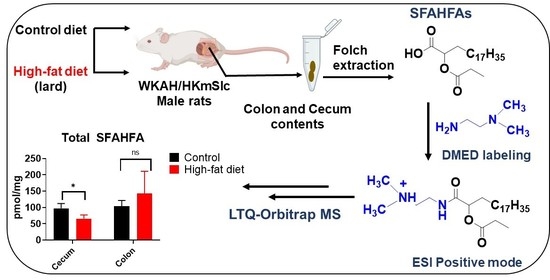
Chemical Labeling Assisted Detection and Identification of Short Chain Fatty Acid Esters of Hydroxy Fatty Acid in Rat Colon and Cecum Contents
Abstract
:1. Introduction
2. Results and Discussion
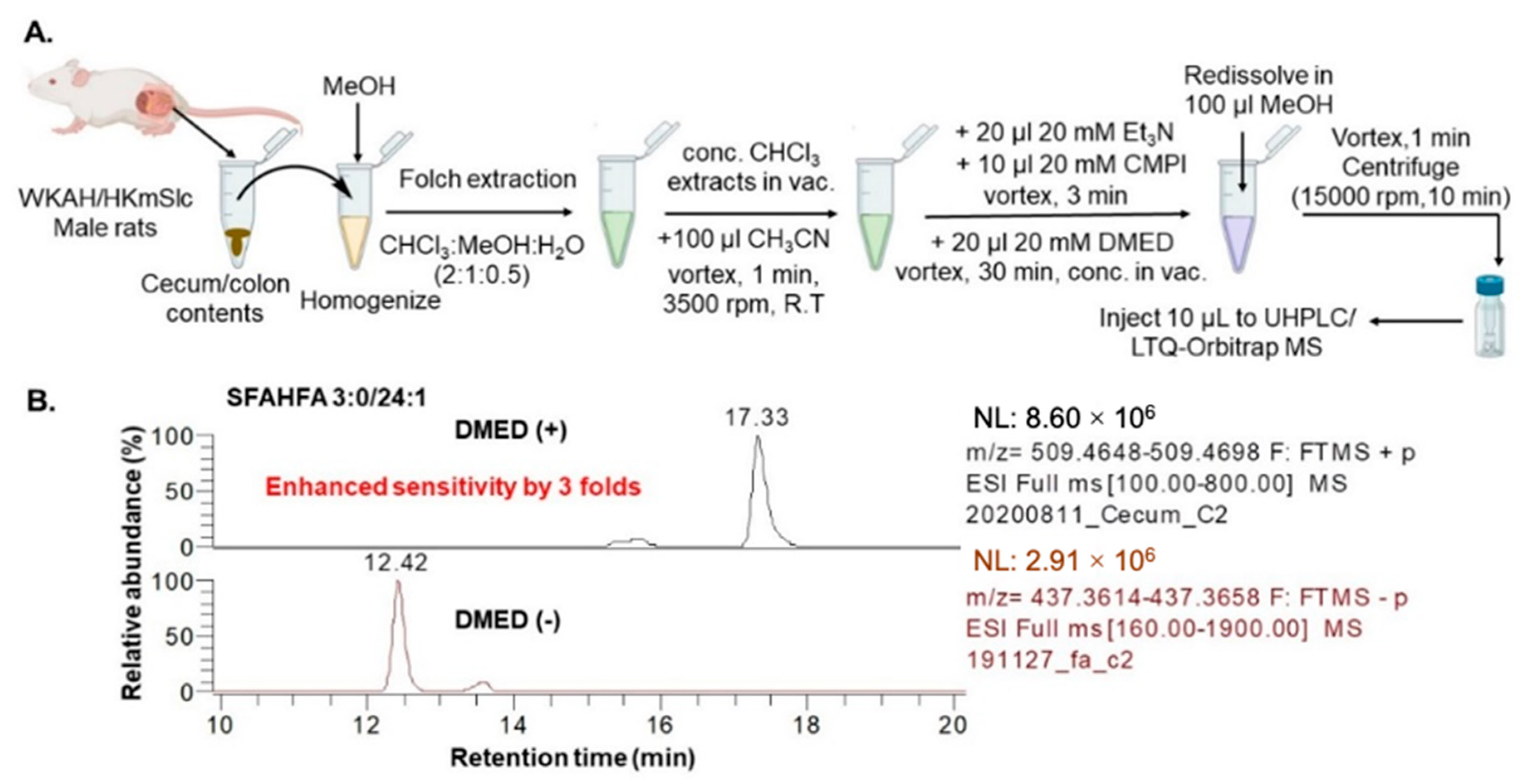
2.1. Chemical Labeling Assisted Detection and Identification of SFAHFAs
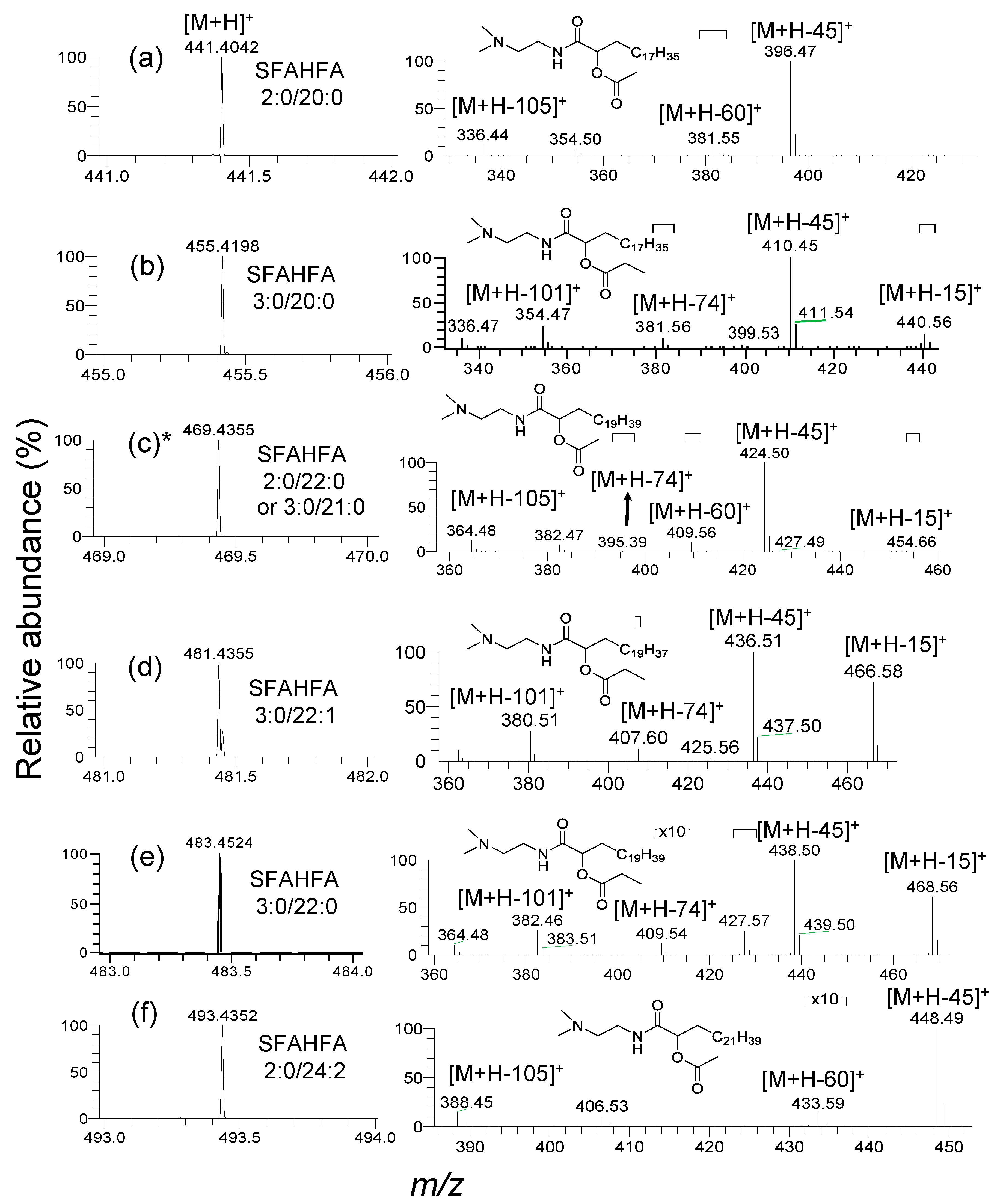
2.2. Annotation of DMED-labeled-SFAHFAs by MS/MS Analysis
2.3. Fragmentation Pattern of DMED-labeled SFAHFAs
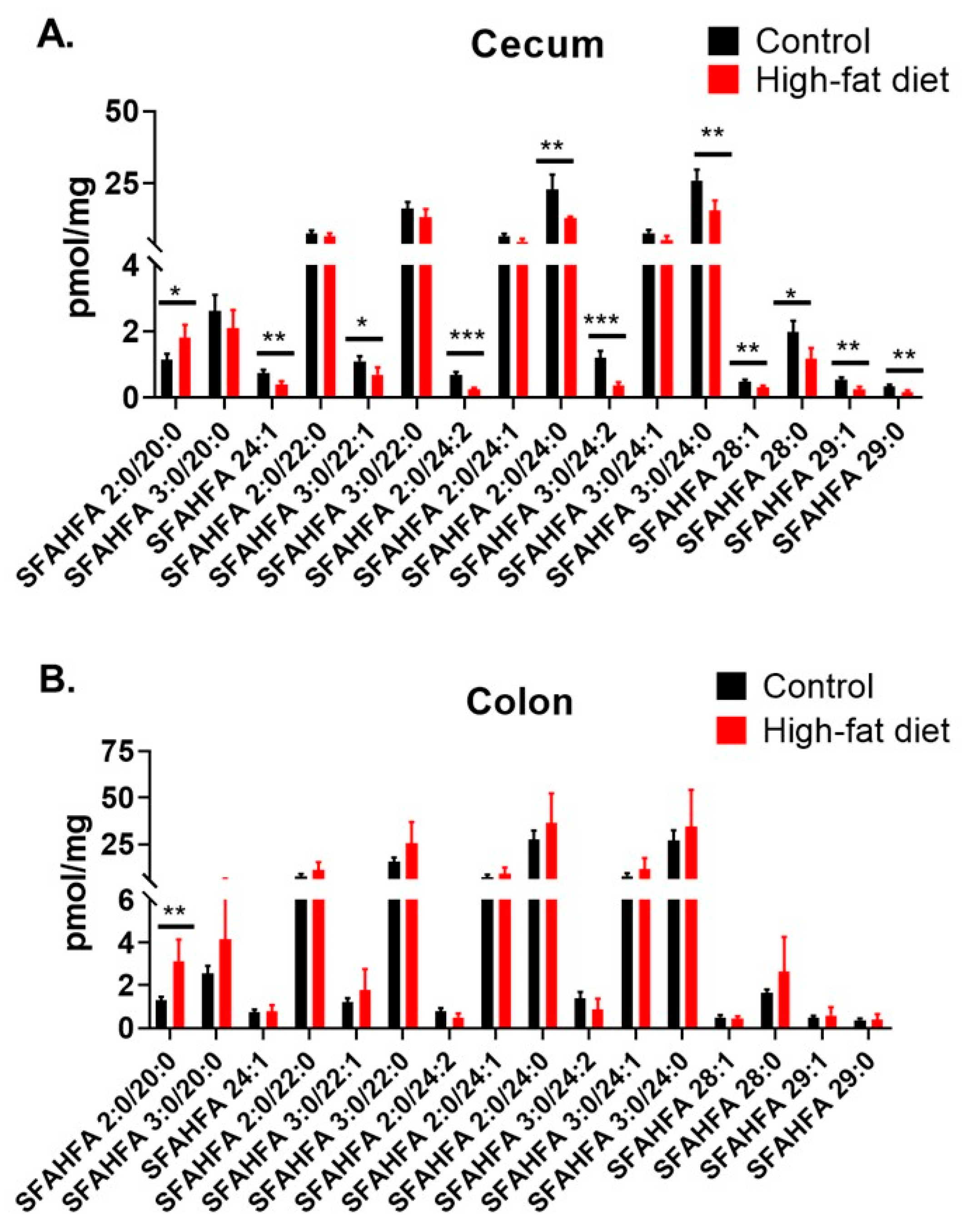
2.4. Altered Levels of SFAHFAs as Possible Biomarkers of Obesity
3. Materials and Methods
3.1. Chemicals
3.2. Animal Samples
3.3. Sample Preparation for LC-MS
3.4. LC-MS Analysis
3.5. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Boden, G. Obesity and Free Fatty Acids. Endocrinol. Metab. Clin. N. Am. 2008, 37, 635–646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halade, G.V.; Kain, V. Obesity and cardiometabolic defects in heart failure pathology. Compr. Physiol. 2017, 7, 1463–1477. [Google Scholar] [CrossRef] [PubMed]
- Jaramillo, M.G.; Lytle, K.A.; Spooner, M.H.; Jump, D.B. A lipidomic analysis of docosahexaenoic acid (22:6, ω3) mediated attenuation of western diet induced nonalcoholic steatohepatitis in male ldlr -/- mice. Metabolites 2019, 9, 252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smedley, S.R.; Schroeder, F.C.; Weibel, D.B.; Meinwald, J.; Lafleur, K.A.; Renwick, J.A.; Rutowski, R.; Eisner, T. Mayolenes: Labile defensive lipids from the glandular hairs of a caterpillar (Pieris rapae). Proc. Natl. Acad. Sci. USA 2002, 99, 6822–6827. [Google Scholar] [CrossRef] [Green Version]
- Yore, M.M.; Syed, I.; Moraes-Vieira, P.M.; Zhang, T.; Herman, M.A.; Homan, E.A.; Patel, R.T.; Lee, J.; Chen, S.; Peroni, O.D.; et al. Discovery of a class of endogenous mammalian lipids with anti-diabetic and anti-inflammatory effects. Cell 2014, 159, 318–332. [Google Scholar] [CrossRef] [Green Version]
- Kuda, O.; Brezinova, M.; Rombaldova, M.; Slavikova, B.; Posta, M.; Beier, P.; Janovska, P.; Veleba, J.; Kopecky, J.; Kudova, E.; et al. Docosahexaenoic acid-derived fatty acid esters of hydroxy fatty acids (FAHFAS) with anti-inflammatory properties. Diabetes 2016, 65, 2580–2590. [Google Scholar] [CrossRef] [Green Version]
- Moraes-Vieira, P.M.; Saghatelian, A.; Kahn, B.B. GLUT4 expression in adipocytes regulates de novo lipogenesis and levels of a novel class of lipids with antidiabetic and anti-inflammatory effects. Diabetes 2016, 65, 1808–1815. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Moraes-Vieira, P.M.; Castoldi, A.; Aryal, P.; Yee, E.U.; Vickers, C.; Parnas, O.; Donaldson, C.J.; Saghatelian, A.; Kahn, B.B. Branched fatty acid esters of hydroxy fatty acids (FAHFAs) protect against colitis by regulating gut innate and adaptive immune responses. J. Biol. Chem. 2016, 291, 22207–22217. [Google Scholar] [CrossRef] [Green Version]
- Gowda, S.G.B.; Fuda, H.; Tsukui, T.; Chiba, H.; Hui, S.P. Discovery of eicosapentaenoic acid esters of hydroxy fatty acids as potent nrf2 activators. Antioxidants 2020, 9, 397. [Google Scholar] [CrossRef]
- Zhu, Q.F.; Yan, J.W.; Gao, Y.; Zhang, J.W.; Yuan, B.F.; Feng, Y.Q. Highly sensitive determination of fatty acid esters of hydroxyl fatty acids by liquid chromatography-mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2017, 1061, 34–40. [Google Scholar] [CrossRef]
- Zhu, Q.F.; Yan, J.W.; Zhang, T.Y.; Xiao, H.M.; Feng, Y.Q. Comprehensive Screening and Identification of Fatty Acid Esters of Hydroxy Fatty Acids in Plant Tissues by Chemical Isotope Labeling-Assisted Liquid Chromatography-Mass Spectrometry. Anal. Chem. 2018, 90, 10056–10063. [Google Scholar] [CrossRef] [PubMed]
- Gowda, S.G.B.; Liang, C.; Gowda, D.; Hou, F.; Kawakami, K.; Fukiya, S.; Yokota, A.; Chiba, H.; Hui, S.P. Identification of short-chain fatty acid esters of hydroxy fatty acids (SFAHFAs) in a murine model by nontargeted analysis using ultra-high-performance liquid chromatography/linear ion trap quadrupole-Orbitrap mass spectrometry. Rapid Commun. Mass Spectrom. 2020, 34, e8831. [Google Scholar] [CrossRef] [PubMed]
- Kokotou, M.G. Analytical methods for the determination of fatty acid esters of hydroxy fatty acids (FAHFAs) in biological samples, plants and foods. Biomolecules 2020, 10, 1092. [Google Scholar] [CrossRef] [PubMed]
- Kolar, M.J.; Nelson, A.T.; Chang, T.; Ertunc, M.E.; Christy, M.P.; Ohlsson, L.; Härröd, M.; Kahn, B.B.; Siegel, D.; Saghatelian, A. Faster Protocol for Endogenous Fatty Acid Esters of Hydroxy Fatty Acid (FAHFA) Measurements. Anal. Chem. 2018, 90, 5358–5365. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Wang, M.; Duan, Q.; Han, X. Sensitive analysis of fatty acid esters of hydroxy fatty acids in biological lipid extracts by shotgun lipidomics after one-step derivatization. Anal. Chim. Acta 2020, 1105, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Kind, T.; Zhu, Q.F.; Wang, Y.; Yan, J.W.; Fiehn, O.; Feng, Y.Q. In-Silico-Generated Library for Sensitive Detection of 2-Dimethylaminoethylamine Derivatized FAHFA Lipids Using High-Resolution Tandem Mass Spectrometry. Anal. Chem. 2020, 92, 5960–5968. [Google Scholar] [CrossRef] [PubMed]
- Pfeuffer, M.; Jaudszus, A. Pentadecanoic and Heptadecanoic Acids: Multifaceted Odd-Chain Fatty Acids. Adv. Nutr. Int. Rev. J. 2016, 15, 730–734. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.L.; Leung, E.L.H.; Zhou, H.; Liu, L.; Li, N. Metabolite analysis of toosendanin by an ultra-high performance liquid chromatography-quadrupole-time of flight mass spectrometry technique. Molecules 2013, 18, 12144–12153. [Google Scholar] [CrossRef]
- Li, J.; Xu, J.; Zhang, R.; Hao, Y.; He, J.; Chen, Y.; Jiao, G.; Abliz, Z. Strategy for Global Profiling and Identification of 2- and 3-Hydroxy Fatty Acids in Plasma by UPLC–MS/MS. Anal. Chem. 2020, 92, 5143–5151. [Google Scholar] [CrossRef]
- Hiraki, T.; Kubota, M.; Hirano, T.; Niwa, H.; Ohashi, M. FAB-CID Mass Spectrometry of Imidazolone Derivatives Related to the Chromophore of Aequorea Green Fluorescent Protein. J. Mass Spectrom. Soc. Jpn. 1998, 46, 37–43. [Google Scholar] [CrossRef] [Green Version]
- Hirata, S.I.; Kunisawa, J. Gut microbiome, metabolome, and allergic diseases. Allergol. Int. 2017, 66, 523–528. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.Y.; Leelasinjaroen, P.; Gillilland, M.G.; Zhou, H.; Owyang, C. Su2125 High fat diet induced activation of autophagy in paneth cells is responsible for gut dysbiosis and intestinal inflammation in rats. Gastroenterology 2013, 144, S564. [Google Scholar] [CrossRef]
- Araújo, J.R.; Tomas, J.; Brenner, C.; Sansonetti, P.J. Impact of high-fat diet on the intestinal microbiota and small intestinal physiology before and after the onset of obesity. Biochimie 2017, 141, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Li, J.; Tang, R.; Zhang, G.; Zeng, H.; Wood, R.J.; Liu, Z. High Fat Diet Alters Gut Microbiota and the Expression of Paneth Cell-Antimicrobial Peptides Preceding Changes of Circulating Inflammatory Cytokines. Mediat. Inflamm. 2017, 2017, 9474896. [Google Scholar] [CrossRef] [PubMed]
- Jakobsdottir, G.; Xu, J.; Molin, G.; Ahrné, S.; Nyman, M. High-fat diet reduces the formation of butyrate, but increases succinate, inflammation, liver fat and cholesterol in rats, while dietary fibre counteracts these effects. PLoS ONE 2013, 8, e80476. [Google Scholar] [CrossRef] [Green Version]
- Brezinova, M.; Kuda, O.; Hansikova, J.; Rombaldova, M.; Balas, L.; Bardova, K.; Durand, T.; Rossmeisl, M.; Cerna, M.; Stranak, Z.; et al. Levels of palmitic acid ester of hydroxystearic acid (PAHSA) are reduced in the breast milk of obese mothers. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 126–131. [Google Scholar] [CrossRef]
- Mallett, A.K.; Rowland, I.R.; Wise, A. Dietary fat and cecal microbial activity in the rat. Nutr. Cancer 2009, 6, 86–91. [Google Scholar] [CrossRef]
- Gowda, S.G.B.; Gao, Z.-J.; Chen, Z.; Abe, T.; Hori, S.; Fukiya, S.; Ishizuka, S.; Yokota, A.; Chiba, H.; Hui, S.-P. Untargeted lipidomic analysis of plasma from high fat diet-induced obese rats using UHPLC- linear trap quadrupole -Orbitrap MS. Anal. Sci. 2020, 36, 821–828. [Google Scholar] [CrossRef] [Green Version]
- Folch, J.; Lees, M.; Sloane Stanley, G.H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar]
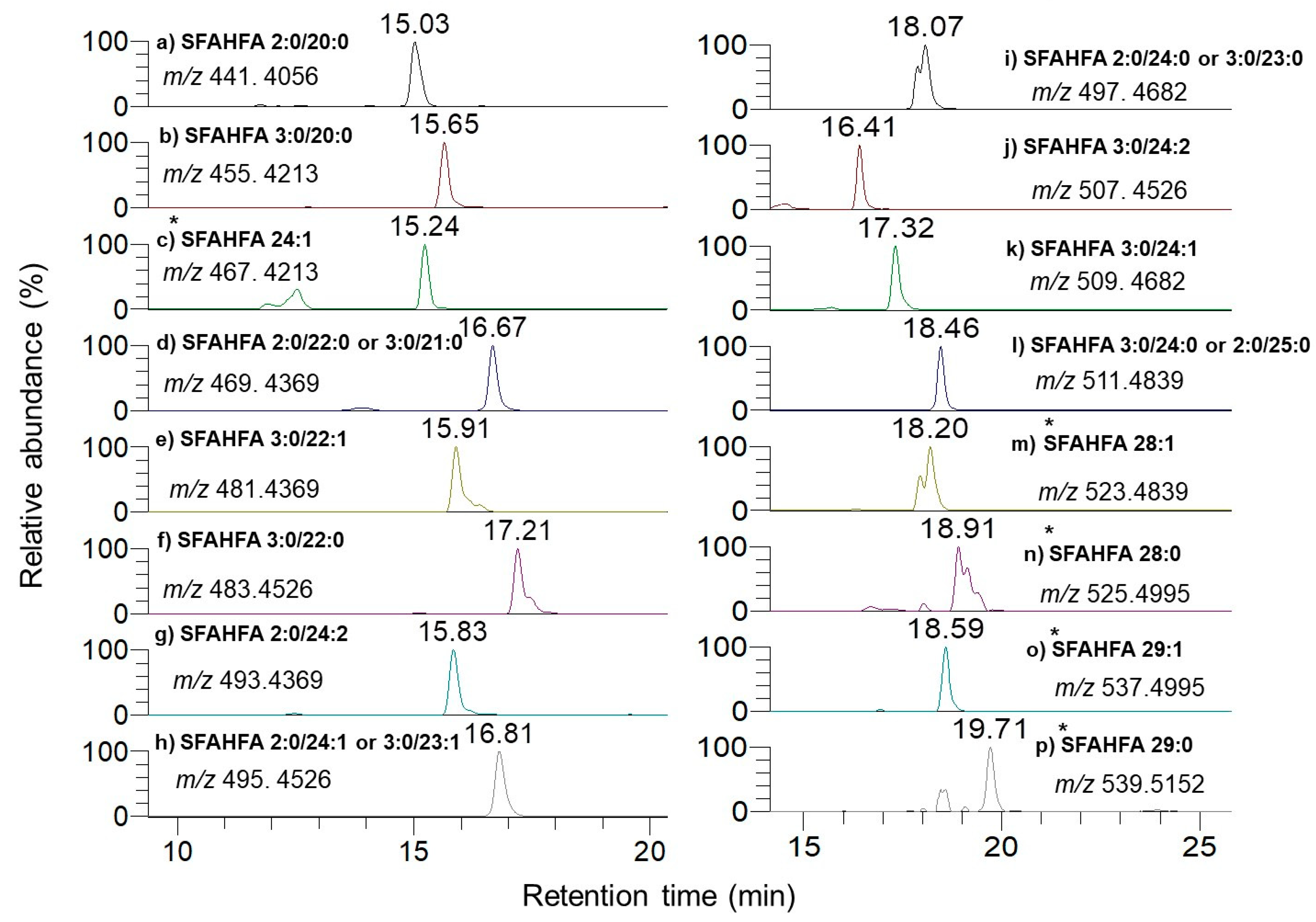

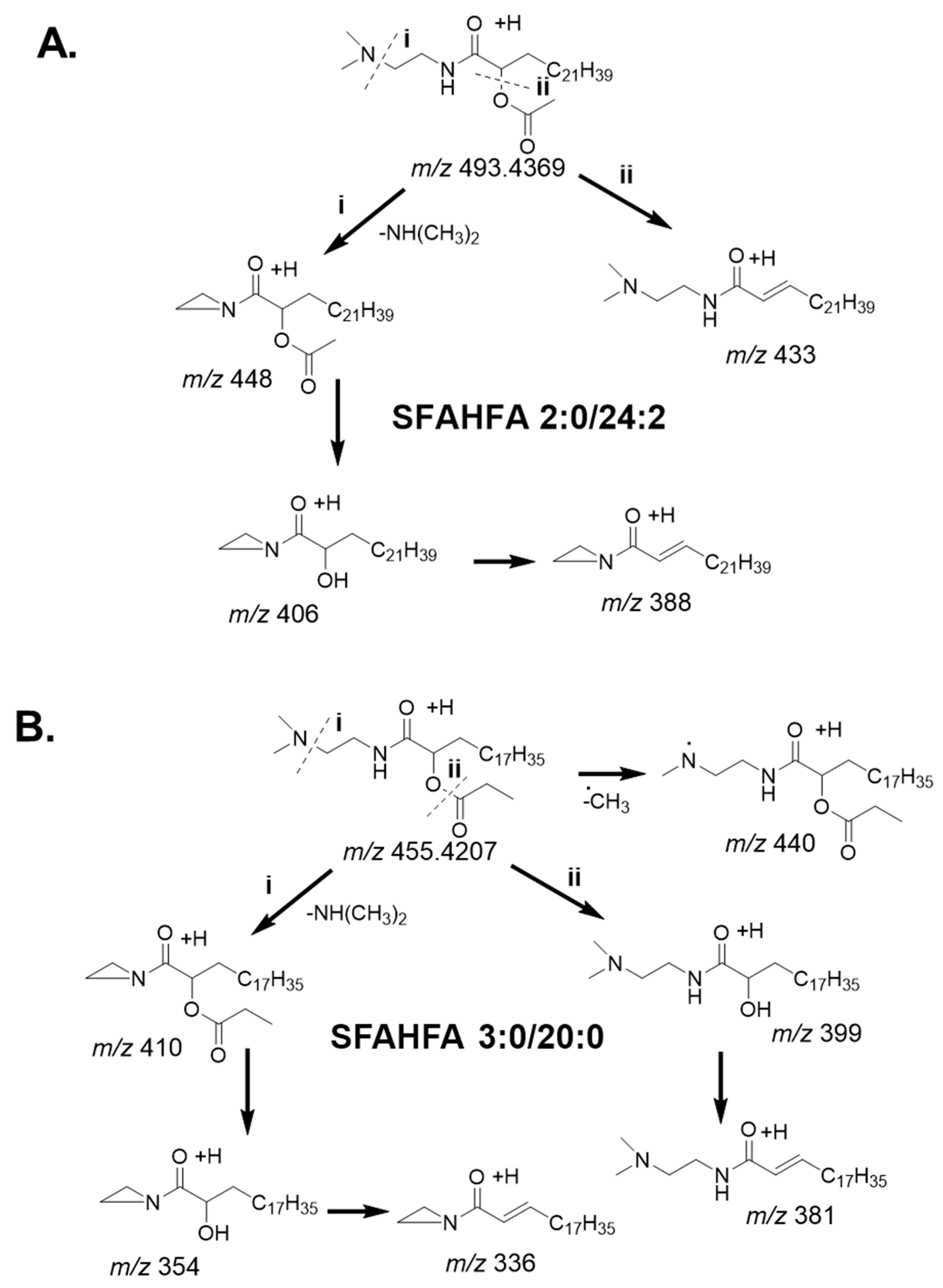
| SI No | Type of SFAHFA | Theoretical m/z | Experimental m/z | Mass Error ppm | Identified |
|---|---|---|---|---|---|
| 1 | SFAHFA 2:0/20:0 | 441.4056 | 441.4042 | −3.1 | [12] |
| 2 | SFAHFA 3:0/20:0 | 455.4213 | 455.4198 | −3.2 | This study |
| 3 | SFAHFA 24:1 | 467.4213 | 467.4197 | −3.4 | [12] |
| 4 | SFAHFA 2:0/22:0 or 3:0/21:0 | 469.4369 | 469.4355 | −2.9 | [12] |
| 5 | SFAHFA 3:0/22:1 | 481.4369 | 481.4355 | −2.9 | This study |
| 6 | SFAHFA 3:0/22:0 | 483.4526 | 483.4524 | −0.4 | [12] |
| 7 | SFAHFA 2:0/24:2 | 493.4369 | 493.4352 | −3.4 | This study |
| 8 | SFAHFA 2:0/24:1 or 3:0/23:1 | 495.4526 | 495.4507 | −3.8 | [12] |
| 9 | SFAHFA 2:0/24:0 or 3:0/23:0 | 497.4682 | 497.4677 | −1.0 | [12] |
| 10 | SFAHFA 3:0/24:2 | 507.4526 | 507.4514 | −2.3 | [12] |
| 11 | SFAHFA 3:0/24:1 | 509.4682 | 509.4673 | −1.7 | [12] |
| 12 | SFAHFA 3:0/24:0 or 2:0/25:0 | 511.4839 | 511.4833 | −1.1 | [12] |
| 13 | SFAHFA 28:1 | 523.4839 | 523.4830 | −1.7 | This study |
| 14 | SFAHFA 28:0 | 525.4995 | 525.4989 | −1.1 | [12] |
| 15 | SFAHFA 29:1 | 537.4995 | 537.4990 | −0.9 | This study |
| 16 | SFAHFA 29:0 | 539.5152 | 539.5150 | −0.4 | This study |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
B. Gowda, S.G.; Gowda, D.; Liang, C.; Li, Y.; Kawakami, K.; Fukiya, S.; Yokota, A.; Chiba, H.; Hui, S.-P. Chemical Labeling Assisted Detection and Identification of Short Chain Fatty Acid Esters of Hydroxy Fatty Acid in Rat Colon and Cecum Contents. Metabolites 2020, 10, 398. https://doi.org/10.3390/metabo10100398
B. Gowda SG, Gowda D, Liang C, Li Y, Kawakami K, Fukiya S, Yokota A, Chiba H, Hui S-P. Chemical Labeling Assisted Detection and Identification of Short Chain Fatty Acid Esters of Hydroxy Fatty Acid in Rat Colon and Cecum Contents. Metabolites. 2020; 10(10):398. https://doi.org/10.3390/metabo10100398
Chicago/Turabian StyleB. Gowda, Siddabasave Gowda, Divyavani Gowda, Chongsheng Liang, Yonghan Li, Kentaro Kawakami, Satoru Fukiya, Atsushi Yokota, Hitoshi Chiba, and Shu-Ping Hui. 2020. "Chemical Labeling Assisted Detection and Identification of Short Chain Fatty Acid Esters of Hydroxy Fatty Acid in Rat Colon and Cecum Contents" Metabolites 10, no. 10: 398. https://doi.org/10.3390/metabo10100398
APA StyleB. Gowda, S. G., Gowda, D., Liang, C., Li, Y., Kawakami, K., Fukiya, S., Yokota, A., Chiba, H., & Hui, S.-P. (2020). Chemical Labeling Assisted Detection and Identification of Short Chain Fatty Acid Esters of Hydroxy Fatty Acid in Rat Colon and Cecum Contents. Metabolites, 10(10), 398. https://doi.org/10.3390/metabo10100398