Computational Assessment of Cannflavin A as a TAK1 Inhibitor: Implication as a Potential Therapeutic Target for Anti-Inflammation
Abstract
1. Introduction
2. Materials and Methods
2.1. Drug-Likeness and ADMET Prediction
2.2. Protein Preparation
2.3. Ligand Preparation
2.4. Molecular Docking
2.5. Molecular Dynamics Simulations and Free Energy Calculations
3. Results and Discussion
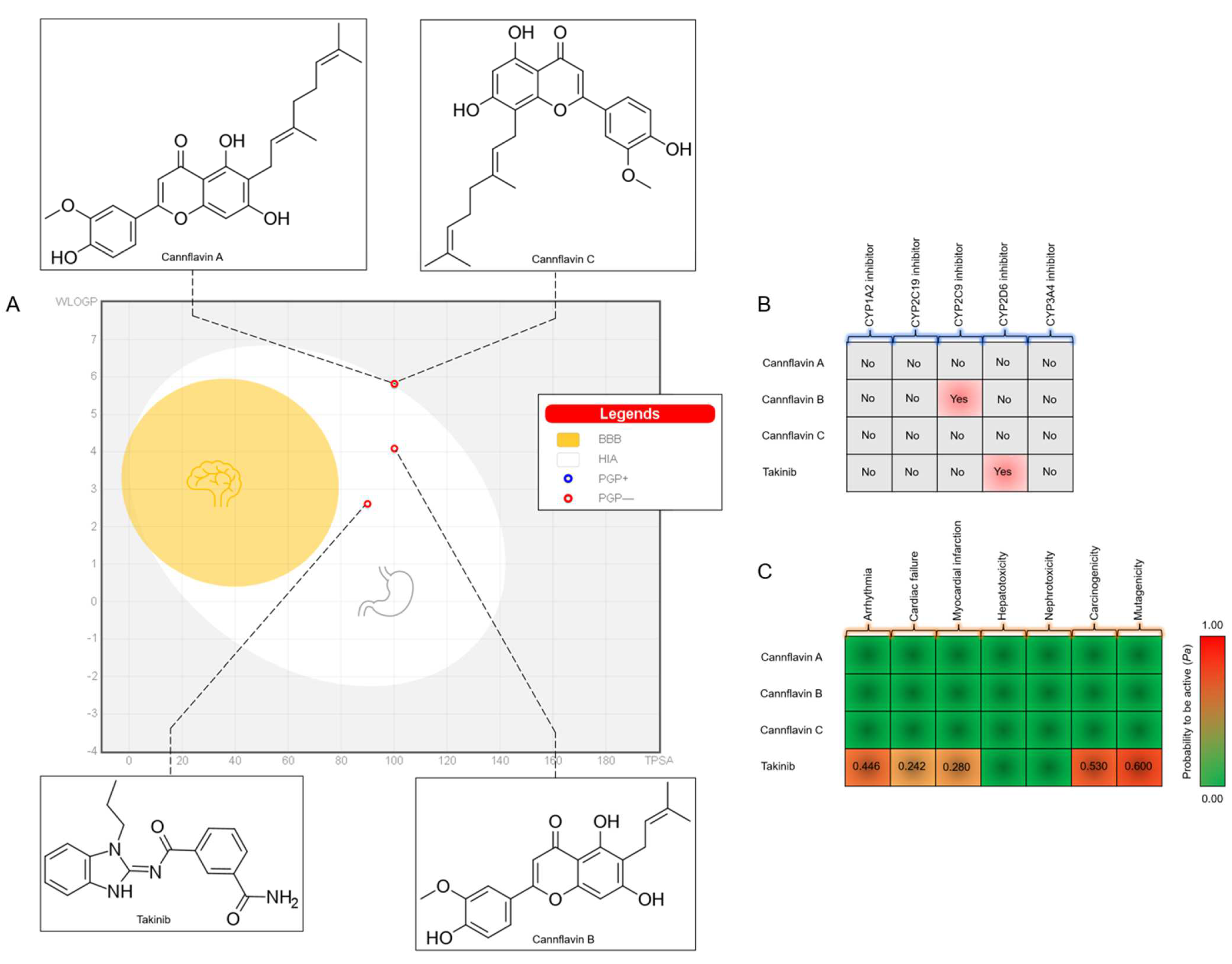
3.1. Drug-Likeness and ADMET Prediction
3.2. Molecular Docking
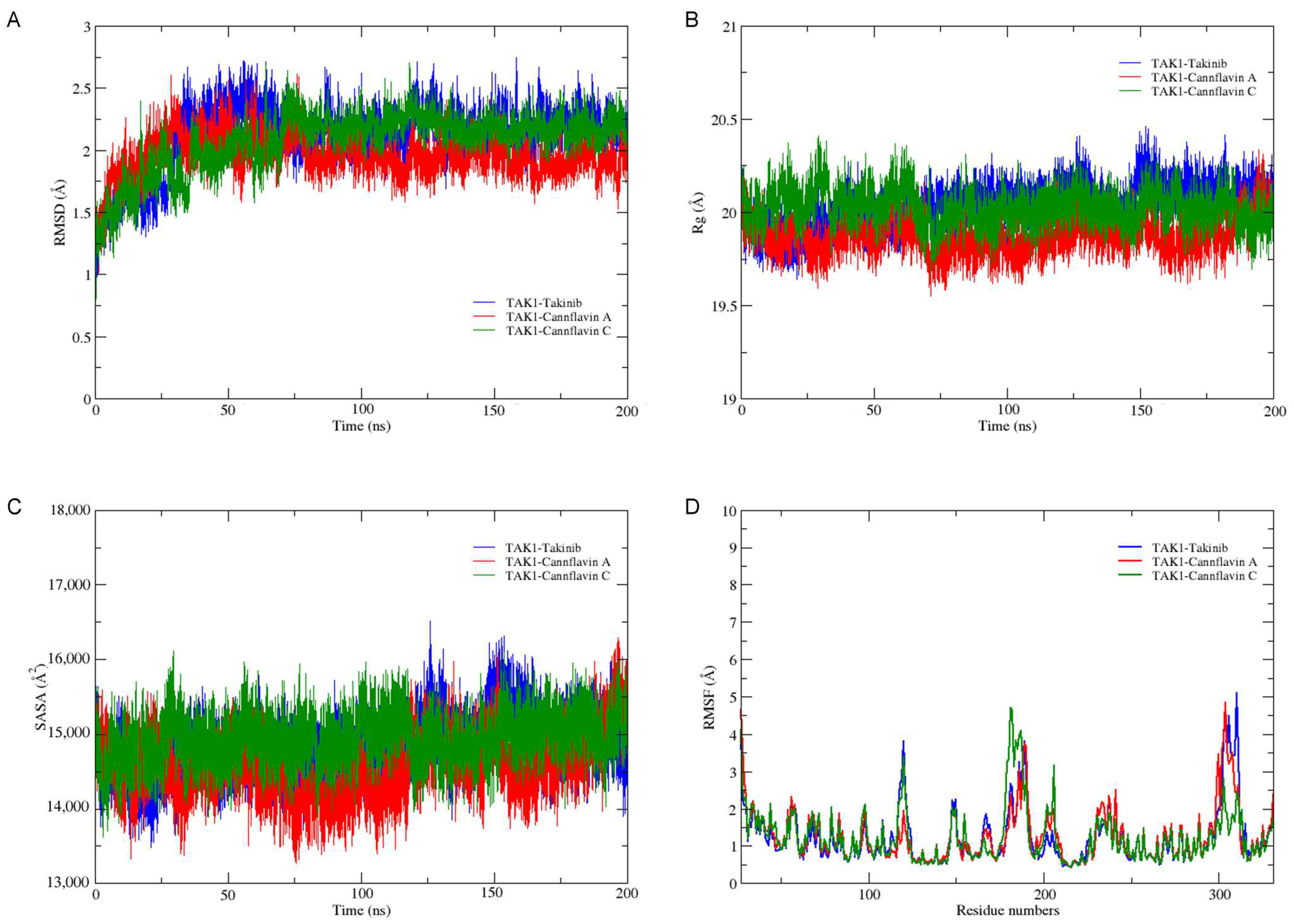
3.3. Molecular Dynamics Simulations and Free Energy Calculations
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Almutairi, K.; Nossent, J.; Preen, D.; Keen, H.; Inderjeeth, C. The global prevalence of rheumatoid arthritis: A meta-analysis based on a systematic review. Rheumatol. Int. 2021, 41, 863–877. [Google Scholar] [CrossRef]
- Manara, M.; Sinigaglia, L. Bone and TNF in rheumatoid arthritis: Clinical implications. RMD Open. 2015, 1, e000065. [Google Scholar] [CrossRef] [PubMed]
- Karampetsou, M.P.; Liossis, S.N.; Sfikakis, P.P. TNF-α antagonists beyond approved indications: Stories of success and prospects for the future. QJM 2010, 103, 917–928. [Google Scholar] [CrossRef] [PubMed]
- Croft, M.; Siegel, R.M. Beyond TNF: TNF superfamily cytokines as targets for the treatment of rheumatic diseases. Nat. Rev. Rheumatol. 2017, 13, 217–233. [Google Scholar] [CrossRef]
- Van Schouwenburg, P.A.; Rispens, T.; Wolbink, G.J. Immunogenicity of anti-TNF biologic therapies for rheumatoid arthritis. Nat. Rev. Rheumatol. 2013, 9, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Ding, T.; Ledingham, J.; Luqmani, R.; Westlake, S.; Hyrich, K.; Lunt, M.; Kiely, P.; Bukhari, M.; Abernethy, R.; Bosworth, A.; et al. BSR and BHPR rheumatoid arthritis guidelines on safety of anti-TNF therapies. Rheumatology 2010, 49, 2217–2219. [Google Scholar] [CrossRef] [PubMed]
- Roda, G.; Jharap, B.; Neeraj, N.; Colombel, J.F. Loss of Response to Anti-TNFs: Definition, Epidemiology, and Management. Clin. Transl. Gastroenterol. 2016, 7, e135. [Google Scholar] [CrossRef] [PubMed]
- Scarneo, S.A.; Eibschutz, L.S.; Bendele, P.J.; Yang, K.W.; Totzke, J.; Hughes, P.; Fox, D.A.; Haystead, T.A.J. Pharmacological inhibition of TAK1, with the selective inhibitor takinib, alleviates clinical manifestation of arthritis in CIA mice. Arthritis Res. Ther. 2019, 21, 292. [Google Scholar] [CrossRef]
- Probert, L. TNF and its receptors in the CNS: The essential, the desirable and the deleterious effects. Neuroscience 2015, 302, 2–22. [Google Scholar] [CrossRef]
- Javaid, N.; Patra, M.C.; Cho, D.E.; Batool, M.; Kim, Y.; Choi, G.M.; Kim, M.S.; Hahm, D.H.; Choi, S. An orally active, small-molecule TNF inhibitor that disrupts the homotrimerization interface improves inflammatory arthritis in mice. Sci. Signal. 2022, 15, eabi8713. [Google Scholar] [CrossRef]
- Rosenblum, H.; Amital, H. Anti-TNF therapy: Safety aspects of taking the risk. Autoimmun. Rev. 2011, 10, 563–568. [Google Scholar] [CrossRef] [PubMed]
- Totzke, J.; Scarneo, S.A.; Yang, K.W.; Haystead, T.A.J. TAK1: A potent tumour necrosis factor inhibitor for the treatment of inflammatory diseases. Open. Biol. 2020, 10, 200099. [Google Scholar] [CrossRef] [PubMed]
- Totzke, J.; Gurbani, D.; Raphemot, R.; Hughes, P.F.; Bodoor, K.; Carlson, D.A.; Loiselle, D.R.; Bera, A.K.; Eibschutz, L.S.; Perkins, M.M.; et al. Takinib, a Selective TAK1 Inhibitor, Broadens the Therapeutic Efficacy of TNF-α Inhibition for Cancer and Autoimmune Disease. Cell. Chem. Biol. 2017, 24, 1029–1039.e1027. [Google Scholar] [CrossRef] [PubMed]
- Scarneo, S.A.; Mansourati, A.; Eibschutz, L.S.; Totzke, J.; Roques, J.R.; Loiselle, D.; Carlson, D.; Hughes, P.; Haystead, T.A.J. Genetic and pharmacological validation of TAK1 inhibition in macrophages as a therapeutic strategy to effectively inhibit TNF secretion. Sci. Rep. 2018, 8, 17058. [Google Scholar] [CrossRef]
- Liu, Z.; Kong, F.; Vallance, J.E.; Harmel-Laws, E.; Amarachintha, S.; Steinbrecher, K.A.; Rosen, M.J.; Bhattacharyya, S. Activation of TGF-β activated kinase 1 promotes colon mucosal pathogenesis in inflammatory bowel disease. Physiol. Rep. 2017, 5, e13181. [Google Scholar] [CrossRef]
- Wu, H.; Zhou, J.; Ou, W.; Li, Y.; Liu, M.; Yang, C. TAK1 as the mediator in the protective effect of propofol on renal interstitial fibrosis induced by ischemia/reperfusion injury. Eur. J. Pharmacol. 2017, 811, 134–140. [Google Scholar] [CrossRef]
- Omori, E.; Morioka, S.; Matsumoto, K.; Ninomiya-Tsuji, J. TAK1 regulates reactive oxygen species and cell death in keratinocytes, which is essential for skin integrity. J. Biol. Chem. 2008, 283, 26161–26168. [Google Scholar] [CrossRef]
- Katsura, H.; Obata, K.; Miyoshi, K.; Kondo, T.; Yamanaka, H.; Kobayashi, K.; Dai, Y.; Fukuoka, T.; Sakagami, M.; Noguchi, K. Transforming growth factor-activated kinase 1 induced in spinal astrocytes contributes to mechanical hypersensitivity after nerve injury. Glia 2008, 56, 723–733. [Google Scholar] [CrossRef]
- Miao, Z.; Zhao, Y.; Chen, M.; He, C. Using flavonoids as a therapeutic intervention against rheumatoid arthritis: The known and unknown. Pharmacol. Res. Mod. Chin. Med. 2022, 3, 100014. [Google Scholar] [CrossRef]
- Erridge, S.; Mangal, N.; Salazar, O.; Pacchetti, B.; Sodergren, M.H. Cannflavins—From plant to patient: A scoping review. Fitoterapia 2020, 146, 104712. [Google Scholar] [CrossRef]
- Radwan, M.M.; Elsohly, M.A.; Slade, D.; Ahmed, S.A.; Wilson, L.; El-Alfy, A.T.; Khan, I.A.; Ross, S.A. Non–cannabinoid constituents from a high potency Cannabis sativa variety. Phytochemistry 2008, 69, 2627–2633. [Google Scholar] [CrossRef] [PubMed]
- Tomko, A.M.; Whynot, E.G.; Dupré, D.J. Anti-cancer properties of cannflavin A and potential synergistic effects with gemcitabine, cisplatin, and cannabinoids in bladder cancer. J. Cannabis Res. 2022, 4, 41. [Google Scholar] [CrossRef] [PubMed]
- Eggers, C.; Fujitani, M.; Kato, R.; Smid, S. Novel cannabis flavonoid, cannflavin A displays both a hormetic and neuroprotective profile against amyloid β-mediated neurotoxicity in PC12 cells: Comparison with geranylated flavonoids, mimulone and diplacone. Biochem. Pharmacol. 2019, 169, 113609. [Google Scholar] [CrossRef]
- Radwan, M.M.; Ross, S.A.; Slade, D.; Ahmed, S.A.; Zulfiqar, F.; Elsohly, M.A. Isolation and characterization of new Cannabis constituents from a high potency variety. Planta Med. 2008, 74, 267–272. [Google Scholar] [CrossRef]
- Byler, K.G.; Ogungbe, I.V.; Setzer, W.N. In-silico screening for anti-Zika virus phytochemicals. J. Mol. Graph. Model. 2016, 69, 78–91. [Google Scholar] [CrossRef] [PubMed]
- Barrett, M.L.; Scutt, A.M.; Evans, F.J. Cannflavin A and B, prenylated flavones from Cannabis sativa L. Experientia 1986, 42, 452–453. [Google Scholar] [CrossRef]
- Werz, O.; Seegers, J.; Schaible, A.M.; Weinigel, C.; Barz, D.; Koeberle, A.; Allegrone, G.; Pollastro, F.; Zampieri, L.; Grassi, G.; et al. Cannflavins from hemp sprouts, a novel cannabinoid-free hemp food product, target microsomal prostaglandin E2 synthase-1 and 5-lipoxygenase. PharmaNutrition 2014, 2, 53–60. [Google Scholar] [CrossRef]
- Barrett, M.L.; Gordon, D.; Evans, F.J. Isolation from Cannabis sativa L. of cannflavin--a novel inhibitor of prostaglandin production. Biochem. Pharmacol. 1985, 34, 2019–2024. [Google Scholar] [CrossRef]
- Bautista, J.L.; Yu, S.; Tian, L. Flavonoids in Cannabis sativa: Biosynthesis, Bioactivities, and Biotechnology. ACS Omega 2021, 6, 5119–5123. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Ivanov, S.M.; Lagunin, A.A.; Rudik, A.V.; Filimonov, D.A.; Poroikov, V.V. ADVERPred–Web Service for Prediction of Adverse Effects of Drugs. J. Chem. Inf. Model. 2018, 58, 8–11. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.; Eckert, A.O.; Schrey, A.K.; Preissner, R. ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018, 46, W257–W263. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 update. Nucleic Acids Res. 2023, 51, D1373–D1380. [Google Scholar] [CrossRef] [PubMed]
- Burley, S.K.; Bhikadiya, C.; Bi, C.; Bittrich, S.; Chao, H.; Chen, L.; Craig, P.A.; Crichlow, G.V.; Dalenberg, K.; Duarte, J.M.; et al. RCSB Protein Data Bank (RCSB.org): Delivery of experimentally-determined PDB structures alongside one million computed structure models of proteins from artificial intelligence/machine learning. Nucleic Acids Res. 2023, 51, D488–D508. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinform. 2014, 47, 5.6.1–5.6.32. [Google Scholar] [CrossRef]
- Frisch, M. Gaussian09. 2009. Available online: http://www.gaussian.com/ (accessed on 1 February 2023).
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Nguyen, N.T.; Nguyen, T.H.; Pham, T.N.H.; Huy, N.T.; Bay, M.V.; Pham, M.Q.; Nam, P.C.; Vu, V.V.; Ngo, S.T. Autodock Vina Adopts More Accurate Binding Poses but Autodock4 Forms Better Binding Affinity. J. Chem. Inf. Model. 2020, 60, 204–211. [Google Scholar] [CrossRef]
- Case, D.A.; Cheatham, T.E., 3rd; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M., Jr.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef]
- Thongsom, S.; Racha, S.; Petsri, K.; Ei, Z.Z.; Visuttijai, K.; Moriue, S.; Yokoya, M.; Chanvorachote, P. Structural modification of resveratrol analogue exhibits anticancer activity against lung cancer stem cells via suppression of Akt signaling pathway. BMC Complement. Med. Ther. 2023, 23, 183. [Google Scholar] [CrossRef] [PubMed]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Woods, R.J.; Chappelle, R. Restrained electrostatic potential atomic partial charges for condensed-phase simulations of carbohydrates. Theochem 2000, 527, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert. Opin. Drug. Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, S.; Lipper, R.A. Role of the development scientist in compound lead selection and optimization. J. Pharm. Sci. 2000, 89, 145–154. [Google Scholar] [CrossRef]
- Daina, A.; Zoete, V. A BOILED-Egg to Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. ChemMedChem 2016, 11, 1117–1121. [Google Scholar] [CrossRef]
- Marconi, A.; Di Forti, M.; Lewis, C.M.; Murray, R.M.; Vassos, E. Meta-analysis of the Association Between the Level of Cannabis Use and Risk of Psychosis. Schizophr. Bull. 2016, 42, 1262–1269. [Google Scholar] [CrossRef]
- Hoosain, F.G.; Choonara, Y.E.; Tomar, L.K.; Kumar, P.; Tyagi, C.; du Toit, L.C.; Pillay, V. Bypassing P-Glycoprotein Drug Efflux Mechanisms: Possible Applications in Pharmacoresistant Schizophrenia Therapy. Biomed. Res. Int. 2015, 2015, 484963. [Google Scholar] [CrossRef]
- Ahmed, J., II; Abdul Hamid, A.A.; Abd Halim, K.B.; Che Has, A.T. P-glycoprotein: New insights into structure, physiological function, regulation and alterations in disease. Heliyon 2022, 8, e09777. [Google Scholar] [CrossRef]
- Zanger, U.M.; Schwab, M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 2013, 138, 103–141. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.H. Pharmacokinetic and pharmacodynamic variability: A daunting challenge in drug therapy. Curr. Drug. Metab. 2007, 8, 109–136. [Google Scholar] [CrossRef] [PubMed]
- Veerman, J.J.N.; Bruseker, Y.B.; Damen, E.; Heijne, E.H.; van Bruggen, W.; Hekking, K.F.W.; Winkel, R.; Hupp, C.D.; Keefe, A.D.; Liu, J.; et al. Discovery of 2,4-1H-Imidazole Carboxamides as Potent and Selective TAK1 Inhibitors. ACS Med. Chem. Lett. 2021, 12, 555–562. [Google Scholar] [CrossRef] [PubMed]



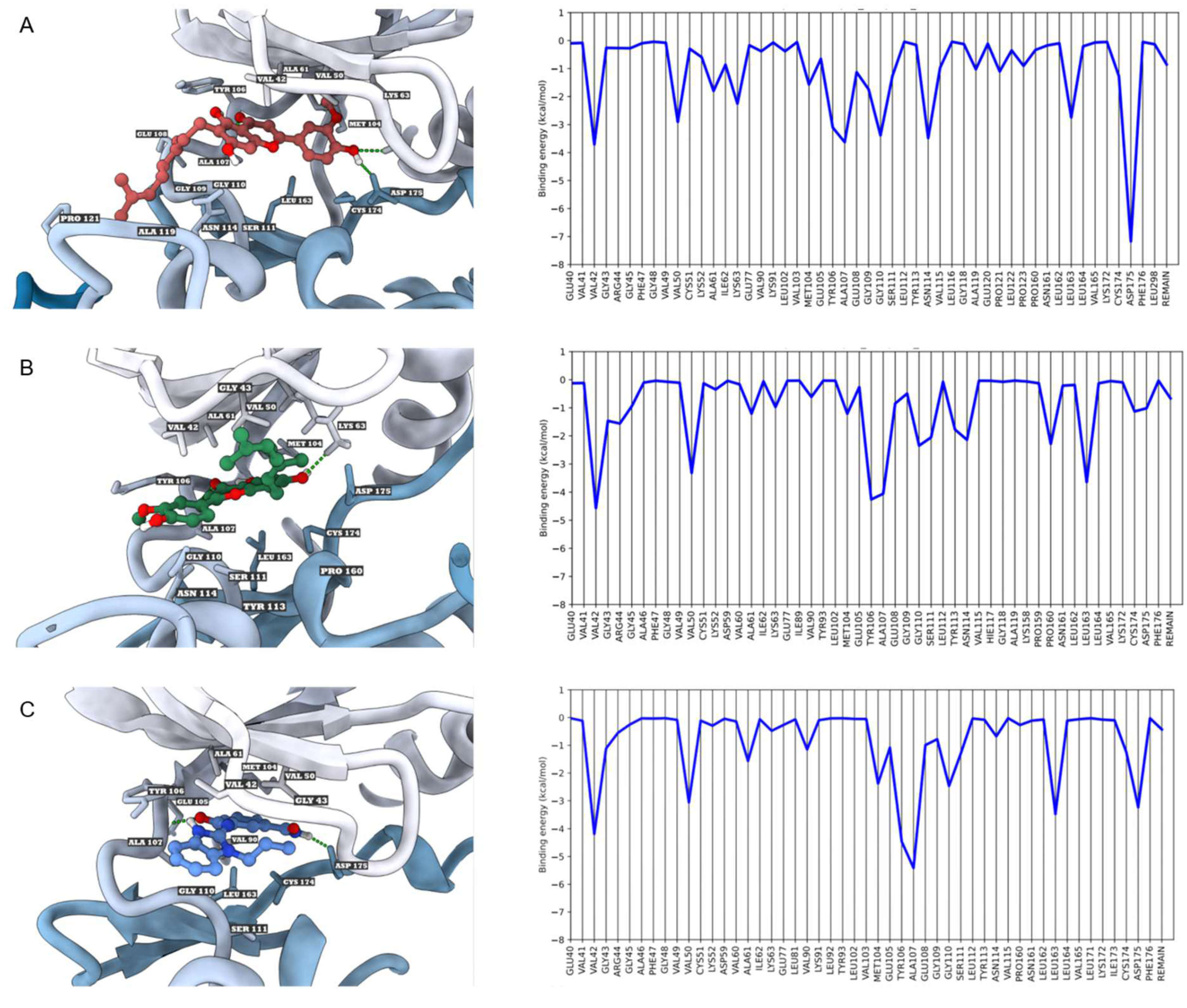
| Compounds | VDW (kcal/mol) | ELE (kcal/mol) | EGB (kcal/mol) | ESURF (kcal/mol) | ΔGgas (kcal/mol) | ΔGsolv (kcal/mol) | ΔGTOTAL (kcal/mol) |
|---|---|---|---|---|---|---|---|
| Cannflavin A | −49.3581 | −22.9419 | 36.3489 | −7.0476 | −72.3000 | 29.3013 | −42.9987 |
| Cannflavin C | −49.2620 | −14.3163 | 32.3529 | −6.5341 | −63.5783 | 25.8188 | −37.7595 |
| Takinib | −42.7576 | −18.2630 | 33.9200 | −5.4303 | −61.0206 | 28.4897 | −32.5310 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chuanphongpanich, S.; Racha, S.; Saengsitthisak, B.; Pirakitikulr, P.; Racha, K. Computational Assessment of Cannflavin A as a TAK1 Inhibitor: Implication as a Potential Therapeutic Target for Anti-Inflammation. Sci. Pharm. 2023, 91, 36. https://doi.org/10.3390/scipharm91030036
Chuanphongpanich S, Racha S, Saengsitthisak B, Pirakitikulr P, Racha K. Computational Assessment of Cannflavin A as a TAK1 Inhibitor: Implication as a Potential Therapeutic Target for Anti-Inflammation. Scientia Pharmaceutica. 2023; 91(3):36. https://doi.org/10.3390/scipharm91030036
Chicago/Turabian StyleChuanphongpanich, Sarunya, Satapat Racha, Banthita Saengsitthisak, Pichai Pirakitikulr, and Kannika Racha. 2023. "Computational Assessment of Cannflavin A as a TAK1 Inhibitor: Implication as a Potential Therapeutic Target for Anti-Inflammation" Scientia Pharmaceutica 91, no. 3: 36. https://doi.org/10.3390/scipharm91030036
APA StyleChuanphongpanich, S., Racha, S., Saengsitthisak, B., Pirakitikulr, P., & Racha, K. (2023). Computational Assessment of Cannflavin A as a TAK1 Inhibitor: Implication as a Potential Therapeutic Target for Anti-Inflammation. Scientia Pharmaceutica, 91(3), 36. https://doi.org/10.3390/scipharm91030036






