Polymorphisms in Drug Transporter and Metabolism Genes Associated with Resistance to Imatinib in Chronic Myeloid Leukemia: A Systematic Review and Meta-Analysis
Abstract
:1. Introduction
State of the Art
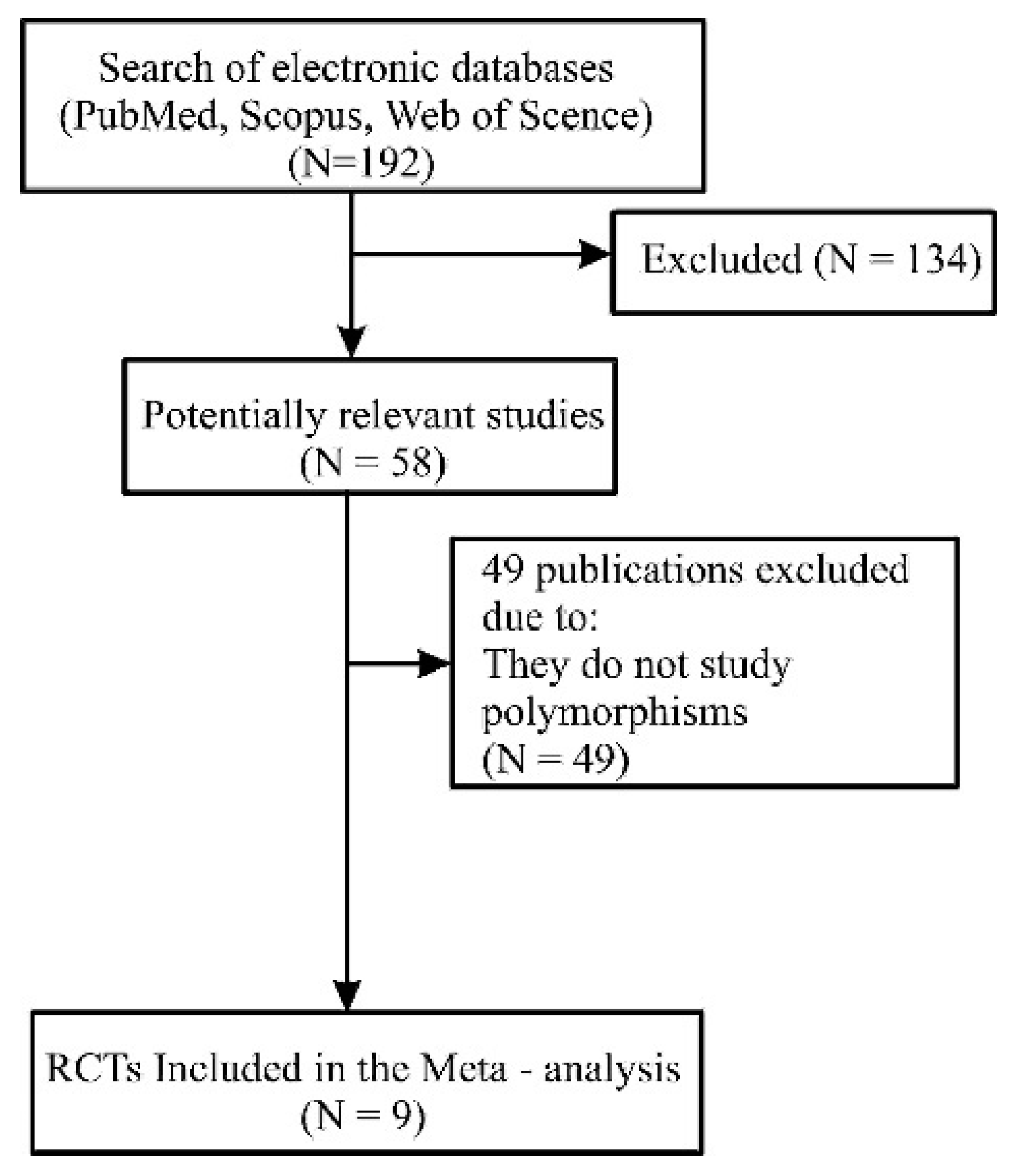
2. Materials and Methods
2.1. Data Collection
2.2. Article Selection
- Published studies related to the use of imatinib in patients diagnosed with chronic myeloid leukemia.
- Studies on polymorphisms in drug transporter and metabolizer genes associated with imatinib resistance.
- Studies with results validated by clinical trials.
- Studies that included odds ratio (OR) as an outcome measure with a 95% confidence interval.
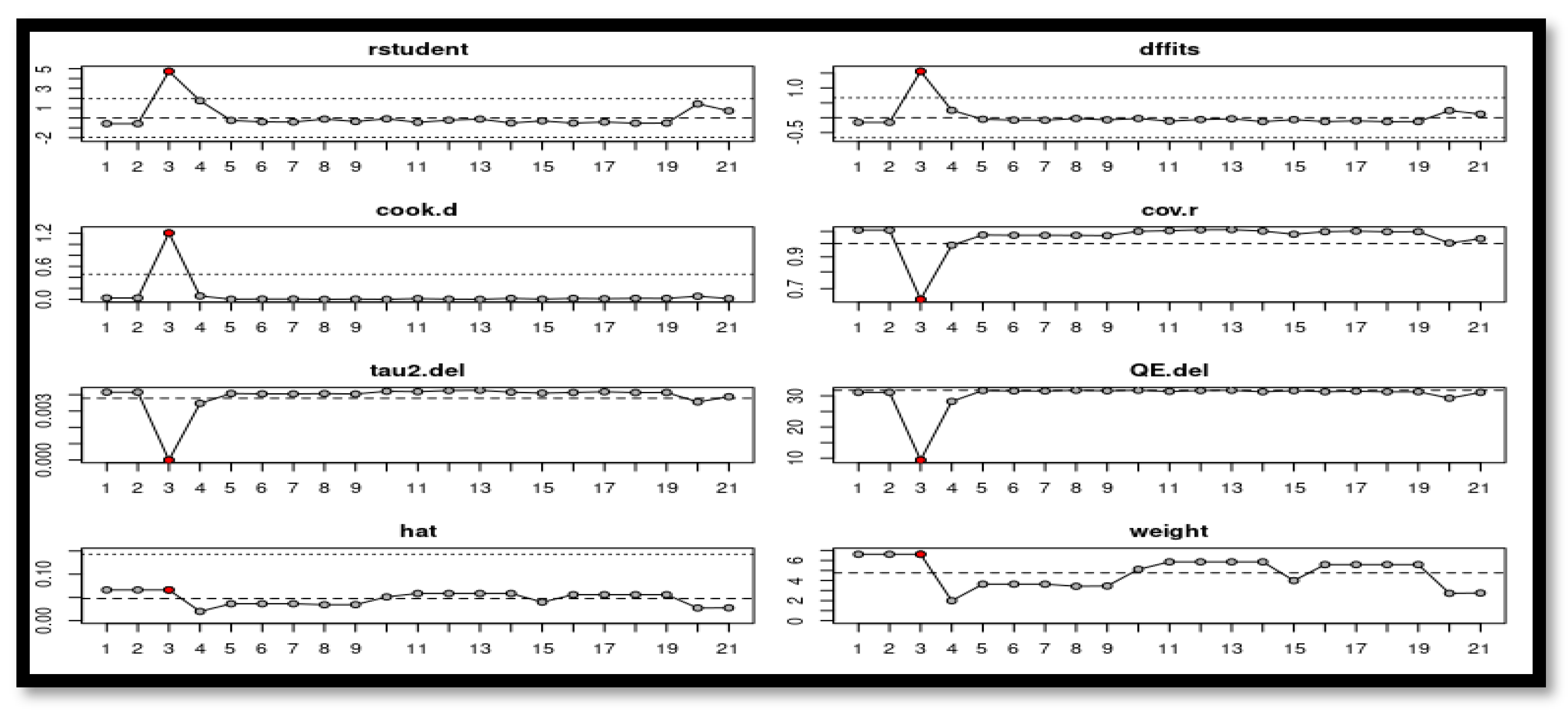
2.3. Data Extraction and Assessment of Quality and Risk of Bias
2.4. Meta-Analysis Methodology
3. Results
3.1. Description of the Studies
3.2. Primary Results
3.3. Meta-Analysis Results

3.4. Publication Bias
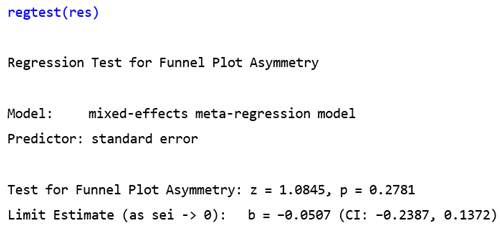



4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abdulmawjood, B.; Costa, B.; Roma-Rodrigues, C.; Baptista, P.V.; Fernandes, A.R. Genetic Biomarkers in Chronic Myeloid Leukemia: What Have We Learned So Far? Natl. Libr. Med. 2021, 22, 12516. [Google Scholar] [CrossRef]
- Thompson, P.A.; Kantarjian, H.M.; Cortes, J.E. Diagnosis and treatment of chronic myeloid leukemia (CML) in 2015. Natl. Libr. Med. 2015, 10, 1440–1454. [Google Scholar]
- Orozco, J.; Valencia, J.E.; Aiello, E.; Ribón, G.; Guerrero, F.; Garcia, R.; Mujica, J.L. Costo-efectividad del dasatinib en el tratamiento de la leucemia mieloide crónica en pacientes resistentes al imatinib. CES Med. 2010, 2, 2215–9177. [Google Scholar]
- Braun, T.P.; Eide, C.A.; Druker, B.J. Response and Resistance to BCR-ABL1-Targeted Therapies. Cancer Cell 2020, 37, 530–542. [Google Scholar] [CrossRef]
- Avilés-Vázquez, S.; Chávez-González, M.; Mayani, H. Inhibidores de cinasas de tirosina (ICT): La nueva revolución en el tratamiento de la leucemia mieloide crónica (LMC). Gac. Méd. México 2013, 149, 646–654. [Google Scholar]
- Jabbour, E.; Kantarjian, H. Chronic myeloid leukemia: 2022 update on diagnosis, therapy, and monitoring. Am. J. Hematol. 2022, 97, 1236–1256. [Google Scholar] [CrossRef]
- González, M.A.C.; Ayala-Sánchez, M.; Mayani, H. La leucemia mieloide crónica en el siglo XXI: Biología y tratamiento. Rev. Investig. Clín. 2009, 61, 221–232. [Google Scholar]
- Delord, M.; Rousselot, P.; Cayuela, J.M.; Sigaux, F.; Guilhot, J.; Preudhomme, C.; Guilhot, F.; Loiseau, P.; Raffoux, E.; Geromin, D.; et al. High imatinib dose overcomes insufficient response associated with ABCG2 haplotype in chronic myelogenous leukemia patients. Oncotarget 2013, 4, 1582–1591. [Google Scholar] [CrossRef]
- Gambacorti-Passerini, C.; Barni, R.; Marchesi, E.; Verga, M.; Rossi, F.; Rossi, F.; Pioltelli, P.; Pogliani, E.; Corneo, G.M. Sensitivity to the abl inhibitor STI571 in fresh leukaemic cells obtained from chronic myelogenous leukaemia patients in different stages of disease. Br. J. Haematol. 2001, 112, 972–974. [Google Scholar] [CrossRef]
- Holtz, M.S.; Slovak, M.L.; Zhang, F.; Sawyers, C.L.; Forman, S.J.; Bhatia, R. Imatinib mesylate (STI571) inhibits growth of primitive malignant progenitors in chronic myelogenous leukemia through reversal of abnormally increased proliferation. Blood 2002, 99, 3792–3800. [Google Scholar] [CrossRef]
- Cohen, P.; Cross, D.; Jänne, P.A. Kinase drug discovery 20 years after imatinib: Progress and future directions. Nat. Rev. Drug Discov. 2021, 20, 551–569. [Google Scholar] [CrossRef] [PubMed]
- Claudiani, S.; Apperley, J.F. The argument for using imatinib in CML. Hematol. Am. Soc. Hematol. Educ. Program. 2018, 1, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, R.; Holtz, M.; Niu, N.; Gray, R.; Snyder, D.S.; Sawyers, C.L.; Arber, D.A.; Slovak, M.L.; Forman, S.J. Persistence of malignant hematopoietic progenitors in chronic myelogenous leukemia patients in complete cytogenetic remission following imatinib mesylate treatment. Blood 2003, 101, 4701–4707. [Google Scholar] [CrossRef] [PubMed]
- Cortes, J.; De Souza, C.; Ayala-Sanchez, M.; Bendit, I.; Best-Aguilera, C.; Enrico, A.; Hamerschlak, N.; Pagnano, K.; Pasquini, R.; Meillon, L. Current patient management of chronic myeloid leukemia in Latin America: A study by the Latin American Leukemia Net (LALNET). Cancer 2010, 116, 4991–5000. [Google Scholar] [CrossRef]
- Hijiya, N.; Schultz, K.R.; Metzler, M.; Millot, F.; Suttorp, M. Pediatric chronic myeloid leukemia is a unique disease that requires a different approach. Blood 2016, 127, 392–399. [Google Scholar] [CrossRef]
- Breccia, M.; Alimena, G. The current role of high-dose imatinib in chronic myeloid leukemia patients, newly diagnosed or resistant to standard dose. Expert. Opin. Pharmacother. 2011, 12, 2075–2087. [Google Scholar] [CrossRef]
- Cruz-Rico, J.; Garrido-Acosta, O.; Anguiano-Robledo, L.; Rodríguez-Wong, U.; Pérez-Cruz, E.; Sánchez-Navarrete, J.; Ruiz-Pérez, N.J.; Montes-Vera, M.D.R. Imatinib: Farmacocinética. Rev. Hosp. Jua. Mex. 2013, 80, 67–72. [Google Scholar]
- Benchikh, S.; Bousfiha, A.; Hamouchi, A.E.; Soro, S.G.C.; Malki, A.; Nassereddine, S. Chronic myeloid leukemia: Cytogenetics and molecular biology’s part in the comprehension and management of the pathology and treatment evolution. Egypt. J. Med. Hum. Genet. Vol. 2022, 29, 1–13. [Google Scholar] [CrossRef]
- Gardner, E.R.; Burger, H.; van Schaik, R.H.; van Oosterom, A.T.; de Bruijn, E.A.; Guetens, G.; Prenen, H.; de Jong, F.A.; Baker, S.D.; Bates, S.E.; et al. Association of enzyme and transporter genotypes with the pharmacokinetics of imatinib. Clin. Pharmacol. Ther. 2006, 80, 192–201. [Google Scholar] [CrossRef]
- Camargo, M.; Soto-Marín, M.I.; Zea, O.; Saavedra, D. Tratamiento con imatinib y el farmacogenotipo CYP3A4 en relación con la expansión clonal Ph(+) en leucemia mieloide crónica (LMC)*. Colombia Médica 2008, 30, 314–322. [Google Scholar]
- Sailaja, K.; Rao, D.N.; Rao, D.R.; Vishnupriya, S. Analysis of CYP3A5*3 and CYP3A5*6 gene polymorphisms in Indian chronic myeloid leukemia patients. Asian Pac. J. Cancer Prev. 2010, 11, 781–784. [Google Scholar]
- Bedewy, A.M.L.; El-Maghraby, S.M. Do SLCO1B3 (T334G) and CYP3A5*3 polymorphisms affect response in Egyptian chronic myeloid leukemia patients receiving imatinib therapy? Hematology 2013, 18, 211–216. [Google Scholar] [CrossRef]
- Vaidya, S.; Ghosh, K.; Shanmukhaiah, C.; Vundint, B.R. Genetic variations of hOCT1 gene and CYP3A4/A5 genes and their association with imatinib response in Chronic Myeloid Leukemia. Eur. J. Pharmacol. 2015, 15, 124–130. [Google Scholar] [CrossRef]
- Maddin, N.; Husin, A.; Gan, S.H.; Aziz, B.A.; Ankathil, R. Impact of CYP3A4*18 and CYP3A5*3 Polymorphisms on Imatinib Mesylate Response Among Chronic Myeloid Leukemia Patients in Malaysia. Oncol. Ther. 2016, 4, 303–314. [Google Scholar] [CrossRef]
- Saiz-Rodríguez, M.; Almenara, S.; Navares-Gómez, M.O.D.; Román, M.; Zubiaur, P.; Koller, D.; Santos, M.; Mejía, G.; Borobia, A.M.; Rodríguez-Antona, C.; et al. Effect of the Most Relevant CYP3A4 and CYP3A5 Polymorphisms on the Pharmacokinetic Parameters of 10 CYP3A Substrates. Biomedicines 2020, 8, 94. [Google Scholar] [CrossRef]
- Harivenkatesh, N.; Kumar, L.; Bakhshi, S.; Sharma, A.; Kabra, M.; Velpandian, T.; Gogia, A.; Shastri, S.S.; Gupta, Y.K. Do polymorphisms in MDR1 and CYP3A5 genes influence the risk of cytogenetic relapse in patients with chronic myeloid leukemia on imatinib therapy? Leuk. Lymphoma 2017, 58, 2218–2226. [Google Scholar] [CrossRef]
- Akram, A.M.; Iqbal, Z.; Akhtar, T.; Khalid, A.M.; Sabar, M.F.; Qazi, M.H.; Aziz, Z.; Sajid, N.; Aleem, A.; Rasool, M.; et al. Presence of novel compound BCR-ABL mutations in late chronic and advanced phase imatinib sensitive CML patients indicates their possible role in CML progression. Cancer Biol. Ther. 2017, 18, 214–221. [Google Scholar] [CrossRef]
- Hoemberger, M.; Pitsawong, W.; Kern, D. Cumulative mechanism of several major imatinib-resistant mutations in Abl kinase. Proc. Natl. Acad. Sci. USA 2020, 117, 19221–19227. [Google Scholar] [CrossRef]
- Minciacchi, V.R.; Kumar, R.; Krause, D.S. Chronic Myeloid Leukemia: A Model Disease of the Past, Present and Future. Cells 2021, 10, 117. [Google Scholar] [CrossRef]
- Molloy, G.; O’Carroll, R.; Ferguson, E. Conscientiousness and Medication Adherence: A Meta-analysis. Ann. F Behav. Med. 2014, 47, 92–101. [Google Scholar] [CrossRef]
- Duval, S.; Tweedie, R. A Nonparametric Trim and Fill” Method of Accounting for Publication Bias in Meta-Analysis. J. Am. Stat. Assoc. 2000, 95, 89–98. [Google Scholar]
- Vevea, J.L.; Woods, C.M. Publication Bias in Research Synthesis: Sensitivity Analysis Using a Priori Weight Functions. Psychol. Methods 2005, 10, 428–443. [Google Scholar] [CrossRef]
- Egger, M.; Schneider, S.G.D.M.; Minder, C. Bias in Meta-Analysis Detected by a Simple, Graphical Test. BMJ 1997, 315, 619. [Google Scholar] [CrossRef]
- Shadish, W.R.; Haddock, C.K. Combining Estimates of Effect Size. In The Handbook of Research Synthesis and Meta-Analysis, 2nd ed.; Russell Sage Foundation: London, UK, 2009; pp. 257–277. [Google Scholar]
- Hamed, N.A.M.; Neanea, H.; Ghanem, A.M.; Elgammal, M.M.A.; Samir, Y. Polymorphism of Human Organic Cationic Transporter1 (C480G) in Egyptian Chronic Myeloid Leukemia Patients on Imatinib. Am. J. Mol. Biol. 2018, 8, 83–91. [Google Scholar] [CrossRef]
- Elghannam, D.M.; Ibrahim, L.; Ebrahim, M.A.; Azmy, H.H.E. Association of MDR1 gene polymorphism(G2677T) with imatinib response in Egyptianchronic myeloid leukemia patients. Hematology 2013, 19, 123–128. [Google Scholar] [CrossRef]
- Wang, Q.; Jiang, Z.-P.; Zeng, J.; Zhu, Y.; Cai, H.-L.; Xiang, D.-X.; He, X.-L.S.Q.; Zhong, X.-L.; Xu, Q. Effects of Trough Concentration and Solute Carrier Polymorphisms on Imatinib Efficacy in Chinese Patients with Chronic Myeloid Leukemia. J. Pharm. Pharm. Sci. 2020, 23, 1–205. [Google Scholar] [CrossRef]
- Au, A.; Baba, A.A.; Goh, A.S.; Fadilah, S.A.W.; Teh, H.R.A.; Ankathil, R. Association of genotypes and haplotypes of multi-drug transporter genes ABCB1 and ABCG2 with clinical response to imatinib mesylate in chronic myeloid leukemia patients. Biomed. Pharmacother. 2014, 6, 343–349. [Google Scholar] [CrossRef]
- De Lima, T.L.; Vivona, D.; Bueno, C.T.; Hirata, R.; Hirata, M.; Luchessi, A.; de Castro, F.A.; Chauffaille, M.D.L.; Zanichelli, M.; Chiattone, C.; et al. Reduced ABCG2 and increased SLC22A1 mRNA expression are associated with imatinib response in chronic myeloid leukemia. Med. Oncol. 2014, 31, 851. [Google Scholar] [CrossRef]
- Alves, R.; Gonçalves, A.C.; Jorge, J.; Marques, G.; Ribeiro, A.B.; Tenreiro, R.; Coucelo, M.; Diamond, J.; Oliveiros, B.; Pereira, A.; et al. Genetic Variants of ABC and SLC Transporter Genes and Chronic Myeloid Leukaemia: Impact on Susceptibility and Prognosis. Int. J. Mol. Sci. 2022, 23, 9815. [Google Scholar] [CrossRef]
- Nouri, N.; Mehrzad, V.; Khalaj, Z.; Zaker, E.; Zare, F.; Abbasi, E.; Khosravi, M.; Kalanta, S.M.; Salehi, M. Effects of ABCG2 C421A and ABCG2 G34A genetic polymorphisms on clinical outcome and response to imatinib mesylate, in Iranian chronic myeloid leukemia patients. Egypt. J. Med. Hum. Genet. 2023, 24, 1–7. [Google Scholar] [CrossRef]
- Leongómez, J.D. Meta-análisis de correlaciones y meta-regresión en R: Guía práctica. MetaArXiv 2023, 4, 1–61. [Google Scholar] [CrossRef]
- Borenstein, M.; Higgns, H.L.V.J.P.T.; Rothstein, H.R. Identifying and Quantifying Heterogeneity. In Introduction to Meta-Analysis; Wiley: Hoboken, NJ, USA, 2009; pp. 107–125. [Google Scholar]
- Takahashi, N.; Miura, M.; Scott, H.; Kameoka, Y.; Tagawa, H.; Saitoh, H.; Fujishima, N.; Yoshioka, T.; Hirokawa, M.; Sawada, K. Influence of CYP3A5 and drug transporter polymorphisms on imatinib trough concentration and clinical response among patients with chronic phase chronic myeloid leukemia. J. Human Gen. 2010, 55, 731–737. [Google Scholar] [CrossRef]
- Vine, J.; Cohen, S.; Ruchlemer, R.; Goldschmidt, N.; Levin, M.; Libster, D.; Gural, A.; Gatt, M.; Lavie, D.; Ben-Yehuda, D.; et al. Polymorphisms in the human organic cation transporter and the multidrug resistance gene: Correlation with imatinib levels and clinical course in patients with chronic myeloid leukemia. Leuk. Lymphoma 2014, 55, 2525–2531. [Google Scholar] [CrossRef]
- Kim, D.; Sriharsha, L.; Xu, W.; Kamel-Reid, S.; Liu, X.; Siminovitch, K.; Messner, H.A.; Lipton, J.H. Clinical Relevance of a Pharmacogenetic Approach Using Multiple Candidate Genes to Predict Response and Resistance to Imatinib Therapy in Chronic Myeloid Leukemia. Clin. Cancer Res. 2009, 15, 4750–4758. [Google Scholar] [CrossRef]
- Van Den, M.; Heuvel-Eibrink, E.; Weimer, M.; De Boevere, B.; Van Der Holt, P.; Vossebeld, R.; Pieters, R.S. MDR1 gene related clonal selection and P-glycoprotein function and expression in relapsed or refractory acute myeloid leukemia. Blood 2001, 97, 3605–3611. [Google Scholar] [CrossRef]
- Khorashad, J.; de Lavallade, H.; Apperley, J.; Milojkovic, D.; Reid, A.; Bua, M.; Szydlo, R.; Olavarria, E.; Kaeda, J.; Goldman, J.; et al. Finding of kinase domain mutations in patients with chronic phase chronic myeloid leukemia responding to imatinib may identify those at high risk of disease progression. J. Clin. Oncol. 2008, 26, 4806–4813. [Google Scholar] [CrossRef]
- Gurney, H.; Wong, M.; Balleine, R.L.; Rivory, L.P.; McLachlan, A.J.; Hoskins, J.M.; Wilcken, N.; Clarke, C.L.; Mann, G.J.; Collins, M.; et al. Imatinib disposition and ABCB1 (MDR1, P-glycoprotein) genotype. Clin. Pharmacol. Ther. 2007, 82, 33–40. [Google Scholar] [CrossRef]
| Field | Content |
|---|---|
| Id_Study ni | Number assigned to the study |
| Title | Title of the article |
| Authors | Authors |
| Year | Year of publication of the article |
| Country | Country where the clinical trial was applied |
| ni | Number of patients |
| Age | Average age of patients |
| Treatment Phase | Disease stage (Chronic, Blast, Accelerated) |
| Intervention | Imatinib dose |
| Duration (Months) | Duration of treatment in months |
| Polymorphism | Type of polymorphism studied |
| Nucleotide | Type of nucleotide studied |
| Gene | Type of gene studied |
| Genotyping Technique | Genotyping technique used in the study |
| Genotyping | Associated genotype |
| Evaluation of response to treatment | Evaluation technique, in this case odds ratio (OR) was chosen as an indicator. |
| OR | OR value for each |
| CI (%) | 95% confidence interval |
| CI: Lower Limit | Lower limit of the confidence interval |
| CI: Upper Limit | Upper limit of the confidence Interval |
| p | Pearson correlation coefficient |
| Association Found (polymorphisms vs. resistance to Im) | Establishes the association found between polymorphisms and resistance to imatinib. |
| Id_Study | Title | References | Year | Country | Database | Journal | Journal Categorization | Country Journal |
|---|---|---|---|---|---|---|---|---|
| 1 | Impact of CYP3A4*18 and CYP3A5*3 Polymorphisms on Imatinib Mesylate Response Among Chronic Myeloid Leukemia Patients in Malaysia | [24] | 2016 | Malasia | PubMed | Oncology and Therapy | Q2 | Switzerland |
| 2 | Polymorphism of Human Organic Cationic Transporter1 (C480G) in Egyptian Chronic Myeloid Leukemia Patients on Imatinib | [35] | 2018 | Egypt | WoS | American Journal of Molecular Biology | Not categorized | USA |
| 3 | Do polymorphisms in MDR1 and CYP3A5 genes influence the risk of cytogenetic relapse in patients with chronic myeloid leukemia on imatinib therapy? | [26] | 2017 | India | PubMed | Leukemia and Lymphoma | Q2 | United Kingdom |
| 4 | Association of MDR1 gene polymorphism(G2677T) with imatinib response in Egyptianchronic myeloid leukemia patients | [36] | 2013 | Egypt | PubMed | Hematology | Q3 | United Kingdom |
| 5 | Effects of Trough Concentration and Solute Carrier Polymorphisms on Imatinib Efficacy in Chinese Patients with Chronic Myeloid Leukemia | [37] | 2020 | Chinese | PubMed | Journal of Pharmacy and Pharmaceutical Sciences | Q2 | Canada |
| 6 | Association of genotypes and haplotypes of multi-drug transporter genes ABCB1 and ABCG2 with clinical response to imatinib mesylate in chronic myeloid leukemia patients | [38] | 2014 | Malasia | PubMed | Biomedicine & Pharmacotherapy | Q1 | France |
| 7 | Reduced ABCG2 and increased SLC22A1 mRNA expression are associated with imatinib response in chronic myeloid leukemia | [39] | 2014 | Brazil | Scopus | Medical Oncology | Q2 | USA |
| 8 | Genetic Variants of ABC and SLC Transporter Genes and Chronic Myeloid Leukaemia: Impact on Susceptibility and Prognosis | [40] | 2022 | Portugal | PubMed | International Journal of Molecular Sciences | Q1 | Switzerland |
| 9 | Effects of ABCG2 C421A and ABCG2 G34A genetic polymorphisms on clinical outcome and response to imatinib mesylate, in Iranian chronic myeloid leukemia patients | [41] | 2023 | Irán | Scopus | Egyptian Journal of Medical Human Genetics | Q4 | Egypt |
| References | ni | Age | Treatment Phase | Intervention | Duration (Months) | Polymorphism | Nucleotide | Gene | Genotyping Technique | Genotyping | Statistical Calculation |
|---|---|---|---|---|---|---|---|---|---|---|---|
| [24] | 270 | 42.515 | Chronicle (220) Accelerated (36) Blastic (14) | Imatinib (400 mg/day) | 12 | CYP3A5*3 | SNP (1) | CYP3A5*3 | Polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP). | Heterozygous (AG) | Binary logistic regression |
| Imatinib (400 mg/day) | CYP3A5*3 | CYP3A5*3 | Homozygous (GG) | ||||||||
| Imatinib (400 mg/day) | CYP3A4*18 | CYP3A4*18 | Heterozygous (TC) | ||||||||
| [35] | 50 | 46 | Chronicle | Response criteria were assessed according to the NCCN guidelines Accesed 12/09/2023 https://www.nccn.org/professionals/physician_gls/PDF/cml.pdf | 10 | C480G | C480G | Real-time PCR using TaqmanTM assays | Homozygous (CC) | N/E | |
| [26] | 104 | 36 | Chronicle | Imatinib (400 mg/día) | 60 | C1236T | SNP (1) | MDR1 | PCR-RFLP method and validated by direct gene sequencing. | CC | N/E |
| C3435T | TT | ||||||||||
| G2677T/A | |||||||||||
| [36] | 96 | 44.44 ± 12.37 | Chronicle (66) Accelerated (18) Blastic (12) | Imatinib 400 a 600 mg | 12 | G2677T | SNP (1) | MDR1 | Polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP). | GT | Unconditional logistic regression. |
| TT | |||||||||||
| [37] | 171 | 43.61 ± 12.27 | Chronicle | Imatinib (400 mg/día) | 12 | SLCO1A2 | SNP (1) | 361G>A | Polymerase chain reaction (PCR) and Sanger sequencing | GA | Unconditional logistic regression. |
| [38] | 215 | 41.5 | Chronicle | Imatinib (400 mg/día) | N/E | T1236C | SNP (1) | ABCB1 | Polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP). | Homozygous CC | N/E |
| G2677T/A | TT/AT/AA variant | ||||||||||
| 2677G>T/A | |||||||||||
| C421>A | SNP (2) | ||||||||||
| [39] | 118 | 50.3 | Chronicle | Imatinib (400 mg/día) | N/E | ARNm | SNP (1) | ABCG2 | PCR–RFLP and real-time PCR using TaqmanTM assays | N/E | N/E |
| [40] | 198 | 54 | Chronicle | Imatinib (400 mg/día) | N/E | ABCB1 | SNP (1) | ABCB1 | PCR method and validated by direct gene sequencing. | Unconditional logistic regression. | |
| rs2231142 | ABCG2 | CC | |||||||||
| rs683369 | SLC22A1 | ||||||||||
| rs2631365 | SLC22A5 | TT | |||||||||
| [41] | 72 | 50.27 ± 12.72 | Chronicle | Imatinib (400 mg/día) | 21 | ABCG2 | SNP (1) | G34A | PCR-RFLP method and validated by direct gene sequencing. | AG | Logistic regression. |
| C421A | California |
| References | Polymorphism | Genotyping | OR | CI 95% | p | Association Found (Polymorphisms vs. Resistance to Im) | |
|---|---|---|---|---|---|---|---|
| Lower Limit | Upper Lim | ||||||
| [24] | CYP3A5*3 | Heterozygous (AG) | 0.171 | 0.09 | 0.324 | 0.001 | Significant minor risk |
| CYP3A5*3 | Homozygous (GG) | 0.257 | 0.126 | 0.525 | 0.001 | Significant minor risk | |
| CYP3A4*18 | Heterozygous (TC) | 0.648 | 0.277 | 1.515 | 0.316 | Negative | |
| [35] | C480G | Homozygous (CC) | 0.089 | 0.312 | Negative | ||
| [26] | C1236T | CC | 4.382 | 1.145 | 16.774 | 0.022 | Significantly increased risk of cytogenetic relapse. |
| C3435T | TT | 0.309 | 0.134 | 0.708 | 0.005 | Significantly lower risk of cytogenetic relapse. | |
| G2677T/A | A | 0.266 | 0.111 | 0.636 | 0.003 | Significantly lower risk of cytogenetic relapse. | |
| [36] | G2677T | GT | 2.519 | 1.059 | 5.99 | 0.037 | May be useful in predicting response to treatment |
| TT | 0.166 | 0.044 | 0.627 | 0.008 | |||
| [37] | SLCO1A2 | GA | 4.32 | 0.924 | 20.206 | 0.042 | Significantly affects |
| [38] | T1236C | Homozygous CC | 2.79 | 1.217 | 6.374 | 0.01 | Significant association with treatment efficacy |
| G2677T/A | TT/AT/AA variant | 0.48 | 0.239 | 0.957 | 0.03 | Significant association with treatment efficacy | |
| ABCB1 | 0.49 | 0.248 | 0.974 | 0.04 | Can influence with resistance | ||
| C421>A | 2.2 | 1.273 | 3.811 | 0.004 | Can influence with resistance | ||
| [39] | ABCG2 | N/E | 24 | 1.74 | 330.8 | 0.018 | May be associated with resistance |
| [40] | ABCB1 | 1.483 | 1.154 | 1.906 | 0.002 | Findings on SNVs may be a useful tool for understanding interindividual variability and improving therapeutic decisions, including treatment selection. | |
| rs2231142 | CC | 0.589 | 0.388 | 0.892 | 0.012 | ||
| rs683369 | 0.598 | 0.469 | 0.762 | <0.001 | |||
| rs2631365 | TT | 0.682 | 0.534 | 0.869 | 0.002 | ||
| [41] | ABCG2 | AG | 1.89 | 0.66 | 5.39 | 0.235 | Not useful for predicting IM response |
| CYP3A5*3 | California | 2.78 | 0.7 | 11.02 | 0.146 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gómez, A.M.A.; Díaz-Mendoza, M.A.; Lemus, Y.B.; León-Mejía, G.; Benitez, M.L.R. Polymorphisms in Drug Transporter and Metabolism Genes Associated with Resistance to Imatinib in Chronic Myeloid Leukemia: A Systematic Review and Meta-Analysis. Sci. Pharm. 2024, 92, 2. https://doi.org/10.3390/scipharm92010002
Gómez AMA, Díaz-Mendoza MA, Lemus YB, León-Mejía G, Benitez MLR. Polymorphisms in Drug Transporter and Metabolism Genes Associated with Resistance to Imatinib in Chronic Myeloid Leukemia: A Systematic Review and Meta-Analysis. Scientia Pharmaceutica. 2024; 92(1):2. https://doi.org/10.3390/scipharm92010002
Chicago/Turabian StyleGómez, Ana Marcela Arrieta, María Antonia Díaz-Mendoza, Yesit Bello Lemus, Grethel León-Mejía, and Martha Lucia Ruiz Benitez. 2024. "Polymorphisms in Drug Transporter and Metabolism Genes Associated with Resistance to Imatinib in Chronic Myeloid Leukemia: A Systematic Review and Meta-Analysis" Scientia Pharmaceutica 92, no. 1: 2. https://doi.org/10.3390/scipharm92010002
APA StyleGómez, A. M. A., Díaz-Mendoza, M. A., Lemus, Y. B., León-Mejía, G., & Benitez, M. L. R. (2024). Polymorphisms in Drug Transporter and Metabolism Genes Associated with Resistance to Imatinib in Chronic Myeloid Leukemia: A Systematic Review and Meta-Analysis. Scientia Pharmaceutica, 92(1), 2. https://doi.org/10.3390/scipharm92010002







