 1979 versus 2000. J. Geophys. Res. Biogeosci. 2008, 113. [Google Scholar] [CrossRef]
- Koehler, B.; Wachenfeldt, E.V.; Kothawala, D.; Tranvik, L.J. Reactivity continuum of dissolved organic carbon decomposition in lake water. J. Geophys. Res. Biogeosci. 2012, 117. [Google Scholar] [CrossRef]
- Berggren, M.; Giorgio, P.A. del Distinct patterns of microbial metabolism associated to riverine dissolved organic carbon of different source and quality. J. Geophys. Res. Biogeosci. 2015, 120, 989–999. [Google Scholar] [CrossRef]
- Stegen, J.C.; Johnson, T.; Fredrickson, J.K.; Wilkins, M.J.; Konopka, A.E.; Nelson, W.C.; Arntzen, E.V.; Chrisler, W.B.; Chu, R.K.; Fansler, S.J.; et al. Influences of organic carbon speciation on hyporheic corridor biogeochemistry and microbial ecology. Nat. Commun. 2018, 9, 1034. [Google Scholar] [CrossRef]
- Garayburu-Caruso, V.A.; Stegen, J.C.; Song, H.-S.; Renteria, L.; Wells, J.; Garcia, W.; Resch, C.T.; Goldman, A.E.; Chu, R.K.; Toyoda, J.; et al. Carbon Limitation Leads to Thermodynamic Regulation of Aerobic Metabolism. Environ. Sci. Technol. Lett. 2020, 7, 517–524. [Google Scholar] [CrossRef]
- Graham, E.B.; Crump, A.R.; Kennedy, D.W.; Arntzen, E.; Fansler, S.; Purvine, S.O.; Nicora, C.D.; Nelson, W.; Tfaily, M.M.; Stegen, J.C. Multi ’omics comparison reveals metabolome biochemistry, not microbiome composition or gene expression, corresponds to elevated biogeochemical function in the hyporheic zone. Sci. Total Environ. 2018, 642, 742–753. [Google Scholar] [CrossRef]
- Graham, E.B.; Tfaily, M.M.; Crump, A.R.; Goldman, A.E.; Bramer, L.M.; Arntzen, E.; Romero, E.; Resch, C.T.; Kennedy, D.W.; Stegen, J.C. Carbon Inputs From Riparian Vegetation Limit Oxidation of Physically Bound Organic Carbon Via Biochemical and Thermodynamic Processes. J. Geophys. Res. Biogeosci. 2017, 122, 3188–3205. [Google Scholar] [CrossRef]
- Bundy, J.G.; Davey, M.P.; Viant, M.R. Environmental metabolomics: A critical review and future perspectives. Metabolomics 2008, 5, 3. [Google Scholar] [CrossRef]
- Rue, G.P.; Rock, N.D.; Gabor, R.S.; Pitlick, J.; Tfaily, M.; McKnight, D.M. Concentration-discharge relationships during an extreme event: Contrasting behavior of solutes and changes to chemical quality of dissolved organic material in the Boulder Creek Watershed during the September 2013 flood. Water Resour. Res. 2017, 53, 5276–5297. [Google Scholar] [CrossRef]
- Wilson, R.M.; Tfaily, M.M. Advanced Molecular Techniques Provide New Rigorous Tools for Characterizing Organic Matter Quality in Complex Systems. J. Geophys. Res. Biogeosci. 2018, 123, 1790–1795. [Google Scholar] [CrossRef]
- Li, L.; He, Z.L.; Tfaily, M.M.; Inglett, P.; Stoffella, P.J. Spatial-temporal variations of dissolved organic nitrogen molecular composition in agricultural runoff water. Water Res. 2018, 137, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Walker, L.R.; Tfaily, M.M.; Shaw, J.B.; Hess, N.J.; Paša-Tolić, L.; Koppenaal, D.W. Unambiguous identification and discovery of bacterial siderophores by direct injection 21 Tesla Fourier transform ion cyclotron resonance mass spectrometry. Metallomics 2017, 9, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Hodgkins, S.B.; Tfaily, M.M.; McCalley, C.K.; Logan, T.A.; Crill, P.M.; Saleska, S.R.; Rich, V.I.; Chanton, J.P. Changes in peat chemistry associated with permafrost thaw increase greenhouse gas production. Proc. Natl. Acad. Sci. USA 2014, 111, 5819–5824. [Google Scholar] [CrossRef] [PubMed]
- Jones, O.A.H.; Lear, G.; Welji, A.M.; Collins, G.; Quince, C. Community Metabolomics in Environmental Microbiology. In Microbial Metabolomics: Applications in Clinical, Environmental, and Industrial Microbiology; Beale, D.J., Kouremenos, K.A., Palombo, E.A., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 199–224. ISBN 978-3-319-46326-1. [Google Scholar]
- Jones, O.A.H.; Sdepanian, S.; Lofts, S.; Svendsen, C.; Spurgeon, D.J.; Maguire, M.L.; Griffin, J.L. Metabolomic analysis of soil communities can be used for pollution assessment. Environ. Toxicol. Chem. 2014, 33, 61–64. [Google Scholar] [CrossRef]
- Beale, D.J.; Crosswell, J.; Karpe, A.V.; Metcalfe, S.S.; Morrison, P.D.; Staley, C.; Ahmed, W.; Sadowsky, M.J.; Palombo, E.A.; Steven, A.D.L. Seasonal metabolic analysis of marine sediments collected from Moreton Bay in South East Queensland, Australia, using a multi-omics-based approach. Sci. Total Environ. 2018, 631–632, 1328–1341. [Google Scholar] [CrossRef]
- Beale, D.J.; Crosswell, J.; Karpe, A.V.; Ahmed, W.; Williams, M.; Morrison, P.D.; Metcalfe, S.; Staley, C.; Sadowsky, M.J.; Palombo, E.A.; et al. A multi-omics based ecological analysis of coastal marine sediments from Gladstone, in Australia’s Central Queensland, and Heron Island, a nearby fringing platform reef. Sci. Total Environ. 2017, 609, 842–853. [Google Scholar] [CrossRef]
- Shah, R.M.; Crosswell, J.; Metcalfe, S.S.; Carlin, G.; Morrison, P.D.; Karpe, A.V.; Palombo, E.A.; Steven, A.D.L.; Beale, D.J. Influence of Human Activities on Broad-Scale Estuarine-Marine Habitats Using Omics-Based Approaches Applied to Marine Sediments. Microorganisms 2019, 7, 419. [Google Scholar] [CrossRef]
- Kimes, N.E.; Callaghan, A.V.; Aktas, D.F.; Smith, W.L.; Sunner, J.; Golding, B.; Drozdowska, M.; Hazen, T.C.; Suflita, J.M.; Morris, P.J. Metagenomic analysis and metabolite profiling of deep-sea sediments from the Gulf of Mexico following the Deepwater Horizon oil spill. Front. Microbiol. 2013, 4, 50. [Google Scholar] [CrossRef]
- Lam, B.; Baer, A.; Alaee, M.; Lefebvre, B.; Moser, A.; Williams, A.; Simpson, A.J. Major Structural Components in Freshwater Dissolved Organic Matter. Environ. Sci. Technol. 2007, 41, 8240–8247. [Google Scholar] [CrossRef]
- Jaffé, R.; McKnight, D.; Maie, N.; Cory, R.; McDowell, W.H.; Campbell, J.L. Spatial and temporal variations in DOM composition in ecosystems: The importance of long-term monitoring of optical properties. J. Geophys. Res. Biogeosci. 2008, 113. [Google Scholar] [CrossRef]
- Gonsior, M.; Peake, B.M.; Cooper, W.T.; Podgorski, D.; D’Andrilli, J.; Cooper, W.J. Photochemically induced changes in dissolved organic matter identified by ultrahigh resolution fourier transform ion cyclotron resonance mass spectrometry. Environ. Sci. Technol. 2009, 43, 698–703. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Li, X.; Mesfioui, R.; Bauer, J.E.; Chambers, R.M.; Canuel, E.A.; Hatcher, P.G. Use of ESI-FTICR-MS to Characterize Dissolved Organic Matter in Headwater Streams Draining Forest-Dominated and Pasture-Dominated Watersheds. PLoS ONE 2015, 10, e0145639. [Google Scholar] [CrossRef] [PubMed]
- Stegen, J.C.; Goldman, A.E. WHONDRS: A Community Resource for Studying Dynamic River Corridors. mSystems 2018, 3. [Google Scholar] [CrossRef]
- Jaffé, R.; Yamashita, Y.; Maie, N.; Cooper, W.T.; Dittmar, T.; Dodds, W.K.; Jones, J.B.; Myoshi, T.; Ortiz-Zayas, J.R.; Podgorski, D.C.; et al. Dissolved Organic Matter in Headwater Streams: Compositional Variability across Climatic Regions of North America. Geochim. Cosmochim. Acta 2012, 94, 95–108. [Google Scholar] [CrossRef]
- Wagner, S.; Riedel, T.; Niggemann, J.; Vähätalo, A.V.; Dittmar, T.; Jaffé, R. Linking the Molecular Signature of Heteroatomic Dissolved Organic Matter to Watershed Characteristics in World Rivers. Environ. Sci. Technol. 2015, 49, 13798–13806. [Google Scholar] [CrossRef]
- Danczak, R.E.; Goldman, A.E.; Chu, R.K.; Toyoda, J.G.; Garayburu-Caruso, V.A.; Tolić, N.; Graham, E.B.; Morad, J.W.; Renteria, L.; Wells, J.R.; et al. Ecological theory applied to environmental metabolomes reveals compositional divergence despite conserved molecular properties. bioRxiv 2020. [Google Scholar] [CrossRef]
- Steefel, C.I.; Appelo, C.A.J.; Arora, B.; Jacques, D.; Kalbacher, T.; Kolditz, O.; Lagneau, V.; Lichtner, P.C.; Mayer, K.U.; Meeussen, J.C.L.; et al. Reactive transport codes for subsurface environmental simulation. Comput. Geosci. 2015, 19, 445–478. [Google Scholar] [CrossRef]
- Song, H.-S.; Stegen, J.C.; Graham, E.B.; Lee, J.-Y.; Garayburu-Caruso, V.A.; Nelson, W.C.; Chen, X.; Moulton, J.D.; Scheibe, T.D. Representing Organic Matter Thermodynamics in Biogeochemical Reactions via Substrate-Explicit Modeling. bioRxiv 2020. [Google Scholar] [CrossRef]
- Stegen, J.; Brodie, E.; Wrighton, K.; Bayer, P.; Lesmes, D.; Emani, S.; Moerman, J. Open Watershed Science by Design: Leveraging Distributed Research Networks to Understand Watershed Systems: Workshop Report, DOE/SC-0200; USDOE Office of Science (SC): Washington, DC, USA, 2019; Available online: https://doesbr.org/openwatersheds/Open_Watersheds_Low_Res.pdf (accessed on 12 June 2019).
- Uhlmann, E.L.; Ebersole, C.R.; Chartier, C.R.; Errington, T.M.; Kidwell, M.C.; Lai, C.K.; McCarthy, R.J.; Riegelman, A.; Silberzahn, R.; Nosek, B.A. Scientific Utopia III: Crowdsourcing Science. Perspect. Psychol. Sci. 2019. [Google Scholar] [CrossRef]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.-W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef] [PubMed]
- Toyoda, J.G.; Goldman, A.E.; Chu, R.K.; Danczak, R.E.; Daly, R.A.; Garayburu-Caruso, V.A.; Graham, E.B.; Lin, X.; Moran, J.J.; Ren, H. WHONDRS Summer 2019 Sampling Campaign: Global River Corridor Surface Water FTICR-MS and Stable Isotopes. Environmental System Science Data Infrastructure for a Virtual Ecosystem, Worldwide Hydrobiogeochemistry Observation Network for Dynamic River Systems (WHONDRS). 2020. Available online: https://data.ess-dive.lbl.gov/view/doi:10.15485/1603775 (accessed on 12 June 2019). [CrossRef]
- Koch, B.P.; Dittmar, T. From mass to structure: An aromaticity index for high-resolution mass data of natural organic matter. Rapid Commun. Mass Spectrom. 2016, 20, 926–932, Erratum in 2016, 30, 250–250. [Google Scholar] [CrossRef]
- Willoughby, A.S.; Wozniak, A.S.; Hatcher, P.G. A molecular-level approach for characterizing water-insoluble components of ambient organic aerosol particulates using ultrahigh-resolution mass spectrometry. Atmos. Chem. Phys. 2014, 14, 10299–10314. [Google Scholar] [CrossRef]
- LaRowe, D.E.; van Cappellen, P. Degradation of natural organic matter: A thermodynamic analysis. Geochim. Cosmochim. Acta 2011, 75, 2030–2042. [Google Scholar] [CrossRef]
- Boye, K.; Noël, V.; Tfaily, M.M.; Bone, S.E.; Williams, K.H.; Bargar, J.R.; Fendorf, S. Thermodynamically controlled preservation of organic carbon in floodplains. Nat. Geosci. 2017, 10, 415–419. [Google Scholar] [CrossRef]
- Kim, S.; Kramer, R.W.; Hatcher, P.G. Graphical Method for Analysis of Ultrahigh-Resolution Broadband Mass Spectra of Natural Organic Matter, the Van Krevelen Diagram. Anal. Chem. 2003, 75, 5336–5344. [Google Scholar] [CrossRef]
- Hedges, J.I.; Mann, D.C. The lignin geochemistry of marine sediments from the southern Washington coast. Geochim. Cosmochim. Acta 1979, 43, 1809–1818. [Google Scholar] [CrossRef]
- Stegen, J.C.; Fredrickson, J.K.; Wilkins, M.J.; Konopka, A.E.; Nelson, W.C.; Arntzen, E.V.; Chrisler, W.B.; Chu, R.K.; Danczak, R.E.; Fansler, S.J.; et al. Groundwater–surface water mixing shifts ecological assembly processes and stimulates organic carbon turnover. Nat. Commun. 2016, 7, 11237. [Google Scholar] [CrossRef]
- Pracht, L.E.; Tfaily, M.M.; Ardissono, R.J.; Neumann, R.B. Molecular characterization of organic matter mobilized from Bangladeshi aquifer sediment: Tracking carbon compositional change during microbial utilization. Biogeosciences 2018, 15, 1733–1747. [Google Scholar] [CrossRef]
- Valle, J.; Harir, M.; Gonsior, M.; Enrich-Prast, A.; Schmitt-Kopplin, P.; Bastviken, D.; Hertkorn, N. Molecular differences between water column and sediment pore water SPE-DOM in ten Swedish boreal lakes. Water Res. 2020, 170, 115320. [Google Scholar] [CrossRef]
- Boulton, A.J.; Findlay, S.; Marmonier, P.; Stanley, E.H.; Valett, H.M. The functional significance of the hyporheic zone in streams and rivers. Annu. Rev. Ecol. Syst. 1998, 29, 59–81. [Google Scholar] [CrossRef]
- Boano, F.; Harvey, J.W.; Marion, A.; Packman, A.I.; Revelli, R.; Ridolfi, L.; Wörman, A. Hyporheic flow and transport processes: Mechanisms, models, and biogeochemical implications. Rev. Geophys. 2014, 52, 603–679. [Google Scholar] [CrossRef]
- Harvey, J.; Gooseff, M. River corridor science: Hydrologic exchange and ecological consequences from bedforms to basins. Water Resour. Res. 2015, 51, 6893–6922. [Google Scholar] [CrossRef]
- Gomez-Velez, J.D.; Harvey, J.W.; Cardenas, M.B.; Kiel, B. Denitrification in the Mississippi River network controlled by flow through river bedforms. Nat. Geosci. 2015, 8, 941–945. [Google Scholar] [CrossRef]
- Knapp, J.L.A.; González-Pinzón, R.; Drummond, J.D.; Larsen, L.G.; Cirpka, O.A.; Harvey, J.W. Tracer-based characterization of hyporheic exchange and benthic biolayers in streams. Water Resour. Res. 2017, 53, 1575–1594. [Google Scholar] [CrossRef]
- Fischer, H.; Kloep, F.; Wilzcek, S.; Pusch, M.T. A river’s liver–microbial processes within the hyporheic zone of a large lowland river. Biogeochemistry 2005, 76, 349–371. [Google Scholar] [CrossRef]
- Battin, T.J.; Besemer, K.; Bengtsson, M.M.; Romani, A.M.; Packmann, A.I. The ecology and biogeochemistry of stream biofilms. Nat. Rev. Microbiol. 2016, 14, 251. [Google Scholar] [CrossRef]
- Bailey, V.L.; Smith, A.P.; Tfaily, M.; Fansler, S.J.; Bond-Lamberty, B. Differences in soluble organic carbon chemistry in pore waters sampled from different pore size domains. Soil Biol. Biochem. 2017, 107, 133–143. [Google Scholar] [CrossRef]
- Cory, R.M.; Kling, G.W. Interactions between sunlight and microorganisms influence dissolved organic matter degradation along the aquatic continuum. Limnol. Oceanogr. Lett. 2018, 3, 102–116. [Google Scholar] [CrossRef]
- Bao, H.; Niggemann, J.; Huang, D.; Dittmar, T.; Kao, S.-J. Different Responses of Dissolved Black Carbon and Dissolved Lignin to Seasonal Hydrological Changes and an Extreme Rain Event. J. Geophys. Res. Biogeosci. 2019, 124, 479–493. [Google Scholar] [CrossRef]
- Fellman, J.B.; Hood, E.; Edwards, R.T.; D’Amore, D.V. Changes in the concentration, biodegradability, and fluorescent properties of dissolved organic matter during stormflows in coastal temperate watersheds. J. Geophys. Res. Biogeosci. 2009, 114. [Google Scholar] [CrossRef]
- Hood, E.; Gooseff, M.N.; Johnson, S.L. Changes in the character of stream water dissolved organic carbon during flushing in three small watersheds, Oregon. J. Geophys. Res. Biogeosci. 2006, 111. [Google Scholar] [CrossRef]
- Ward, N.D.; Richey, J.E.; Keil, R.G. Temporal variation in river nutrient and dissolved lignin phenol concentrations and the impact of storm events on nutrient loading to Hood Canal, Washington, USA. Biogeochemistry 2012, 111, 629–645. [Google Scholar] [CrossRef]
- Tfaily, M.M.; Chu, R.K.; Toyoda, J.; Tolić, N.; Robinson, E.W.; Paša-Tolić, L.; Hess, N.J. Sequential extraction protocol for organic matter from soils and sediments using high resolution mass spectrometry. Anal. Chim. Acta 2017, 972, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Kaling, M.; Schmidt, A.; Moritz, F.; Rosenkranz, M.; Witting, M.; Kasper, K.; Janz, D.; Schmitt-Kopplin, P.; Schnitzler, J.-P.; Polle, A. Mycorrhiza-Triggered Transcriptomic and Metabolomic Networks Impinge on Herbivore Fitness. Plant Physiol. 2018, 176, 2639–2656. [Google Scholar] [CrossRef]
- Moritz, F.; Kaling, M.; Schnitzler, J.-P.; Schmitt-Kopplin, P. Characterization of poplar metabotypes via mass difference enrichment analysis. Plant Cell Environ. 2017, 40, 1057–1073. [Google Scholar] [CrossRef]
- Craine, J.M.; Morrow, C.; Fierer, N. Microbial Nitrogen Limitation Increases Decomposition. Ecology 2007, 88, 2105–2113. [Google Scholar] [CrossRef]
- Moorhead, D.L.; Sinsabaugh, R.L. A Theoretical Model of Litter Decay and Microbial Interaction. Ecol. Monogr. 2006, 76, 151–174. [Google Scholar] [CrossRef]
- Zhao, F.J.; Wu, J.; McGrath, S.P. Chapter 12—Soil Organic Sulphur and its Turnover. In Humic Substances in Terrestrial Ecosystems; Piccolo, A., Ed.; Elsevier Science B.V.: Amsterdam, The Netherlands, 1996; pp. 467–506. ISBN 978-0-444-81516-3. [Google Scholar]
- Reddy, K.R.; Kadlec, R.H.; Flaig, E.; Gale, P.M. Phosphorus Retention in Streams and Wetlands: A Review. Crit. Rev. Environ. Sci. Technol. 1999, 29, 83–146. [Google Scholar] [CrossRef]
- Freixa, A.; Ejarque, E.; Crognale, S.; Amalfitano, S.; Fazi, S.; Butturini, A.; Romaní, A.M. Sediment microbial communities rely on different dissolved organic matter sources along a Mediterranean river continuum. Limnol. Oceanogr. 2016, 61, 1389–1405. [Google Scholar] [CrossRef]
- Vazquez, E.; Amalfitano, S.; Fazi, S.; Butturini, A. Dissolved organic matter composition in a fragmented Mediterranean fluvial system under severe drought conditions. Biogeochemistry 2011, 102, 59–72. [Google Scholar] [CrossRef]
- Butturini, A.; Guarch, A.; Romaní, A.M.; Freixa, A.; Amalfitano, S.; Fazi, S.; Ejarque, E. Hydrological conditions control in situ DOM retention and release along a Mediterranean river. Water Res. 2016, 99, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Ejarque, E.; Freixa, A.; Vazquez, E.; Guarch, A.; Amalfitano, S.; Fazi, S.; Romaní, A.M.; Butturini, A. Quality and reactivity of dissolved organic matter in a Mediterranean river across hydrological and spatial gradients. Sci. Total Environ. 2017, 599–600, 1802–1812. [Google Scholar] [CrossRef]
- Robinson, N.P.; Allred, B.W.; Jones, M.O.; Moreno, A.; Kimball, J.S.; Naugle, D.E.; Erickson, T.A.; Richardson, A.D. A dynamic Landsat derived normalized difference vegetation index (NDVI) product for the conterminous United States. Remote Sens. 2017, 9, 863. [Google Scholar] [CrossRef]
- Sayre, R. (Ed.) A New Map of Standardized Terrestrial Ecosystems of the Conterminous United States; U.S. Department of the Interior, U.S. Geological Survey: Reston, VA, USA, 2009; ISBN 978-1-4113-2432-9.
- Fierer, N.; Ladau, J.; Clemente, J.C.; Leff, J.W.; Owens, S.M.; Pollard, K.S.; Knight, R.; Gilbert, J.A.; McCulley, R.L. Reconstructing the Microbial Diversity and Function of Pre-Agricultural Tallgrass Prairie Soils in the United States. Science 2013, 342, 621–624. [Google Scholar] [CrossRef] [PubMed]
- Horton, R.E. Erosional Development of Streams and Their Drainage Basins; Hydrophysical Approach to Quantitative Morphology. GSA Bull. 1945, 56, 275–370. [Google Scholar] [CrossRef]
- Strahler, A.N. Dimensional Analysis Applied to Fluvially Eroded Landforms. GSA Bull. 1958, 69, 279–300. [Google Scholar] [CrossRef]
- Jensen, B. AOS Protocol and Procedure: Sediment Chemistry Sampling in Wadeable Streams (NEON.DOC.001193). Available online: http://data.neonscience.org/documents (accessed on 12 June 2019).
- Dittmar, T.; Koch, B.; Hertkorn, N.; Kattner, G. A simple and efficient method for the solid-phase extraction of dissolved organic matter (SPE-DOM) from seawater: SPE-DOM from seawater. Limnol. Oceanogr. Methods 2008, 6, 230–235. [Google Scholar] [CrossRef]
- Tolić, N.; Liu, Y.; Liyu, A.; Shen, Y.; Tfaily, M.M.; Kujawinski, E.B.; Longnecker, K.; Kuo, L.-J.; Robinson, E.W.; Paša-Tolić, L.; et al. Formularity: Software for Automated Formula Assignment of Natural and Other Organic Matter from Ultrahigh-Resolution Mass Spectra. Anal. Chem. 2017, 89, 12659–12665. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Bramer, L.M.; White, A.M.; Stratton, K.G.; Thompson, A.M.; Claborne, D.; Hofmockel, K.; McCue, L.A. ftmsRanalysis: An R package for exploratory data analysis and interactive visualization of FT-MS data. PLoS Comput. Biol. 2020, 16, e1007654. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’hara, R.; Simpson, G.L.; Solymos, P. vegan: Community Ecology Package. R package version 2.5-6. Vienna R Found. Stat. Comput. Sch. 2019. [Google Scholar]
- Hawkes, J.A.; D’Andrilli, J.; Agar, J.N.; Barrow, M.P.; Berg, S.M.; Catalán, N.; Chen, H.; Chu, R.K.; Cole, R.B.; Dittmar, T.; et al. An international laboratory comparison of dissolved organic matter composition by high resolution mass spectrometry: Are we getting the same answer? Limnol. Oceanogr. Methods 2020, 18, 235–258. [Google Scholar] [CrossRef]
- He, C.; Zhang, Y.; Li, Y.; Zhuo, X.; Li, Y.; Zhang, C.; Shi, Q. In-House Standard Method for Molecular Characterization of Dissolved Organic Matter by FT-ICR Mass Spectrometry. ACS Omega 2020, 5, 11730–11736. [Google Scholar] [CrossRef] [PubMed]
- Goldman, A.E.; Chu, R.K.; Danczak, R.E.; Daly, R.A.; Fansler, S.; Garayburu-Caruso, V.A.; Graham, E.B.; McCall, M.L.; Ren, H.; Renteria, L. WHONDRS Summer 2019 Sampling Campaign: Global River Corridor Sediment FTICR-MS, NPOC, and Aerobic Respiration. Environmental System Science Data Infrastructure for a Virtual Ecosystem, Worldwide Hydrobiogeochemistry Observation Network for Dynamic River Systems (WHONDRS). 2020. Available online: https://data.ess-dive.lbl.gov/view/doi:10.15485/1729719 (accessed on 12 June 2019). [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garayburu-Caruso, V.A.; Danczak, R.E.; Stegen, J.C.; Renteria, L.; Mccall, M.; Goldman, A.E.; Chu, R.K.; Toyoda, J.; Resch, C.T.; Torgeson, J.M.; et al. Using Community Science to Reveal the Global Chemogeography of River Metabolomes. Metabolites 2020, 10, 518. https://doi.org/10.3390/metabo10120518
Garayburu-Caruso VA, Danczak RE, Stegen JC, Renteria L, Mccall M, Goldman AE, Chu RK, Toyoda J, Resch CT, Torgeson JM, et al. Using Community Science to Reveal the Global Chemogeography of River Metabolomes. Metabolites. 2020; 10(12):518. https://doi.org/10.3390/metabo10120518
Chicago/Turabian StyleGarayburu-Caruso, Vanessa A., Robert E. Danczak, James C. Stegen, Lupita Renteria, Marcy Mccall, Amy E. Goldman, Rosalie K. Chu, Jason Toyoda, Charles T. Resch, Joshua M. Torgeson, and et al. 2020. "Using Community Science to Reveal the Global Chemogeography of River Metabolomes" Metabolites 10, no. 12: 518. https://doi.org/10.3390/metabo10120518
APA StyleGarayburu-Caruso, V. A., Danczak, R. E., Stegen, J. C., Renteria, L., Mccall, M., Goldman, A. E., Chu, R. K., Toyoda, J., Resch, C. T., Torgeson, J. M., Wells, J., Fansler, S., Kumar, S., & Graham, E. B. (2020). Using Community Science to Reveal the Global Chemogeography of River Metabolomes. Metabolites, 10(12), 518. https://doi.org/10.3390/metabo10120518