Abstract
Antibiotic resistance is a major health concern worldwide. In our previous study, some bacterial isolates exhibited antibacterial activity against Staphylococcus aureus and methicillin-resistant S. aureus (MRSA). However, the production of antibacterial substances by native microorganisms is limited by biosynthetic genes. This study aimed to improve the antibacterial activity of SPR19 using atmospheric and room temperature plasma mutagenesis (ARTP). The results showed that SPR19 belonged to the Brevibacillus genus. The growth curves and production kinetics of antibacterial substances were investigated. Argon-based ARTP was applied to SPR19, and the 469 mutants were preliminarily screened using agar overlay method. The remaining 25 mutants were confirmed by agar well diffusion assay against S. aureus TISTR 517 and MRSA isolates 142, 1096, and 2468. M285 exhibited the highest activity compared to the wild-type strain (10.34–13.59%) and this mutant was stable to produce the active substances throughout 15 generations consistently. The antibacterial substances from M285 were tolerant to various conditions (heat, enzyme, surfactant, and pH) while retaining more than 90% of their activities. Therefore, Brevibacillus sp. SPR19 is a potential source of antibacterial substances. ARTP mutagenesis is a powerful method for strain improvement that can be utilized to treat MRSA infection in the future.
1. Introduction
Infectious diseases caused by pathogenic microorganisms such as bacteria, viruses, protozoa, and fungi are a major global health problem [1]. Antibiotics are secondary metabolites produced by some microorganisms useful for treating these harmful infections [2]. Staphylococcus aureus is a pathogenic bacterium that causes severe infections. The advent of antibiotics such as penicillin and methicillin has revolutionized the treatment against them. However, these drugs rapidly become ineffective as the pathogens develop into resistant strains, methicillin-resistant S. aureus (MRSA), rendering them difficult to cure [3]. In 2019, at least 2.8 million people in the US were infected with antibiotic-resistant bacteria or fungi, and more than 35,000 people died as a result [4]. The increase in antibiotic resistance is a consequence of multiple factors, including the irrational use of antibiotics in agriculture, veterinary, and human medication and the evolution of pathogen tolerance to drugs [5]. However, the number of novel antibiotics approved for commercialization is relatively small [5,6].
Most commercial antibiotic drugs such as streptomycin, kanamycin, and polymyxin are natural products. They are produced by soil bacteria originating from Streptomyces sp. and Bacillus sp. [7,8]. Generally, bacteria have a limited ability to produce antibacterial compounds due to the restricted gene regulation of the relevant biosynthetic pathways. Thus, bacterial utilization in medical and industrial uses is necessary to increase the efficiency or productivity of bioactive agents by optimizing the culture media to be suitable for microbial growth or using genetic engineering to express the active substances effectively. Mutagenesis of microbial genes is also useful for enhancing the production of active compounds. There are several mutagenesis methods for strain improvement, including physical (e.g., γ-ray and UV radiation) and chemical mutagens (e.g., ethyl methanesulfonate and N-methyl-N′-nitro-N-nitrosoguanidine (MNNG)) [9,10].
Random mutagenesis is advantageous when the location of the desired gene is unknown or when a complex gene regulation system is present. Almost all tools used for random mutagenesis carry inherent risks. Chemical mutagens (e.g., ethidium bromide and sodium azide) expose users to high or extremely high levels of toxicity. Radiation (e.g., X-rays and γ-rays) exposes the user to permanent danger, necessitating additional control measures such as radiation badges and radiation barriers. Additionally, radiation generation requires the use of large and expensive equipment, specialized knowledge, and other security measures to restrict access.
Atmospheric and room temperature plasma (ARTP) is a random mutagenesis technique that has led to the development of physical methods, causing greater DNA damage with a higher mutation rate than UV and certain chemical mutagens. Several studies have shown that ARTP successfully improves genetic enhancement [10]. A previous study showed that the ARTP mutagenesis of Bacillus amyloliquefaciens caused a lethality rate of approximately 95% at 100 s, where a positive mutant increased the production of menaquinone-7 by 4.25 times [11]. Another study used helium-based ARTP on Bacillus subtilis for 150 s, where a colorimetric response above 26% was used as the criterion to select the high-yield mutants for surfactin production. They screened 27,000 mutants and identified 37 high-yielding isolates. One mutant increased surfactin synthesis by 5.4 times, and a C15 surfactin variant was highly produced compared with the parent [12]. Although the exact reason for the mutagenesis mechanism of the ARTP system has not yet been fully clarified, the generation of UV radiation and reactive species have been reported as possible causes [13]. Reactive chemical species alter DNA bases and induce single- or double-strand breaks [14].
SPR19 strain was isolated from soil at a botanical garden in Nakhon Si Thammarat, Thailand, while searching for bacterial isolates with antibacterial activity. Our preliminary results revealed that it possesses potent antibacterial activity against S. aureus and MRSA, suggesting that improving the activity of this strain would benefit further studies. In this study, the argon-based ARTP mutation system was applied to SPR19, and surviving mutants with enhanced antibacterial activity against S. aureus and MRSA were selected. The mutant that exhibited the highest activity was used for determining the strain stability. Additionally, the stability properties of the antibacterial substances under stressed conditions were characterized.
2. Materials and Methods
2.1. Microorganisms and Culture Conditions
Brevibacillus sp. SPR19 (GenBank accession no. MZ298491) and the indicator strains (S. aureus TISTR 517 from Thailand Institute of Scientific and Technological Research, Thailand, MRSA clinical isolates 142, 1096, and 2468 from Maharaj Nakhon Si Thammarat Hospital, Nakhon Si Thammarat, Thailand) were maintained in 40% glycerol solution at −80 °C [15]. The bacteria were thawed and then streaked on Mueller Hinton (MH) agar (Beef, infusion form 300 g/L, casein acid hydrolysate 17.5 g/L, agar 17.0 g/L, and starch 1.5 g/L) (Titan Biotech Ltd., Rajasthan, India). The plates were incubated at 30 °C and 37 °C for 24 h for SPR19 and indicator strains, respectively (Memmert GmbH+ Co., Schwabach, Germany). The single colony of SPR19 was inoculated in half formula of Luria Bertani (half LB) broth (Casein enzymic hydrolysate 5.0 g/L, sodium chloride 5.0 g/L, and yeast extract 2.5 g/L) (Titan Biotech Ltd., Rajasthan, India) at 30 °C, 150 rpm for 24 h.
2.2. Strain Identification
SPR19 was subjected to Gram and spore staining and subsequently visualized under a light microscope (Carl Zeiss, Oberkochen, Germany) at 1000× magnification [16,17]. Genomic DNA was extracted, and the 16s rRNA sequencing was performed using the forward primer (27F; 5′-AGAGTTTGATCCTGGCTCAG-3′) and reverse primer (1492r; 3′-GGTTACCTTGTTACGACTT-5′) [18]. The sequence was analyzed using NCBI BLAST, and a phylogenetic tree was constructed using MEGA X software with neighbor-joining analysis (1000 bootstrap confidence values) [19,20].
2.3. Growth Curve and Production Kinetics of Antibacterial Substances
The single colonies of the wild-type strain of SPR19 were inoculated in 50 mL of half LB broth at 30 °C, 150 rpm for 24 h. The culture was adjusted to a turbidity equivalent to 0.5 McFarland, and an aliquot of 500 µL was transferred to 49.50 mL (1%) of half LB broth at 30 °C, 150 rpm. The samples were collected at the specified time intervals and centrifuged at 10,000× g at 4 °C for 15 min (Sigma-Aldrich Co., St. Louis, MO, USA). The cell-free supernatant (CFS) was used to determine the antibacterial activity against the indicator strains (S. aureus TISTR 517 and MRSA isolates 142, 1096, and 2468) using the agar well diffusion method. Bacterial growth was measured using a spectrophotometer at an optical density (OD) of 600 nm (Thermo Fisher Scientific, Waltham, MA, USA) [21]. The incubation time of the culture that exhibited the highest antibacterial activity was used for further assays.
2.4. Atmospheric and Room Temperature Plasma (ARTP) Mutagenesis
The argon-based ARTP was designed and constructed by a team of plasma scientists at the Center of Excellence in Plasma Science and Electromagnetic Waves at Walailak University, Thailand. A single colony of the wild-type strain of SPR19 was inoculated in half LB broth, and the culture was shaken at 30 °C, for about 6 h to achieve the mid-logarithmic phase. The culture (100 mL) was centrifuged at 10,000× g at 4 °C for 5 min, washed three times with 0.85% NaCl (RCI Labscan Ltd., Bangkok, Thailand), and then resuspended in 1 mL of 0.85% NaCl. Subsequently, the turbidity of SPR19 cells was adjusted until the OD 600 nm reached 0.60. Ten microliters of SPR19 cell suspension were transferred to a PCR tube cap and positioned under a plasma nozzle. The distance between the sample and the plasma nozzle was constant at 2 mm, and the input power was approximately 4 W. The argon gas flow rate was 10 standard liter per minute (SLPM), and the mutagenic exposure time was varied between 0 and 150 s. After ARTP treatment, the sample was withdrawn from the tube cap and diluted to 1 mL with 0.85% NaCl. One hundred microliters of the diluted sample were spread on MH agar to count the surviving cells [10]. Three replicate experiments were performed, and the lethality rate was calculated as follows:
where NT and NU are the colony numbers in the treated and untreated groups, respectively [22].
Lethality rate = [1−(NT/NU)] × 100
2.5. Determination of Reactive Oxygen Species (ROS) and Reactive Nitrogen Species (RNS) Concentration
Hydrogen peroxide radical content was measured using a fluorometric hydrogen peroxide assay kit (Sigma-Aldrich Co., St. Louis, MO, USA). After the ARTP treatment, 50 µL of the diluted sample was transferred to a 96-well plate. Fifty microliters of the reaction solution, including the red peroxidase substrate, peroxidase, and assay buffer, were added to each well. The plate was mixed and incubated at room temperature for 30 min under light protection. Fluorescence intensity at 590 nm was measured using a fluorescence plate reader (BioTek, Winooski, VT, USA) at an excitation wavelength of 540 nm. The reaction solution without the sample was used as the negative control. The concentration of hydrogen peroxide in the sample was compared to the calibration curve of the hydrogen peroxide standard in the range of 0.01–10.00 µM. Furthermore, the concentration of nitrite radicals was determined using the Griess reagent (Sigma-Aldrich Co., St. Louis, MO, USA). Fifty microliters of the diluted individual time sample after ARTP treatment were transferred to each well of a 96-well plate and 50 µL of Griess reagent was added. The plates were mixed and incubated at room temperature for 15 min. The absorbance at 540 nm was measured using a microplate reader (BioTek, Winooski, VT, USA). Nitrite-free water with the Griess reagent was used as the negative control. The nitrite concentration was determined from the calibration curve using a sodium nitrite standard in the range of 0.509–8.125 µM. In addition, pH after ARTP treatment was determined using universal indicators (Merck KGaA, Darmstadt, Germany).
2.6. Antibacterial Assay by Agar Overlay and Agar Well Diffusion Methods
After plasma treatment, surviving mutants were preliminarily screened for antibacterial activity using the agar overlay method [23]. The culture of S. aureus TISTR 517 was adjusted to a turbidity equivalent to 0.5 McFarland. The mutant colonies were spotted on MH agar, and the plates were then incubated at 30 °C for 24 h. The soft MH agar (0.7% agar) containing a cell suspension of S. aureus TISTR 517 was overlaid on the spotted plates. The plates were incubated at 37 °C overnight. The ratio between the inhibition zone and colony diameter was measured, and the results were compared with those of the wild-type strain. The mutants that showed significantly higher antibacterial activity than the wild-type strain were further used to confirm the agar well diffusion method against all indicator strains (S. aureus TISTR 517 and MRSA isolates 142, 1096, and 2468) [21]. One hundred microliters of CFS of the selected mutants were introduced into each well (diameter 9 mm) of MH agar that was spread with the indicator strains. The plates were then incubated at 37 °C overnight. The inhibition zone was determined and compared with that of the wild-type strain. The mutants which exhibited significantly higher activity than the wild-type strain were used to study strain stability. In addition, the antibacterial activity spectrum of CFS of the wild-type and mutant strains was compared to that of antibiotic drugs, including vancomycin (30 µg), cefoxitin (30 µg), and oxacillin (1 µg) (Sigma-Aldrich Co., St. Louis, MO, USA) by agar well diffusion assay. Each experiment was performed in triplicate.
2.7. Strain Stability
The stability of the wild-type and mutant strains to produce constant antibacterial activity was examined by consecutively streaking them on MH agar for 15 generations [24]. Each generation was inoculated in half LB broth, and turbidity was adjusted equivalent to 0.5 McFarland before transferring 1% of the preculture into 49.50 mL of fresh half LB broth. The culture was incubated at 30 °C, 150 rpm for 24 h, and the supernatants of the 5th, 10th, and 15th generations were collected by centrifugation at 10,000× g at 4 °C for 15 min. The antibacterial activity was assessed by agar well diffusion methods, and the inhibition zone was measured using the wild-type strain for comparison.
2.8. Drug Susceptibility Study
The susceptibility of the wild-type and mutant strains to standard antibiotics was determined using the disk diffusion method [25]. The wild-type and mutant strains were grown on MH agar at 30 °C for 24 h. The turbidity of a single colony was adjusted equivalent to 0.5 McFarland standard before spreading on MH agar. The antibiotic disks; cefoxitin (30 µg), ceftriaxone (30 µg), ciprofloxacin (5 µg), doxycycline (30 µg), erythromycin (15 µg), gentamycin (10 µg), imipenem (10 µg), and vancomycin (30 µg) (Oxoid Ltd., Hampshire, UK) were placed on the surface of MH agar and then incubated at 30 °C for 24 h. The inhibition zone was measured, and the results are presented as the mean ± SD.
2.9. Scanning Electron Microscope (SEM)
The cell morphologies of the wild-type and mutant strains were captured by SEM [22]. Briefly, the bacterial cells were cultured on MH agar at 30 °C for 8 d. The cells were dispersed on a glass slide and fixed with 2.5% glutaraldehyde in 0.1 M phosphate buffer pH 7.2 for 24 h. The fixed samples were washed twice with phosphate buffer. The samples were fixed by 1.0% OsO4 at 4 °C for 1 h and washed with deionized water. The post-fixed samples were dehydrated stepwise by increasing the ethanol concentration gradually from 20% to 100%. Residual ethanol was removed from the sample using a critical point dryer (Quorum Technologies Ltd., Lewes, UK) before coating it with a gold sputter coater (Cressington Scientific Instrument Ltd., Watford, UK). SEM micrographs (Carl Zeiss, Oberkochen, Germany) were captured at a magnification of 10,000×.
2.10. Comparison of Growth Curve and Production Kinetics of Antibacterial Substances
The preculture of the wild-type and mutant strains was adjusted equivalent to turbidity equivalently to 0.5 McFarland. Five hundred microliters of the preculture were transferred to 49.50 mL of half LB broth, further incubating the culture at 30 °C, 150 rpm. The samples were collected at the specified time intervals and centrifuged at 10,000× g at 4 °C for 15 min. CFS was collected and used to determine antibacterial activity against S. aureus TISTR 517 and MRSA isolate 2468 using the agar well diffusion method. The growth curve of the bacteria was measured using a spectrophotometer at OD 600 nm (Thermo Fisher Scientific, Waltham, MA, USA) [21].
2.11. Determination of Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC)
The antibacterial activity of SPR19 was proportional to the concentration of its active substances. Therefore, the MIC and MBC of CFS can be determined directly from its activity [26]. After 24 h of incubation of the wild-type and mutant isolates, the MIC and MCB of the CFS against the indicator strains (S. aureus TISTR 517, MRSA isolates 142, 1096, and 2468) were determined by the critical-dilution method in a 96-well plate. Samples (100 µL) after a two-fold dilution of CFS with half LB broth were added to each well containing 10 µL of indicator strains (5 × 105 cells/mL). The plates were then incubated at 37 °C for 24 h. Bacterial growth was monitored by measuring the absorbance at 625 nm using a microplate reader. The medium broth alone and medium broth without CSF were used as the blank and untreated samples, respectively. The highest inhibitory dilution of CFS that resulted in more than 90% of inhibition between the treated and untreated samples was used to calculate MIC. The MBC of CFS was determined by spreading an aliquot (50 µL) of the diluted sample on MH agar to observe colony formation. The highest bactericidal dilution of CFS, which caused no colony growth, was recorded for MBC. The CFS of wild-type is used as the positive control to monitor the mutagenized consequence of M285 on the enhanced antibacterial activity. The result of the highest dilution was expressed as the activity of the respective MIC and MBC in arbitrary units per mL (AU/mL) using the following equation:
where n is the highest two-fold dilution showing antibacterial activity, and V is the volume in µL used to test the antibacterial activity [27].
Arbitrary Unit = (2n × 1000)/V
2.12. Stability of the Antibacterial Substances
The colonies of wild-type and M285 strains were inoculated into 50 mL of half LB broth and incubated at 30 °C, 150 rpm for 24 h. The preculture was dispersed in 0.85% NaCl until the turbidity was equivalent to 0.5 McFarland. The aliquot (1%) was transferred to 100 mL of half LB broth and incubated at 30 °C, 150 rpm for 24 h. The CSF was collected by centrifugation at 10,000× g at 4 °C for 15 min. The stability was determined by the following conditions. The percent of the remaining activity was calculated by the ratio between the activity of the treated and untreated samples and multiplied by 100.
2.12.1. Effect of Temperature
The CFSs of the wild-type and mutant strains were incubated at 60 °C, 80 °C, and 100 °C for 1 h, and 121 °C for 15 min (autoclave). Antibacterial activity was tested against the indicator strains (S. aureus TISTR 517 and MRSA isolate 2468). Half of the LB broth and untreated samples were used as negative and positive controls, respectively.
2.12.2. Effect of Proteolytic Enzyme
The CFS was incubated at 37 °C for 1 h in combination with proteinase K, trypsin, and α-chymotrypsin (1 mg/mL) (Vivantis Technologies Sdn. Bhd., Selangor Darul Ehsan, Malaysia). Proteolytic enzymes alone and the untreated sample were used as negative and positive controls, respectively.
2.12.3. Effect of Surfactant
Triton X-100 and sodium dodecyl sulfate (SDS) (1% v/v) (AppliChem GmbH, Darmstadt, Germany) were added to the CFS of the wild-type and mutant strains. The mixtures were incubated at 37 °C for 1 h. Triton X-100 or SDS alone and the untreated samples were used as the negative and positive controls, respectively.
2.12.4. Effect of pH
The culture supernatant was adjusted to pH, ranging from 1.0–14.0 by using 1.0 N HCl or 1.0 N NaOH. The samples were incubated at 37 °C for 1 h and then neutralized to pH 7.0, before testing the antibacterial activity against the indicator strains. Half of the LB and untreated samples were used as negative and positive controls, respectively [27].
2.13. Statistical Analysis
The results are represented as the mean ± SD of triplicate experiments. The Student’s t-test was used to analyze the significantly different sample with a p-value < 0.05 [15].
3. Results and Discussion
3.1. Characteristic, Growth Curve, and Production Kinetics of SPR19
In our previous study, we isolated SPR19 from terrestrial soil in a botanical garden in Thailand, which showed potential antibacterial activity against S. aureus and MRSA. Microscopic examination revealed that SPR19 is a Gram-positive bacterium with rod-shaped and spore-forming characteristics. The isolate was identified as Brevibacillus sp. SPR19 (GenBank accession number MZ298491) based on 16s rRNA sequencing. This isolate was closely related to Brevibacillus halotolerans LAM0312, with a similarity of 99.71% (Figure 1). Brevibacillus halotolerans LAM0312 is a new species in this genus and was isolated from saline soil. There are few literature reviews of its activities [28]. The CFS of Brevibacillus sp. SPR19 wild-type isolate at the specified incubation time was tested for antibacterial activity against S. aureus TISTR 517, MRSA isolates 142, 1096, and 2468 using the agar well diffusion assay. The SPR19 culture showed activity after 16 h of incubation. The maximum activity was observed at 24 h, corresponding to the early stationary phase of the bacterial growth curve. After 48 h of incubation, antibacterial activity gradually declined until 168 h (Figure 2).
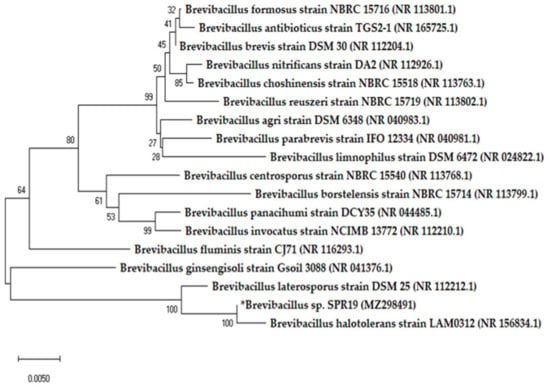
Figure 1.
Phylogenetic tree of Brevibacillus sp. SPR19 based on 16s rRNA sequencing. The tree was constructed by MEGA X software with neighbor-joining analysis.
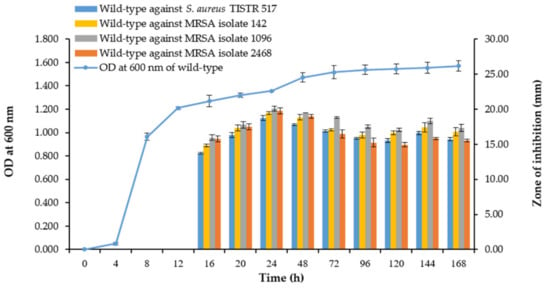
Figure 2.
Growth curve of Brevibacillus sp. SPR19 and its production kinetics of antibacterial substances. The growth curve was measured at OD 600 nm, while the antibacterial activity against S. aureus TISTR 517 and MRSA isolates 142, 1096, and 2468 was determined by the agar well diffusion method. The experiment was performed in triplicate, and the results were expressed as mean ± SD (n = 3).
3.2. ARTP Mutagenesis and Lethality Rate
Normally, natural microorganisms have a limited ability to produce bioactive substances. Thus, medical and industrial applications require the enhancement of these strains by upregulating corresponding genes to achieve high efficiency in producing these bioactive substances [29]. ARTP mutagenesis leads to greater DNA damage, resulting in higher production of bioactive substances [10,30]. In this study, mutagenesis of Brevibacillus sp. SPR19 was accomplished using the ARTP method with argon as the carrier gas. The result showed that the bacterial lethality rate followed a time-dependent profile, and the rate achieved was 95.92 ± 5.31% and 100.00 ± 0.01% when exposed to plasma for 30 and 150 s, respectively (Figure 3). Longer exposure time to plasma radiation resulted in higher bactericidal activity. This result was consistent with other studies, which reported that plasma treatment caused a higher positive genotoxic response. Generally, ARTP mutation is regarded to work effectively when the lethality rate is more than 90% [31]. Therefore, the mutants with a lethality rate of more than 95% were collected in this study for subsequent experiments.
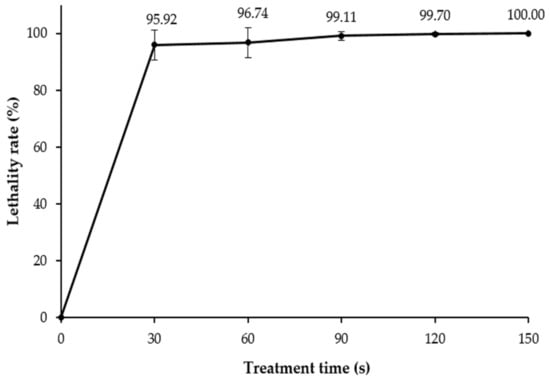
Figure 3.
The lethality rate of Brevibacillus sp. SPR19 by ARTP treatment. The experiment was performed in triplicate, and the results were expressed as mean ± SD (n = 3).
3.3. Determination of ROS and RNS Concentration
The exact mechanism of the ARTP mutagenesis system has not yet been fully understood; however, the generation of UV radiation and ROS have been identified as a possible cause, leading to DNA base alteration and induction of single-or double-strand breaks [13,14]. The amount of UV radiation and composition of the reactive chemical species produced are primarily determined by the carrier gases [10]. Argon plasma emits less UV radiation and produces oxygen-derived species, such as atomic oxygen (O), hydroxyl radicals (•OH), hydrogen peroxide (H2O2), ozone (O3), and superoxide anion radicals (•O2−), which are the most abundant. Nitrogen-containing species, such as nitric oxide (NO) and nitrite/nitrate (NO2−/NO3−), are less abundant but are present in these [10,32]. Plasma produces short-lived ROS as a stable product during treatment [33]. The amount of hydrogen peroxide (H2O2) was directly correlated with the treatment time. The hydrogen peroxide (H2O2) concentration increased from 1.69 ± 0.13 to 14.50 ± 1.37 μM after 150 s of treatment. Furthermore, nitrite is frequently used as an indicator of the chemistry of reactive nitrogen species. As a result, the nitrite (NO2−) levels during treatment were quantified. The results showed that nitrite concentration decreased with treatment time. The nitrite level declined from 2.43 ± 0.00 μM to near-zero after 150 s of treatment. This can be attributed to the conversion of nitrite (NO2−) to nitrate (NO3−) in an acidic environment [34]. This was consistent with our finding that the reaction pH changed from 7 to 5 during the plasma treatment (Figure 4). Therefore, increasing ROS and RNS levels could cause ARTP-induced bacterial mutagenesis. However, one limitation of this technique is random mutagenesis. Achieving mutations may depend on various factors such as treatment conditions (energy input power, gas flow rate, distance between sample and nozzle, and exposure time), type of bacterial culture, and post-treatment conditions [10]. A previous study investigated the influence of carrier gas on the generation of free radical species. It was found that nitrogen- or air-based ionizing gases resulted in the production of more nitrate and nitrite species, while oxygen- or argon-based gases provided a high content of hydrogen peroxide [35]. These free radicals increased as a function of treatment time [36]. Generally, free radicals damage DNA bases and sugar moieties, leading to base modifications and mutagenesis [30,37,38]. A recent study investigated the molecular mechanisms of plasma treatment on structural changes to mononucleotides and oligonucleotides [37]. This study showed that dATP is sensitive to plasma radiation, and the radiation primarily breaks the phosphate backbone. Interestingly, the oligonucleotides (dC8, dA8, and dG8) were susceptible to degradation into small fragments, resulting from cleavage of the heterocyclic rings of purine and pyrimidine bases under ARTP exposure [37]. DNA damage is relieved by complicated repair responses and proceeds to genetic mutations, resulting in cell survival [38,39].
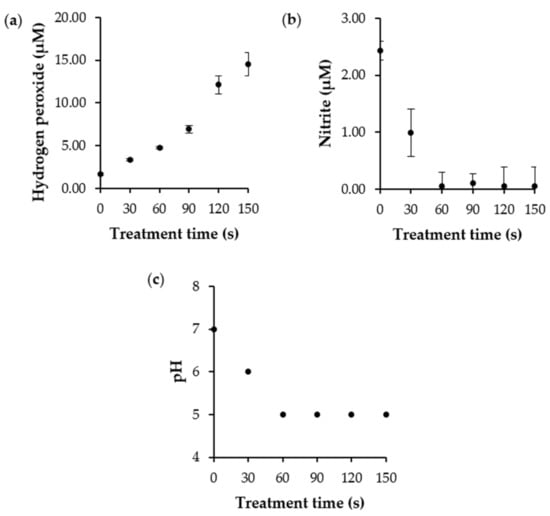
Figure 4.
(a) The hydrogen peroxide, (b) nitrite concentration, and (c) pH after ARTP treatment. The experiments were performed in triplicate, and the results were expressed as mean ± SD (n = 3).
3.4. Determination of the Antibacterial Activity of the Mutants
After ARTP mutagenesis, 469 isolates survived and were screened for antibacterial activity using the agar overlay method. Twenty-five mutants showed a significantly higher ratio between the inhibition zone against S. aureus TISTR 517 and the colony diameter than the wild-type strain (Table 1). The antibacterial activities of these isolates were determined using the agar well diffusion method. Only four mutants (M251, M276, M285, and M403) exhibited significant activity against all the indicator strains. M285 showed the highest activity with the inhibition zone of 20.97 ± 0.39, 21.46 ± 0.58, 22.76 ± 0.89, and 22.48 ± 1.01 mm for S. aureus TISTR 517, as well as MRSA isolates 142, 1096, and 2468, respectively. The wild-type strain showed the lower inhibition zone of 18.72 ± 1.54, 19.45 ± 1.39, 20.07 ± 1.67, and 19.79 ± 1.46 mm for the above indicator bacteria, respectively. This indicated that M285 produced higher antibacterial activity than the wild-type strain (10.34–13.59%). Interestingly, the CFS of these four mutants and vancomycin showed a similar spectrum of antibacterial activity against all the indicator pathogens, while cefoxitin and oxacillin did not inhibit MRSA. Similar results have been observed in other studies. The increased production of avermectin by Streptomyces avermitilis ATCC 31267 was improved by 18.9% after plasma mutagenesis of its spores [40]. Furthermore, other bioactive substances such as enzymes and small molecules were produced using this procedure. Plasma-treated yeast Candida tropicalis had 1.4-fold higher xylose reductase activity compared to the wild-type strain, and the mutants increased the production of xylitol (22%) [41]. This indicates that ARTP is a powerful method for producing unique mutants that enhance antibacterial activity, enabling strain improvement for pharmaceutical and industrial applications.

Table 1.
Comparison of antibacterial activity between the wild-type and its mutants by agar well diffusion (mean ± SD; n = 3).
3.5. Strain Stability
The stability of the mutant strain is indispensable for its use in pharmaceutical and industrial applications [41]. The wild-type and mutant strains (M251, M276, M285, and M403) were evaluated for strain stability by measuring antibacterial activity after consecutive subcultures. All mutants showed higher antibacterial activity against S. aureus TISTR 517 and MRSA isolates 142, 1096, and 2468 than the wild-type strain. Interestingly, only M285 exhibited strain stability, which produced significantly higher and constant antibacterial activity against all bacterial indicators throughout the 15 generations (Figure 5). This implies that M285 is genetically stable to produce antibacterial substances.
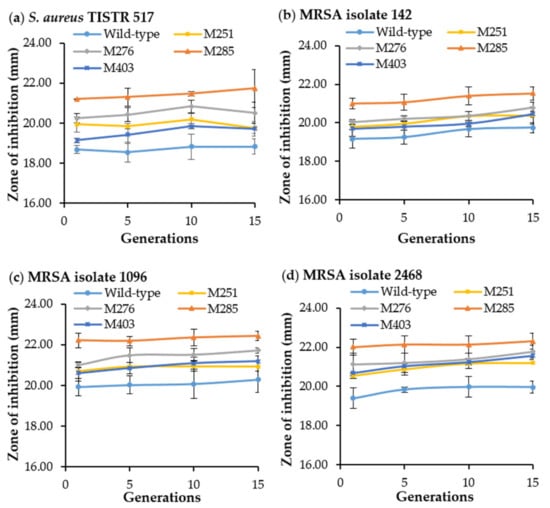
Figure 5.
Antibacterial activity of the wild-type and its mutants (SPR19) after 15 consecutive generations. The CFS of the wild-type and mutants were used to measure the antibacterial activity against (a) S. aureus TISTR 517, (b) MRSA isolate 142, (c) MRSA isolate 1096, and (d) MRSA isolate 2468. The experiments were performed in triplicate, and the results were expressed as mean ± SD (n = 3).
3.6. Drug Susceptibility Study
Susceptibility of the wild-type and mutant strains was evaluated using standard antibiotic disks. M285 was more susceptible to some antibiotics than the wild-type strain by which ceftriaxone, erythromycin, imipenem, and vancomycin showed a significant inhibition zone of 46.57 ± 0.78, 48.60 ± 0.29, 47.92 ± 0.78, and 28.62 ± 0.59 mm, respectively, whereas the wild-type strain had an inhibition zone of 44.03 ± 0.78, 46.57 ± 0.78, 44.20 ± 0.51, and 26.25 ± 0.29 mm. Furthermore, M285 appeared to be more sensitive to the remaining antibiotics (cefoxitin, ciprofloxacin, doxycycline, and gentamycin) than the wild-type strain (Table 2). This indicated the effect of plasma mutagenesis on the drug sensitivity of the mutants, and the underlying mechanisms of these drug susceptibilities require further investigation. Some studies have demonstrated that the generated reactive chemical species affect the drug targets of chemotherapeutic agents and enhance drug sensitivity and cancer inhibition [42].

Table 2.
Antibiotics susceptibility of Brevibacillus sp. SPR19 and its mutant strains (mean ± SD; n = 3).
3.7. Morphological Characteristics by SEM
The wild-type and M285 mutant showed similar morphology, in which the colonies had a white and circular form with convex elevation and curled margin. SEM results revealed that both cells also had similar rod-shaped and spore-forming bacterial characteristics. The size of vegetative cells was in the dimensional range of 0.480 ± 0.040 µm × 2.643 ± 0.719 µm and 0.452 ± 0.058 µm × 3.566 ± 0.859 µm for the wild-type and M285 strains, respectively. The spore sizes of the wild-type and mutant strains were 1.151 ± 0.087 × 2.216 ± 0.231 µm and 1.038 ± 0.142 µm × 2.102 ± 0.210 µm, respectively (Figure 6). This indicated that both wild-type and mutant isolates showed no significant differences in cell surface and shape.
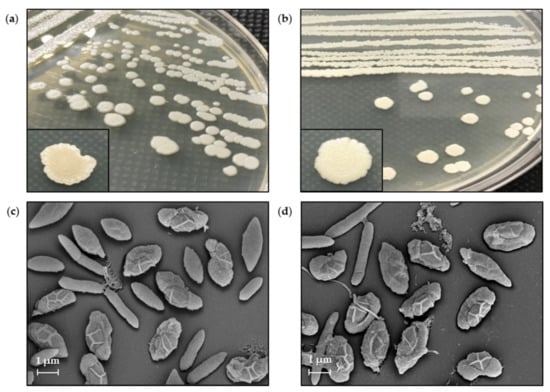
Figure 6.
Colony morphology of Brevibacillus sp. SPR19 of (a) wild-type and (b) M285 strains. The SEM micrograph of vegetative cells and spores of (c) wild-type and (d) M285 strains at magnitude 10,000×. The vegetative cells and spores were obtained by cell incubation on MH agar at 30 °C for 8 d.
3.8. Comparison of Growth Curve and Kinetics of Antibacterial Production
Typically, mutagenesis increases the overall antibacterial activity by promoting bacterial growth. Nevertheless, this result showed that the wild-type and M285 strains showed a similar growth curve pattern and production kinetics of antibacterial substances against S. aureus TISTR 517 and MRSA isolates 2468. The activity of both strains started at 16 h of incubation and reached a maximum activity at 24 h, which was related to the early stationary phase. M285 still produced a higher activity than the wild-type strain. After 48 h of incubation, the antibacterial activities of both strains gradually declined until 144 h (Figure 7). Taken together, ARTP treatment can increase the generation of reactive chemical species and cause genotoxicity, leading to the higher production of the same active substances. Alternatively, it could activate cryptic biosynthetic gene clusters of other antibacterial compounds that are generally not expressed [43]. Consequently, the mutant strains may produce multiple active compounds, resulting in a larger inhibition zone for M285.
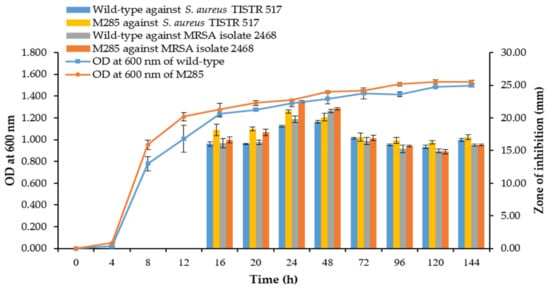
Figure 7.
Growth curve and production kinetics of antibacterial substances of the wild-type and M285 strains.
3.9. Determination of MIC and MBC
The MICs and MBCs of the active substances were directly related to their activities. M285 showed a MIC of 160.00 AU/mL, which was a two-fold higher antibacterial activity when compared to that of the wild-type strain (80.00 AU/mL). The MBC of M285 also exhibited a higher potency in the range of 40.00–160.00 AU/mL, except for the MRSA isolate 142 (Table 3). These results indicated that M285 exhibited enhanced antibacterial activity due to plasma mutagenesis.

Table 3.
Susceptibility testing of S. aureus TISTR 517 and its resistant strains (MRSA) to antibacterial substances of Brevibacillus sp. SPR19 and M285 (mean ± SD; n = 3).
3.10. Stability of the Antibacterial Substances
It was shown that the antibacterial activity of CFS for both wild-type and M285 variant had significantly decreased after being exposed to high temperatures. Interestingly, the lowest activity was found at 100 °C for 1 h condition. At this temperature, the wild-type retained 91.13 ± 0.20% and 88.21 ± 0.77% activity, and M285 retained 90.61 ± 1.63% and 91.18 ± 3.23% activity against S. aureus TISTR 517 and MRSA isolate 2468, respectively (Table 4). The activity after treatment with proteolytic enzymes individually decreased toward the indicator pathogens. The active substances of M285 were significantly sensitive to proteinase K (92.13 ± 1.60%), trypsin (95.83 ± 1.39%), and α-chymotrypsin (93.98 ± 2.12%) for the inhibition of S. aureus TISTR 517. It indicated that the antibacterial substances were peptides or proteins. It was consistent with several previous studies, showing Brevibacillus sp. mostly produced antimicrobial peptides through ribosomal and non-ribosomal syntheses [44]. In addition, the activity of both the strains increased in the presence of Triton X-100 and SDS. A 1% concentration of SDS alone had antibacterial activity against all indicators, while 1% triton X-100 showed no effect. This result supported the stability of the active substance when combined under surfactant conditions. In addition, the antibacterial activity of the wild-type and M285 strains was stable over a pH range of 1–14, except that the activity of the wild-type strain was considerably reduced at a pH of more than 9. However, the antibacterial substances from wild-type and M285 were stable to several conditions (heat, enzyme, surfactant, and pH). The stability profile of antibacterial substances from the wild-type and M285 strains were consistent with a previous study of antimicrobial peptides from Bacillus paralicheniformis in which the activity was tolerated in the treatments of high temperature (below 100 °C) and wide range of pH (2–9). Nevertheless, the activity of that strain was attenuated more than 20% when treated with higher temperature and beyond that pH range. The activity of antimicrobial peptides from B. paralicheniformis was also reduced as it was degraded in the presence of proteolytic enzymes such as proteinase K, trypsin, and α-chymotrypsin. In addition, the wild-type and M285 also showed the enhanced antibacterial activity when combined with Triton X-100, whereas the activity from B. paralicheniformis was decreased [45]. These results showed that M285 strain could produce anti-S. aureus and anti-MRSA compounds with proteinaceous characteristics, and it exhibited a stable and broad spectrum of antibacterial activity.

Table 4.
Stability study of antibacterial substances of the wild-type and M285 strains. The percent of the remaining activity was calculated, and expressed as mean ± SD (n = 3).
4. Conclusions
Brevibacillus sp. SPR19, isolated from a soil sample, displayed potent antibacterial activity against indicator pathogens (S. aureus and MRSA). The maximum activity was observed after 24 h of cell culture. Interestingly, the ARTP technique increased the lethality of SPR19 by more than 95% within 30 s, and M285 exhibited enhanced antibacterial activity against the above indicators compared to the wild-type strain. The active compounds from M285 were more stable under various stress conditions, including temperature, proteolytic enzymes, surfactants, and pH. Both wild-type and M285 isolates proved to be stable strains, and there was no difference in cell morphology and growth. M285 was more sensitive to some antibiotics than the wild-type strain. ARTP is a powerful method for strain improvement. Identification and characterization of bioactive substances from Brevibacillus sp. SPR19 and its mutant strains are necessary for future study to elucidate its structure and function that may develop the potential antibacterial substances against antibiotic-resistant infections. However, there are some limitations in the study that require further investigation of whole genome sequencing to define the nucleotide alterations by plasma mutagenesis. It will provide an insight to monitor subsequent target modification of its genes, regulating the production of the same active substances or cryptic biosynthetic gene clusters of other antibacterial compounds.
Author Contributions
Conceptualization, N.S., M.N., A.A., T.W. and T.C.; methodology, N.S., M.N., A.A., T.W. and T.C.; investigation, N.S., M.N., A.A., T.W. and T.C.; writing—original draft preparation, N.S., M.N., A.A., T.W. and T.C.; writing—review and editing, N.S., M.N., A.A., T.W. and T.C.; supervision, T.C.; project administration, T.C.; and funding acquisition, T.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Walailak University, grant number WU-IRG-63-039.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article.
Acknowledgments
The work was approved by Institutional Biosafety Committee (WU-IBC-63-008; 20 April 2019). In addition, we would like to thank the Center of Scientific and Technological Equipments, Walailak University, for its research facilities.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kotra, L.P. xPharm: The Comprehensive Pharmacology Reference Infectious Diseases; Elsevier: Amsterdam, The Netherlands, 2007; pp. 1–2. [Google Scholar] [CrossRef]
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, present and future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef] [PubMed]
- McGuinness, W.A.; Malachowa, N.; DeLeo, F.R. Vancomycin resistance in Staphylococcus aureus. Yale J. Biol. Med. 2017, 90, 269–281. [Google Scholar] [PubMed]
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States. 2019. Available online: https://stacks.cdc.gov/view/cdc/82532/ (accessed on 19 February 2022).
- Ventola, C.L. The Antibiotic Resistance Crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Da Cunha, B.R.; Fonseca, L.P.; Calado, C.R.C. Antibiotic Discovery: Where Have We Come from, Where Do We Go? Antibiotics 2019, 8, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Procópio, R.E.D.L.; da Silva, I.R.; Martins, M.K.; de Azevedo, J.L.; de Araújo, J.M. Antibiotics produced by Streptomyces. Braz. J. Infect. Dis. 2012, 16, 466–471. [Google Scholar] [CrossRef] [Green Version]
- Sumi, C.D.; Yang, B.W.; Yeo, I.-C.; Hahm, Y.T. Antimicrobial peptides of the genus Bacillus: A new era for antibiotics. Can. J. Microbiol. 2015, 61, 93–103. [Google Scholar] [CrossRef]
- Okafor, N. Screening for productive strains and strain improvement in biotechnological organisms. In Modern Industrial Microbiology and Biotechnology Strain Improvement; Science Publishers: Enfield, NH, USA, 2007; Volume 3, pp. 125–170. [Google Scholar]
- Ottenheim, C.; Nawrath, M.; Wu, J.C. Microbial mutagenesis by atmospheric and room-temperature plasma (ARTP): The latest development. Bioresour. Bioprocess. 2018, 5, 12. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.-Z.; Zhang, W.-G. Menaquinone-7 production from maize meal hydrolysate by Bacillus isolates with diphenylamine and analogue resistance. J. Zhejiang Univ. Sci. B 2017, 18, 462–473. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Xu, Q.; Jiang, L.; Huang, H.; Li, S. Polydiacetylene-Based High-Throughput Screen for Surfactin Producing Strains of Bacillus subtilis. PLoS ONE 2014, 9, e88207. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Li, H.-P.; Wang, L.-Y.; Wang, S.; Zhao, H.-X.; Sun, W.-T.; Xing, X.-H.; Bao, C.-Y. Genetic effects of radio-frequency, atmospheric-pressure glow discharges with helium. Appl. Phys. Lett. 2008, 92, 221504. [Google Scholar] [CrossRef] [Green Version]
- Arjunan, K.P.; Sharma, V.K.; Ptasinska, S. Effects of Atmospheric Pressure Plasmas on Isolated and Cellular DNA—A Review. Int. J. Mol. Sci. 2015, 16, 2971–3016. [Google Scholar] [CrossRef] [Green Version]
- Jiang, H.; Zou, J.; Cheng, H.; Fang, J.; Huang, G. Purification, Characterization, and Mode of Action of Pentocin JL-1, a Novel Bacteriocin Isolated from Lactobacillus pentosus, against Drug-Resistant Staphylococcus aureus. BioMed Res. Int. 2017, 2017, 7657190. [Google Scholar] [CrossRef] [Green Version]
- Tripathi, N.; Sapra, A. Gram Staining, in StatPearls Gram Staining; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Ashby, G.K. Simplified Schaeffer spore stain. Science 1938, 87, 443. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, Y.; Yang, H.; Bu, Y.; Yi, H.; Zhang, L.; Han, X.; Ai, L. Purification and Partial Characterization of Bacteriocin Lac-B23, a Novel Bacteriocin Production by Lactobacillus plantarum J23, Isolated From Chinese Traditional Fermented Milk. Front. Microbiol. 2018, 9, 2165. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Goh, H.F.; Philip, K. Purification and Characterization of Bacteriocin Produced by Weissella confusa A3 of Dairy Origin. PLoS ONE 2015, 10, e0140434. [Google Scholar] [CrossRef]
- Liu, K.; Fang, H.; Cui, F.; Nyabako, B.A.; Tao, T.; Zan, X.; Chen, H.; Sun, W. ARTP mutation and adaptive laboratory evolution improve probiotic performance of Bacillus coagulans. Appl. Microbiol. Biotechnol. 2020, 104, 6363–6373. [Google Scholar] [CrossRef]
- Halder, D.; Mandal, M.; Chatterjee, S.S.; Pal, N.K.; Mandal, S. Indigenous Probiotic Lactobacillus Isolates Presenting Antibiotic like Activity against Human Pathogenic Bacteria. Biomedicines 2017, 5, 31. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Huang, Z.; Li, G.; Zhao, H.; Xing, X.; Sun, W.; Li, H.; Gou, Z.; Bao, C. Novel mutation breeding method for Streptomyces avermitilis using an atmospheric pressure glow discharge plasma. J. Appl. Microbiol. 2010, 108, 851–858. [Google Scholar] [CrossRef]
- Bauer, A.W.; Kirby, W.M.; Sherris, J.C.; Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966, 45, 493–496. [Google Scholar] [CrossRef]
- Daba, H.; Pandian, S.; Gosselin, J.F.; Simard, R.E.; Huang, J.; Lacroix, C. Detection and activity of a bacteriocin produced by Leuconostoc mesenteroides. Appl. Environ. Microbiol. 1991, 57, 3450–3455. [Google Scholar] [CrossRef] [Green Version]
- Chalasani, A.G.; Dhanarajan, G.; Nema, S.; Sen, R.; Roy, U. An Antimicrobial Metabolite from Bacillus sp.: Significant Activity Against Pathogenic Bacteria Including Multidrug-Resistant Clinical Strains. Front. Microbiol. 2015, 6, 1335. [Google Scholar] [CrossRef] [Green Version]
- Song, J.; Wang, Y.; Song, Y.; Zhao, B.; Wang, H.; Zhou, S.; Kong, D.; Guo, X.; Li, Y.; He, M.; et al. Brevibacillus halotolerans sp. nov., isolated from saline soil of a paddy field. Int. J. Syst. Evol. Microbiol. 2017, 67, 772–777. [Google Scholar] [CrossRef]
- Adrio, J.L.; Demain, A.L. Genetic improvement of processes yielding microbial products. FEMS Microbiol. Rev. 2006, 30, 187–214. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, X.-F.; Li, H.-P.; Wang, L.-Y.; Zhang, C.; Xing, X.-H.; Bao, C.-Y. Atmospheric and room temperature plasma (ARTP) as a new powerful mutagenesis tool. Appl. Microbiol. Biotechnol. 2014, 98, 5387–5396. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, C.; Zhou, Q.-Q.; Zhang, X.-F.; Wang, L.-Y.; Chang, H.-B.; Li, H.-P.; Oda, Y.; Xing, X.-H. Quantitative evaluation of DNA damage and mutation rate by atmospheric and room-temperature plasma (ARTP) and conventional mutagenesis. Appl. Microbiol. Biotechnol. 2015, 99, 5639–5646. [Google Scholar] [CrossRef]
- Wende, K.; Bekeschus, S.; Schmidt, A.; Jatsch, L.; Hasse, S.; Weltmann, K.; Masur, K.; von Woedtke, T. Risk assessment of a cold argon plasma jet in respect to its mutagenicity. Mutat. Res. Toxicol. Environ. Mutagen. 2016, 798–799, 48–54. [Google Scholar] [CrossRef]
- Adhikari, B.C.; Lamichhane, P.; Lim, J.S.; Nguyen, L.N.; Choi, E.H. Generation of reactive species by naturally sucked air in the Ar plasma jet. Results Phys. 2021, 30, 104863. [Google Scholar] [CrossRef]
- Oehmigen, K.; Hähnel, M.; Brandenburg, R.; Wilke, C.; Weltmann, K.-D.; Von Woedtke, T. The Role of Acidification for Antimicrobial Activity of Atmospheric Pressure Plasma in Liquids. Plasma Process. Polym. 2010, 7, 250–257. [Google Scholar] [CrossRef]
- Bolouki, N.; Kuan, W.-H.; Huang, Y.-Y.; Hsieh, J.-H. Characterizations of a Plasma-Water System Generated by Repetitive Microsecond Pulsed Discharge with Air, Nitrogen, Oxygen, and Argon Gases Species. Appl. Sci. 2021, 11, 6158. [Google Scholar] [CrossRef]
- Ghimire, B.; Szili, E.J.; Patenall, B.L.; Lamichhane, P.; Gaur, N.; Robson, A.J.; Trivedi, D.; Thet, N.T.; Jenkins, A.T.A.; Choi, E.H.; et al. Enhancement of hydrogen peroxide production from an atmospheric pressure argon plasma jet and implications to the antibacterial activity of plasma activated water. Plasma Sources Sci. Technol. 2021, 30, 035009. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, H.; He, D.; Wu, Y.; Jin, L.; Li, G.; Su, N.; Li, H.; Xing, X.-H. Insights into the molecular-level effects of atmospheric and room-temperature plasma on mononucleotides and single-stranded homo- and hetero-oligonucleotides. Sci. Rep. 2020, 10, 14298. [Google Scholar] [CrossRef] [PubMed]
- Dizdaroglu, M.; Jaruga, P. Mechanisms of free radical-induced damage to DNA. Free Radic. Res. 2012, 46, 382–419. [Google Scholar] [CrossRef]
- Chatterjee, N.; Walker, G.C. Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 2017, 58, 235–263. [Google Scholar] [CrossRef] [Green Version]
- Cao, X.; Luo, Z.; Zeng, W.; Xu, S.; Zhao, L.; Zhou, J. Enhanced avermectin production by Streptomyces avermitilis ATCC 31267 using high-throughput screening aided by fluorescence-activated cell sorting. Appl. Microbiol. Biotechnol. 2018, 102, 703–712. [Google Scholar] [CrossRef]
- Zhang, C.; Qin, J.; Dai, Y.; Mu, W.; Zhang, T. Atmospheric and room temperature plasma (ARTP) mutagenesis enables xylitol over-production with yeast Candida tropicalis. J. Biotechnol. 2019, 296, 7–13. [Google Scholar] [CrossRef]
- Mitra, S.; Nguyen, L.N.; Akter, M.; Park, G.; Choi, E.H.; Kaushik, N.K. Impact of ROS Generated by Chemical, Physical, and Plasma Techniques on Cancer Attenuation. Cancers 2019, 11, 1030. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Huang, Z.; Gui, X.; Xiang, W.; Jin, Y.; Chen, J.; Zhao, J. Multi-omics Comparative Analysis of Streptomyces Mutants Obtained by Iterative Atmosphere and Room-Temperature Plasma Mutagenesis. Front. Microbiol. 2021, 11, 630309. [Google Scholar] [CrossRef]
- Yang, X.; Yousef, A.E. Antimicrobial peptides produced by Brevibacillus spp.: Structure, classification and bioactivity: A mini review. World J. Microbiol. Biotechnol. 2018, 34, 57. [Google Scholar] [CrossRef]
- Choyam, S.; Jain, P.M.; Kammara, R. Characterization of a Potent New-Generation Antimicrobial Peptide of Bacillus. Front. Microbiol. 2021, 12, 710741. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).