Abstract
The emergence of multidrug-resistant Mycobacterium tuberculosis (MTB) has become a major problem in treating tuberculosis (TB) and shows the need to develop new and efficient drugs for better TB control. This study aimed to use in silico techniques to discover potential inhibitors to the Enoyl-[acyl-carrier-protein] reductase (InhA), which controls mycobacterial cell wall construction. Initially, 391 quercetin analogs present in the KNApSAck_3D database were selected, filters were sequentially applied by docking-based virtual screening. After recategorizing the variables (bond energy prediction and molecular interaction, including hydrogen bond and hydrophobic bond), compounds C00013874, C00006532, and C00013887 were selected as hit ligands. These compounds showed great hydrophobic contributions, and for each hit ligand, 100 ns of molecular dynamic simulations were performed, and the binding free energy was calculated. C00013874 demonstrated the greatest capacity for the InhA enzyme inhibition with ΔGbind = −148.651 kcal/mol compare to NAD (native ligand) presented a ΔGbind = −87.570 kcal/mol. These data are preliminary studies and might be a suitable candidate for further experimental analysis.
1. Introduction
Mycobacterium tuberculosis (MTB) causes an infectious disease leading to death in adults worldwide. It has been estimated that one-third of the world’s population is infected with MTB, and 10% of these people will develop active tuberculosis (TB) during their lifetime. Multiple drug combination therapy has generally been applied in TB treatment to prevent the development of drug resistance. However, the inappropriate and long-term treatment allowed the emergence of multidrug-resistant tuberculosis (MDR-TB) and extensively drug-resistant TB (XDR-TB). Treatment for MDR-TB and XDR-TB is lengthy, expensive, and characterized by a high adverse event rate, complicating TB control [1]. Thus, there is an urgent need to discover new anti-TB agents effective in TB treatment capable of sterilizing drug-resistant mycobacteria.
Isoniazid (INH), which is known as front-line anti-TB drugs, has been identified to target the Enoyl-[acyl-carrier-protein] reductase (InhA) protein encoded by inhA [2]. InhA is an enzyme responsible for producing long-chain fatty acid derivatives that are crucial precursors to mycolic acids and are involved in the biogenesis of MTB’s cell wall and physiological functions [3]. The enzyme catalyzes the NADH-dependent reduction of the double bond of 2-trans-enoyl-acyl carrier protein, an essential step in the type II fatty acid synthase (FAS-II) system [4,5]. INH is a prodrug which requires activation, carried out by the enzyme catalase-peroxidase of MTB. Nicotinic acid, an active form of INH, will inhibit the InhA through a covalently bonding to NADH within the protein’s active site forming nicotinamide adenine dinucleotide (NAD). Furthermore, the FAS-II system in MTB is significantly different from the mammalian fatty acid synthesis pathway makes the InhA enzyme an attractive target for drug discovery [6].
Quercetin, 3, 3′,4′,5,7-pentahydroxyflvanone, is a flavonoid widely found in fruits and vegetables and has been reported to have various pharmacological properties [7]. This flavonoid has been shown to affect glutathione enrichment, cytokine regulation, activation of detoxification processes, and a proteasome inhibitor [8,9]. Extensive studies have been done on quercetin against MTB, but quercetin’s effect on the mycobacterial cell wall and detail molecular mechanism is yet to be thoroughly understood. Quercetin was observed to inhibit 99.30 ± 0.268% against MTB H37Rv in luciferase reporter phage assay at the concentration of 200 µg/mL [10]. In silico study demonstrated that this phenolic compound was reported to inhibited MTB glutamate racemase (Murl) that is involved in cell wall peptidoglycan (PG) biosynthesis [11]. In addition, Premalatha et al., (2020) revealed that four quercetin analogs have an excellent binding with panthothenate synthase (PanC), which is necessary for the production of pantothenate (vitamin B5), critical components of fatty acid synthesis of MTB based on the molecular docking analysis [12]. Based on these facts, quercetin has the potential to study the development of anti-TB drugs further. Thus, comprehensive computational approaches are needed against the large set of quercetin analogs to develop novel anti-TB drugs.
This study aimed to identify the novel and potential lead molecules as an inhibitor of InhA, which belongs to MTB’s FAS-II system. Together, the high-throughput docking virtual screening (VS) and dynamic simulation study were performed to quercetin analogs as a promising lead molecule in the InhA inhibitor development.
2. Materials and Methods
2.1. Protein Structure Preparation
We selected the Enoyl-[acyl-carrier-protein] reductase (InhA) crystallographic structure from the Protein Data Bank (PDB ID: 1BVR) based on our previous study [13]. This study determined the protein target structure based on high resolution and bound inhibitor (nicotinamide adenine dinucleotide/NAD as a native ligand). The crystallography structure was cleaned to ensure maximum quality and reliability. The bound ligand, water molecules, and chain B were removed. Polar hydrogen atoms and a Kollman charge were added using methods implemented in Auto Dock Tools v.1.5.6. [14].
2.2. Preparation of Small Molecule Library
Docking-based virtual screening was carried out with the clean lead compounds from the KNApSAck_3D database (http://knapsack3d.sakura.ne.jp/, accessed on 4 November 2020). Three hundred and ninety-one compounds of quercetin analogs were selected and further considered for molecular docking analysis. All ligand structures were prepared by calculating the gasteiger charges, and the hydrogen atoms were added using the DockPrep module implemented at Auto Dock Tools v.1.5.6. The bound inhibitor, NAD, was considered an InhA inhibitor and taken as a standard compound.
2.3. Docking-Based Virtual Screening
The molecular docking was performed using iDock software [15] to determine the protein-ligand interaction’s binding affinity using the Lamarckian genetic algorithm with a population size of 100 individuals. Gasteiger charges were computed, and all the charges of nonpolar hydrogen were assigned [16]. The docking parameters were validated by redocking the native ligand to the protein target, and the validated parameters (root mean square deviation (RMSD) < 2 Å) were used for docking all selected quercetin analogs. The inhibitor binding site, the same as the quercetin analog’s binding site, was used by calculating the cubic docking box of x, y, and z coordinated. The grid map was set at 46 × 40 × 40 Å, and the grid was spaced at 0.375 Å. All the 391 compounds were docked to the inhibitor-binding site, and the different binding affinities were calculated in kilocalorie per mole.
2.4. Analysis of the Protein–Ligand Complex
The protein–ligand complex’s visualization and analysis were performed using Pymol (1.7.4.5 Edu), Molecular Graphics System, and Discovery Studio Visualizer (D.S.V.) v16.1.0.153500 software. The hydrogen bonding, polar, and hydrophobic interaction between the protein and ligands were analyzed and visualized in the same software mentioned above.
2.5. Molecular Dynamic (MD) Simulation
The molecular dynamic simulation of the protein–ligand complex was carried out using Gromacs 2016.3 software [17]. Topology and ligand parameters were build using the antechamber python parser interface (ACPYPE) tools [18]. AMBER99SB-ILDN force fields were assigned to both ligand and protein. The distance electrostatic force was determined by the Ewald Particle Mesh method [19]. The neutralization system was carried out by adding Na+ and Cl- ions, and solvation was carried out using the TIP3P water cube model. The simulation process included the minimization for 10,000 steps using the steepest descent method (5000 steps) and conjugate gradients method (5000 steps). The whole system was heated up for 100 ps with the initial and final temperatures of 0 and 310 K using Langevin dynamic temperature regulation. Lastly, the simulation’s production was performed at both constant temperature and pressure of 310 K and 1 atm, respectively, using the timestep of 2 fs. All hydrogen bonds were constrained with the SHAKE algorithm, and 100 ns (100,000 ps)-long simulation was produced by saving the coordinate trajectories at every 20 ps. The MD simulation result was analyzed based on the RMSD, root mean square fluctuation (RMSF), radius gyration, solvent accessible surface area (SASA), and principal component analysis (PCA).
2.6. Binding Free Energy (MM-PBSA) Analysis
The Molecular Mechanics-Poisson-Boltzmann Surface Area (MM/PBSA) calculations were performed using the g_mmpbsa package [20] integrated with the Gromacs 2016.3 software. The energy was calculated based on the average of 500 snapshots extracted at every ten ps from the MD trajectory between 45 and 50 ns. The polar desolvation energy was calculated using the Poisson–Boltzmann equation with a grid size of 0.5 Å. The dielectric constant of the solvent is set at 80 to represent water as the solvent [21]. The nonpolar contribution calculation was done by calculating the surface area accessed by the solvent with a solvent radius of 1.4 Å [22]. The analysis of protein–ligand free energy was carried out based on complex molecular dynamics simulation results (when the complex interactions were stable).
3. Results
3.1. Docking-Based Virtual Screening
The aim of docking native ligand (NAD) to protein target (InhA) was to validate the software before the targeted molecules were docked to the protein target. We used RMSD as a validation parameter. Ideally, a computational method’s quality of reproduction of binding pose is suitable when the RMSD value less than 2.0 Å [23]. Based on the result, the protein target’s active site had the coordinates of center x = 19,579, center y = 9067, center z = 14,345 as the binding site of the native ligand. The box size was 46 × 40 × 40 Å with a point spacing of 0.375 Å. This grid box parameter for InhA had RMSD 1.849 Å after overlapped the X-ray conformation of NAD and the best conformation resulting from the redocking. The native ligand had a binding affinity of −9.81 kcal/mol and was found to bind to the active site of InhA by interacting with the crucial amino acid Thr196 residue (Figure 1a) [24].
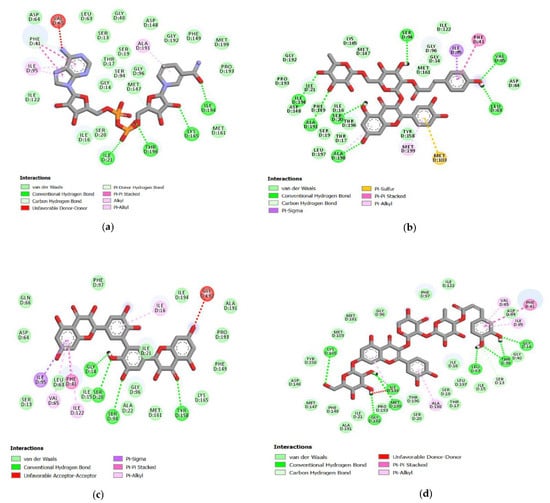
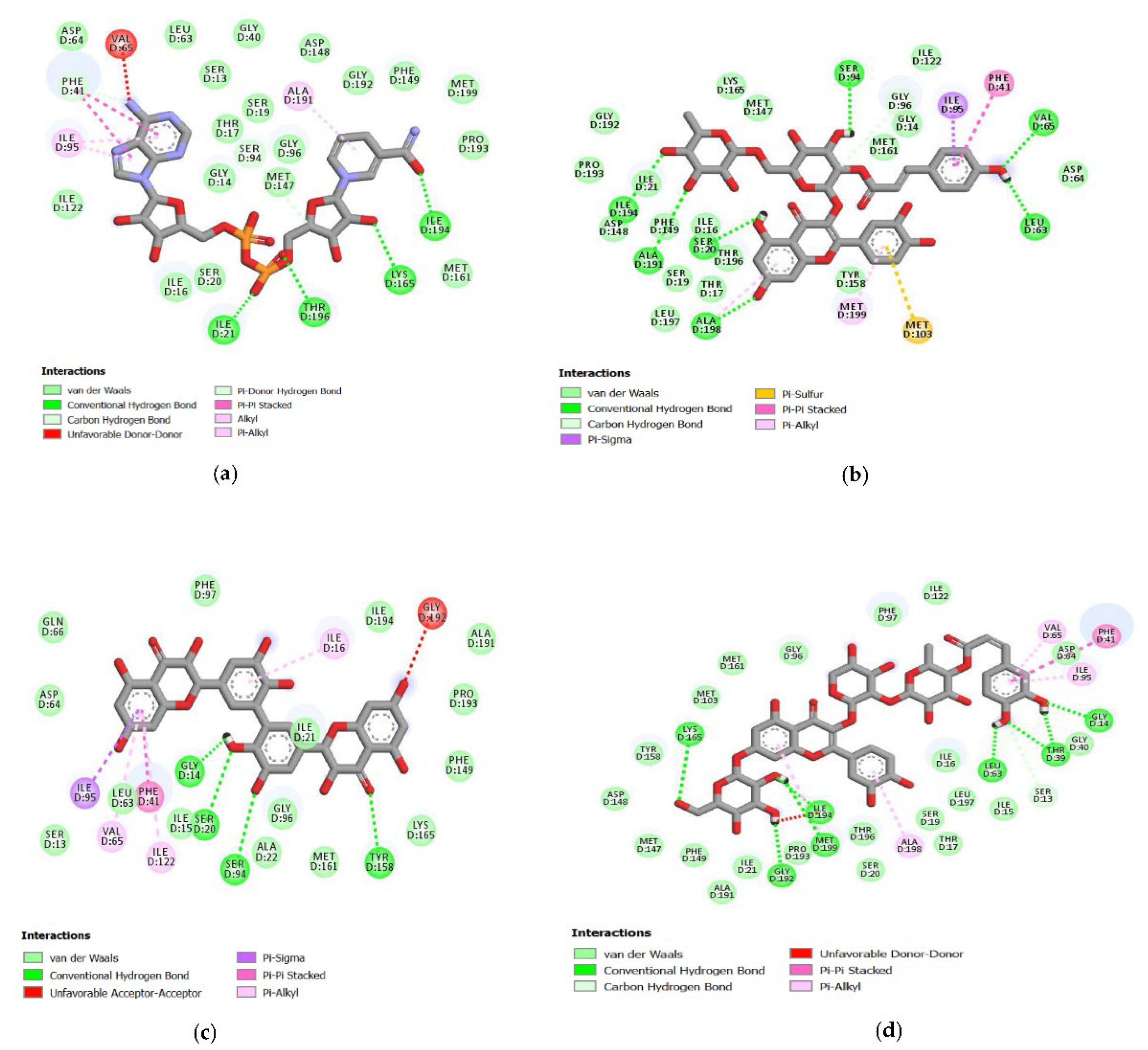
Figure 1.
Molecular interaction between ligand and protein target InhA. (a) NAD; (b) C00013874; (c) C00006532; (d) C00013887.
Molecular docking of 391 quercetin analogs has a binding affinity range from −6.73 to −11.09 kcal/mol. This binding affinity showed the binding stability between the ligand or molecules target and receptor. The best pose results from the NAD compound’s docking were hydrogen bonds in the carbonyl and oxygen groups attached to the phosphate atoms against the amino acids Thr196 and Ile21. In the hydroxy group of tetrahydro-furans, there was also a hydrogen bond with the amino acid Lys165 and the hydroxyl group from the pyridine ring with amino acid Ile194. In the best pose, there was a hydrophobic interaction in the purine and pyridine rings with the amino acids Phe41 and Ala191 (Figure 1a).
Sixteen quercetin analogs had a lower binding affinity than NAD and bound to InhA’s active site with the C00006532 had the lowest binding energy value of −11.09 kcal/mol. This quercetin analog formed four hydrogen bonds with the binding site of InhA. The number 2 carbon atom of the hydroxy group in the benzene ring interacted with Ser20 and Gly14 amino acid residues. The number one carbon atom in this compound’s benzene ring interacted with the amino acid Ser94. Hydrogen bonds of C00006532 were also observed with the amino acid Tyr158. Hydrophobic interactions were formed on the chroman ring’s benzene group between hit ligands and the benzene group of InhA in Phe41 (Figure 1c).
The compound C00013874 has a binding affinity value of −10.95 kcal/mol in the docking result analysis. Further analysis related to its interaction with InhA showed hydrogen bonds in the hydroxyl groups of phenol Leu63 and Val65 amino acid residues. Two chroman ring’s hydroxy groups formed hydrogen bonds with Ser20 and Ala198 amino acid residues. In the tetrahydro-pyran ring, hydrogen bonds were observed by hydroxyl groups with Ala191 and Ile194 amino acid residues. The methyl group also established hydrophobic interactions on the tetrahydro-pyran ring of Ile194 and Ile21 and the benzene ring from phenol with Phe41 amino acid residues (Figure 1b).
The molecular docking result showed hydrogen bonds in the hydroxy group of the benzene ring compound C00013887 with Gly14, Thr39, and Leu63 amino acid residues and the hydroxyl groups on the chroman ring with Met199, Ile194, Gly192, and Lys165 amino acid residues of InhA. It was also observed that there was a hydrophobic interaction with the Phe41 residues of InhA. The binding affinity between C00013887 and InhA was −10.63 kcal/mol (Figure 1d).
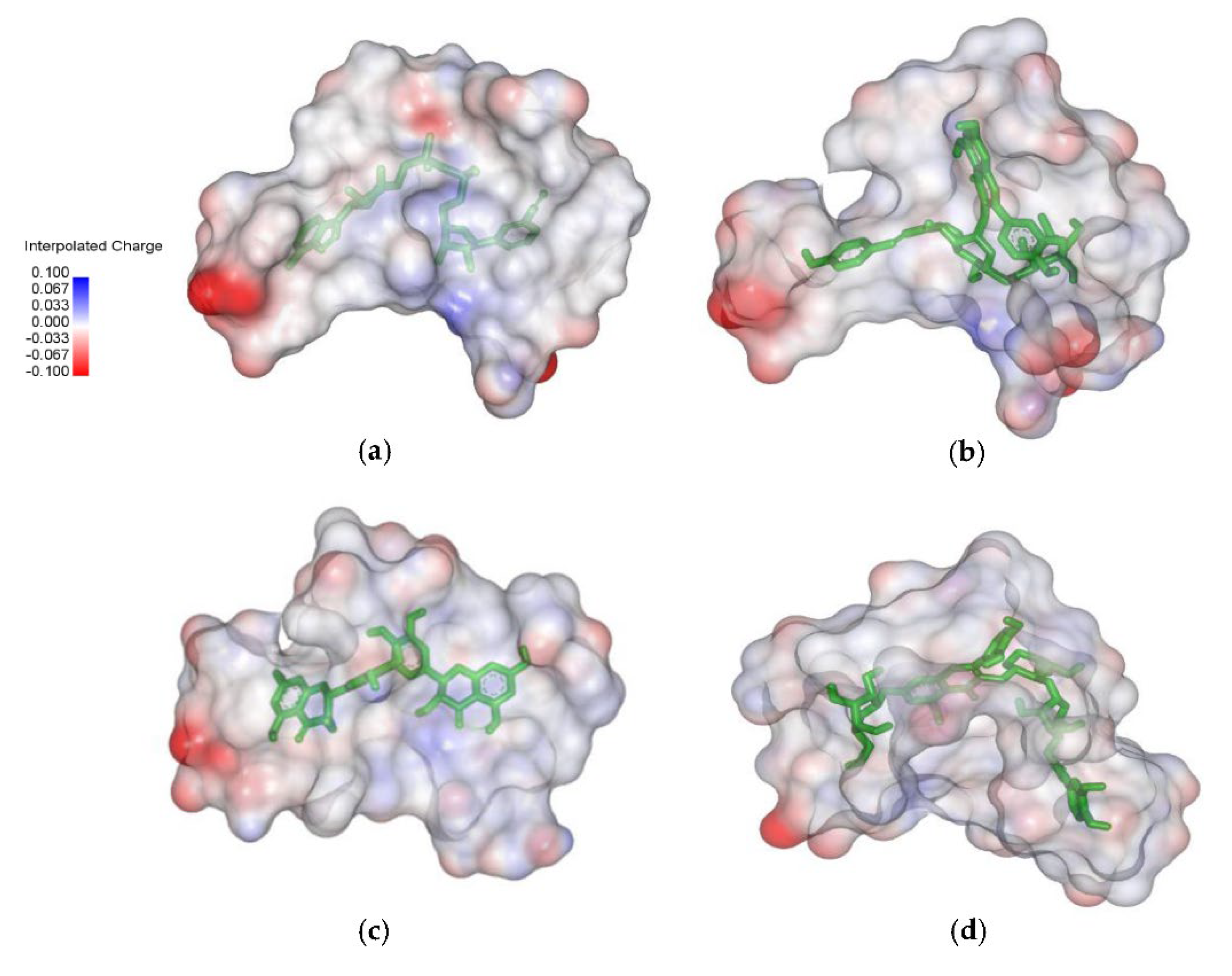
Molecular electrostatic potential (MESP) projection map for native ligand (NAD) and hit ligands (C00013874, C00006532, and C00013887) was also generated using Discovery Studio Visualizer (D.S.V.) v16.1.0.153500 software. MESP provides a three-dimensional visual method to understand the net electrostatic effect of the molecule. It correlates with the partial charges, electronegativity, and chemical reactivity of the molecules [25]. The MESP figure showed that electron-rich hydroxyl groups make native ligand and hit ligands as hydrogen bond donors (negative electrostatic potential showed red color) (Figure 2).
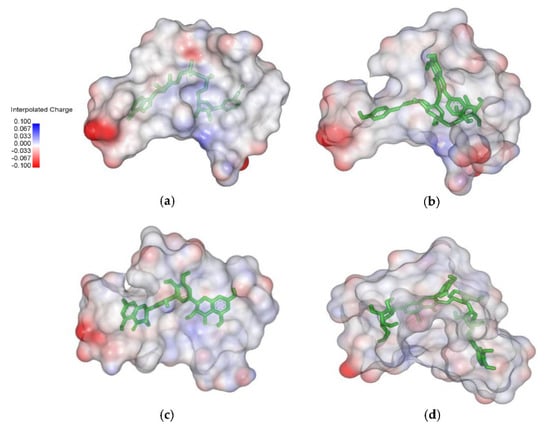
Figure 2.
Molecular electrostatic potential maps between ligand and protein target InhA. (a) NAD; (b) C00013874; (c) C00006532; (d) C00013887.
3.2. Structural Analysis of Systems
3.2.1. Root-Mean-Square Deviation (RMSD) and Root-Mean-Square Fluctuation (RMSF)
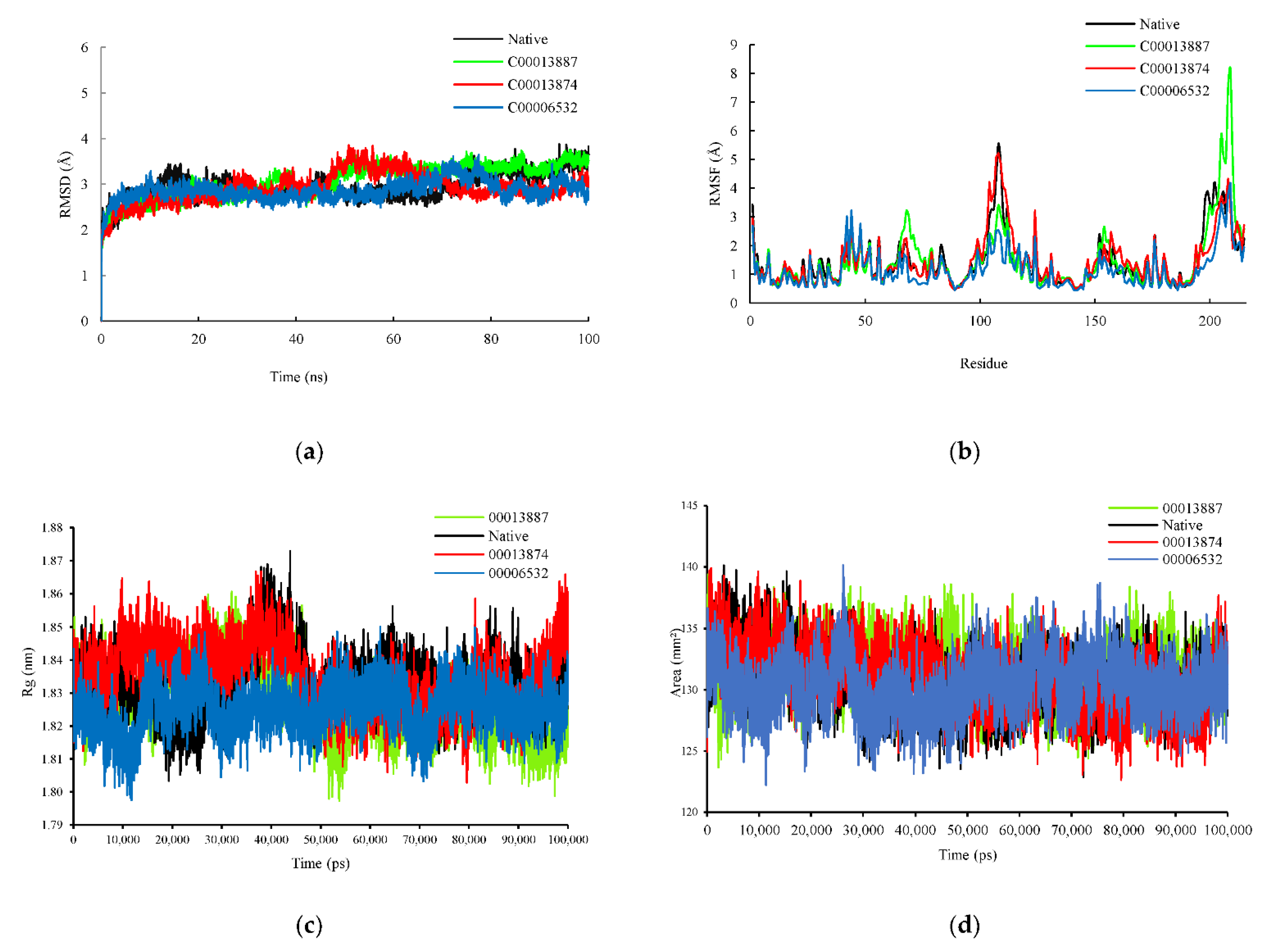
The RMSD plot was analyzed to identify the conformational and structural changes of InhA and ligands (NAD, C00013887, C00013874, C00006532) complexes through 100 ns MD simulation. In the plot in 0–45 ns simulation, all complexes had shown similar fluctuation. Afterward, above 45–70 ns, InhA-C00013887 and InhA-C00013874 complexes showed increased change than the InhA-NAD complex, whereas the InhA-C00006532 complex showed instability, but still similar to the InhA-NAD complex. The InhA-C00013874 and InhA-C00006532 complexes showed lower fluctuations than the InhA-NAD complex observed at the end of the simulation (71–100 ns). However, the InhA-C00013887 complex showed increased instability following the InhA-NAD complex. On average, the RMSD values of InhA-NAD, InhA-C00013887, InhA-C00013874, and InhA-C00006532 complexes were 2.950 Ǻ, 3.070 Ǻ, 2.913 Ǻ, and 2.861 Ǻ, respectively. This result suggests that the average RMSD value between hit ligands and NAD had a similar value (Figure 3a).
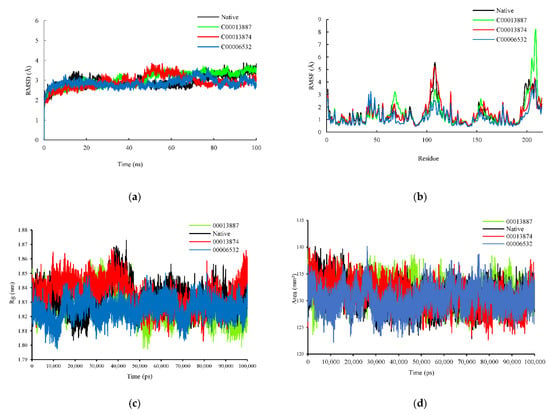
Figure 3.
Evaluation of (a) RMSD, (b) RMSF, (c) radius gyration, and (d) SASA plot plots during MD simulations. The complex of protein-native ligand (NAD) and protein-hit ligands have been represented in different colors. NAD (black), C00013887 (green), C00013874 (red), C00006532 (blue).
The RMSF plots revealed differences in protein flexibility throughout the trajectory. The RMSF plots indicated that each complex observed a similar fluctuation (Figure 3b), especially around the amino acids found on the active site of the InhA protein, such as Gly14, Ser20, Ile21, and Thr39 which previously detected to form a hydrogen bond with ligands. A similar fluctuation feature was shown in the amino acid residues on the sequence 191–199, which are also located on the active site of InhA. However, the difference in fluctuation was also revealed in the amino acid residues which were not in direct contact with the ligands, such as amino acid sequence 68 (InhA-C00013887 complex), 108 (InhA-C00013887 and InhA-C00006532 complexes), 124 (InhA-C00013874), 205 and 209 (InhA-C00013887).
3.2.2. Radius Gyration (Rg) and Solvent Accessible Surface Area (SASA)
The radius gyration (Rg) measurement aims to identify the complex’s stability, whether the complex is stable in folded or unfolded form during the simulation. The Rg plot showed similar characteristics between the complex of protein-native ligand (InhA-NAD) and protein-hit ligands (InhA-C00013887, InhA-C00013874, and InhA-C00006532). The average Rg value for each ligand was 2 nm, 1.83 nm, 1.84 nm, and 1.83 nm. The Rg values of protein-hit ligands complexes showed a significant similarity and stable folded structure compared to the protein-native ligand complex (Figure 3c). Besides, solvent accessible surface area (SASA) was also analyzed to predict the extent to which protein conformation changes during the simulation that water molecules can access. The SASA plot showed similar results between the complex of protein-native ligand and protein-hit ligands. The average value of SASA were 130.85 mm2, 131.46 mm2, 131.01 mm2, 130.27 mm2, respectively for protein-native ligand (InhA-NAD) and protein-hit ligands (InhA-C00013887, InhA-C00013874, and InhA-C00006532) (Figure 3d).
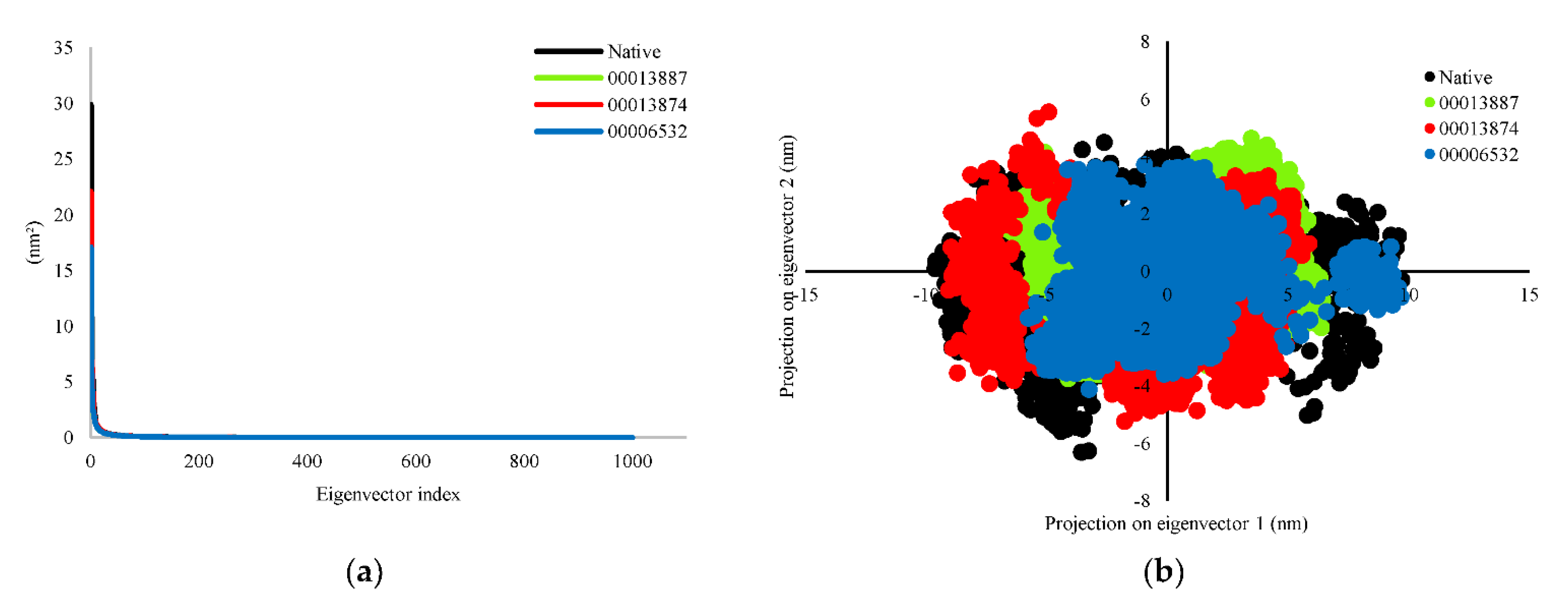
3.2.3. Principal Component Analysis (PCA)
Principal component analysis (PCA) was analyzed for significant fluctuations in protein-ligand complexes. The direction and amplitude of eigenvectors responsible for the motion were analyzed by creating a porcupine plot, which shows a graphical summary of the motion along the trajectories. In this study, all eigenvectors were selected to calculate the motion. The eigenvector showed protein-native ligand (InhA-NAD) complex and protein-hit ligands (InhA-C00013887, InhA-C00013874, and InhA-C00006532) complexes were 54.37%, 38.95%, 43.90%, and 36.41%, respectively for the first 20 ns simulation. The protein-hit ligands complexes formed a stable complex with less motion and showed similar motion than the InhA-NAD complex as a reference compound. We also analyzed the complex dynamics by the 2D projection of trajectory plot generation. The less space occupied by the clusters indicates a more stable complex, whereas those that occupy more space indicate a less stable protein-ligand complex. The protein-hit ligand complexes were found to occupy less space than the protein-native ligand complex. However, the patterns that are formed tend to be similar (Figure 4).
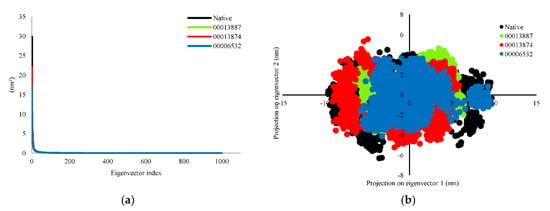
Figure 4.
Evaluation of (a) eigenvalues of the covariance matrix and (b) 2D projection of trajectory during MD simulations. The complex of protein-native ligand (NAD) and protein-hit ligands have been represented in different colors. NAD (black), C00013887 (green), C00013874 (red), C00006532 (blue).
3.3. Binding Free Energy InhA-Ligands
Calculation of MM-PBSA energy was performed to predict each energy’s contribution to the complex binding stability between ligands and protein target [26]. We calculated the MM-PBSA energy from 100 ns from molecular dynamics simulations. It can be found that the three best quercetin analogs had strong predicted binding affinity compared to NAD. The C00013874 had the lowest bond-free energy of −148,651 kJ/mol. The InhA- C00013874 complex had the best binding affinity and stability prediction. The bond-free energy of NAD (native ligand) was −87,570 kJ/mol. Moreover, the solvation polar energy was less favorable to the energy of the complex system. In contrast to this, van der Waals energy and electrostatic energy were very beneficial. They significantly affected the complex bond-free energy’s negative value during molecular dynamics simulations (Table 1).

Table 1.
MM-PBSA energy summary from nature ligand (NAD) and the three best quercetin analogs.
3.4. Hydrogen Bonds Established between Receptor-Ligands
The InhA-ligand complex was also analyzed by looking at the hydrogen bond’s percent occupancy during the simulation. Occupancy is defined as the percentage of time that hydrogen bonding existed during the 100 ns simulation time. The percent occupancy was screened, and only the value above 50% was taken as a reference for assessing the hydrogen bond. The NAD ligand was found to form 169 total hydrogen bonds, followed by the C00006532 with 164 hydrogen bonds, the C00013874 with 188 hydrogen bonds, and the C00013887 301 hydrogen bonds. The NAD showed hydrogen bonds to Gly95 amino acid residue as a donor (85.29%), Gly95 residue as an acceptor (77.45%), and strong hydrogen bonds to Gly13 (50.98%), Ile14 (56.86%), and Thr38 (53.92%) amino acid residues. The C00006532 was found to have hydrogen bonds with a vigorous intensity to the Leu62 (66.67%) and Ala197 (53.92%) amino acid residues. Furthermore, the C00013874 formed strong hydrogen bonds with Gly95 (90.20%), Gly13 (75.49%), Leu62 (98.04%), Ile193 (74.51%), Gly191 (62.75%), and Ser93 (67.65%) amino acid residues. The C00013887 showed strong hydrogen bonds to Leu62 (81.37%), Thr38 (55.88%), and Met97 (69.61%) amino acid residues.
4. Discussion
Virtual drug screening has been proved a successful tool to identify functional lead molecules from a library of compounds within a short period [27]. We performed virtual drug screening and dynamic simulation studies to identify potential lead compounds that may be used as possible TB drug candidates to improve TB treatment. It was noticed that 391 quercetin analogs were docked successfully to MTB-InhA. Three quercetin analogs ranked the highest affinity than the native ligand (NAD) as the most potent inhibitors. Using a computational approach, we demonstrated that three quercetin analogs’ binding to MTB-InhA influenced the protein’s conformation. In silico docking studies revealed that three quercetin analogs interact with critical residues suggesting that these compounds bind strongly at the substrate-binding site. The interacting residues involve Thr196, Ile194, and Tyr158. Thr196 residue in the binding loop between NAD and InhA is a critical amino acid that helps fix the active site’s NAD. The hydroxyl group of Ile194 residue is also responsible for hydrogen bonding between InhA and NAD. Moreover, the Tyr158 residue plays a crucial role in the interaction, and mutation to Ala94 causes MTB to become resistant to INH [24].
A 100 ns molecular dynamics simulation of the docked complexes was then carried out to evaluate the three quercetin analogs’ binding affinity and flexibility (C00013874, C00006532 C00013887) with the MTB-InhA. We observed that the binding patterns obtained after MD simulation were energetically stable throughout the simulation. The effect of ligand conformation changes on proteins during the MD simulation can be observed from the fluctuations in amino acids, represented in Figure 3. The highest fluctuation intensity was seen in the amino acid chain responsible for the loop and C-terminal regions of InhA. The three quercetin analogs showed conformational changes that were similar to NAD in the InhA protein structure. The C00013874 showed the fluctuation most similar to NAD, especially for the 100–115 residue, where the other ligands do not significantly affect the residue. Furthermore, the ligand C00006532 showed less fluctuation towards InhA protein, while C00013887 significantly affected the C-terminal residues.
Multidrug-resistant MTB has resulted in considerably high mortality rates with restricted treatment. Previous studies have shown that MTB-InhA is responsible for developing INH resistance, a primary drug in TB treatment. Approximately 80% of global MTB isolates with phenotypic resistance to INH appeared to contain mutations in codon 315 of the katG gene or position −15 in the inhA promoter [28]. As our previous understanding for catalase-peroxidase enzyme encoded by katG as the drug activator for INH, quercetin analogs do not need an activation step through this enzyme. The compound might be active against the resistance of MTB which occurs due to mutation in katG. Besides, we propose that the decrease in quercetin analogs’ binding affinity caused by perturbation of the hydrogen-binding network in the Ile194 and possibly Tyr158 InhA active sites may directly affect the binding ability as previously described [24].
On the other hand, two quercetin analogs of C00013874 and C00006532 were found to have hydrogen bonds in Ser94 and Ile16 of InhA (Figure 1b,c). Another study has mentioned that S94A and I16T mutation might contribute to a conformational change in the enzyme active site upon ligands binding, resulting in a decrease in the affinity [29]. Based on this finding, the three best quercetin analogs might have binding affinity against MTB-InhA, which have mutations other than those described above. However, in silico combined with the in vitro study needs to be carried out to evaluate these three hit ligands’ effectivity, specifically in the various mutation of the INH resistant strain of MTB.
5. Conclusions
By using computational approaches, we have identified three potential quercetin analogs against MTB-InhA. These lead compounds can provide a platform for further detailed efficacy analysis, leading to discovering and designing new drugs against MTB specifically for further study in multidrug-resistant tuberculosis. Our study provides an impetus for future research in finding novel flavonoids as antimicrobial agents against the resistant microbe. Therefore, in vitro study based on minimal inhibitory concentration (MIC) testing may enlighten the effect of these three quercetin analogs in a better perspective, particularly in the INH resistance that shows a mutation in KatG and inhA genes.
Author Contributions
Conceptualization, D.A.E.P.; methodology, D.S.F.R., A. and T.M.F.; writing—original draft preparation, D.A.E.P., D.S.F.R., A.; writing—review and editing, D.A.E.P. and L.C.; project administration, D.A.E.P.; funding acquisition, D.A.E.P. and L.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Dian Ayu Eka Pitaloka has a postdoctoral fellowship from Universitas Padjadjaran under Rizky Abdulah’s supervision in the Department of Pharmacology and Clinical Pharmacy, Faculty of Pharmacy, Universitas Padjadjaran, Indonesia.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sotgiu, G.; Centis, R.; D’ambrosio, L.; Migliori, G.B. Tuberculosis treatment and drug regimens. Cold Spring Harb. Perspect Med. 2015, 5, a017822. [Google Scholar] [CrossRef] [PubMed]
- Timmins, G.S.; Deretic, V. Mechanisms of action of isoniazid. Mol. Microbiol. 2006, 5, 1220–1227. [Google Scholar] [CrossRef] [PubMed]
- Marrakchi, H.; Lanéelle, G.; Quémard, A. InhA, a target of the antituberculous drug isoniazid, is involved in a mycobacterial fatty acid elongation system, FAS-II. Microbiology 2000, 2, 289–296. [Google Scholar] [CrossRef]
- Takayama, K.; Wang, C.; Besra, G.S. Pathway to Synthesis and Processing of Mycolic Acids in Mycobacterium tuberculosis. Clin. Microbiol. Rev. 2005, 1, 81–101. [Google Scholar] [CrossRef]
- Rotta, M.; Pissinate, K.; Villela, A.D.; Back, D.F.; Timmers, L.F.S.M.; Bachega, J.F.R.; de Souza, O.N.; Santos, D.S.; Basso, L.A.; Machado, P. Piperazine derivatives: Synthesis, inhibition of the Mycobacterium tuberculosis enoyl-acyl carrier protein reductase and SAR studies. Eur. J. Med. Chem. 2015, 1, 436–447. [Google Scholar] [CrossRef]
- Lei, B.; Wei, C.J.; Tu, S.C. Action Mechanism of Antitubercular Isoniazid. J. Biol Chem. 2000, 4, 2520–2526. [Google Scholar] [CrossRef] [PubMed]
- Jäger, S.; Trojan, H.; Kopp, T.; Laszczyk, M.N.; Scheffler, A. Pentacyclic triterpene distribution in various plants—rich sources for a new group of multi-potent plant extracts. Molecules 2009, 6, 2016–2031. [Google Scholar] [CrossRef]
- Zheng, Y.; Jiang, X.; Gao, F.; Song, J.; Sun, J.; Wang, L.; Sun, X.; Lu, Z.; Zhang, H. Identification of plant-derived natural products as potential inhibitors of the Mycobacterium tuberculosis proteasome. BMC Complement. Altern. Med. 2014, 14, 400. [Google Scholar] [CrossRef]
- Cao, R.; Teskey, G.; Islamoglu, H.; Gutierrez, M.; Salaiz, O.; Munjal, S.; Fraix, M.P.; Sathananthan, A.; Nieman, D.C.; Venketaraman, V. Flavonoid Mixture Inhibits Mycobacterium tuberculosis Survival and Infectivity. Molecules 2019, 5, 851. [Google Scholar] [CrossRef]
- Sasikumar, K.; Ghosh, A.R.; Dusthackeer, A. Antimycobacterial potentials of quercetin and rutin against Mycobacterium tuberculosis H37Rv. Biotech 2018, 10, 427. [Google Scholar] [CrossRef]
- Pawar, A.; Jha, P.; Konwar, C.; Chaudhry, U.; Chopra, M.; Saluja, D. Ethambutol targets the glutamate racemase of Mycobacterium tuberculosis—an enzyme involved in peptidoglycan biosynthesis. Appl. Microbiol. Biotechnol. 2019, 2, 843–851. [Google Scholar] [CrossRef]
- Premalatha, E.; Dineshraj, R.; Iyanar, K.; Bhaarath, K.S.; Sharavanan, T.K.V. Molecular docking study on quercetin derivatives as inhibitors of PantothenateSynthetase (PanC) of Mycobacterium tuberculosis. Int. J. Res. Pharm. Sci. 2020, 3, 3684–3690. [Google Scholar] [CrossRef]
- Pitaloka, D.A.E.; Damayanti, S.; Artarini, A.A.; Sukandar, E.Y. Molecular Docking, Dynamics Simulation, and Scanning Electron Microscopy (SEM) Examination of Clinically Isolated Mycobacterium tuberculosis by Ursolic Acid: A Pentacyclic Triterpenes. Indones J. Chem. 2019, 2, 328–336. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Olson, A.J. Using AutoDock for ligand-receptor docking. Curr. Protoc. Bioinform. 2008, 24, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Leung, K.S.; Wong, M.H. idock: A multithreaded virtual screening tool for flexible ligand docking. IEEE Symp. Comput. Intell. Bioinform. Comput. Biol. CIBCB 2012, 77–84. [Google Scholar] [CrossRef]
- Gasteiger, J.; Marsili, M. Iterative partial equalization of orbital electronegativity—a rapid access to atomic charges. Tetrahedron 1980, 22, 3219–3228. [Google Scholar] [CrossRef]
- Petrov, D.; Zagrovic, B. Are Current Atomistic Force Fields Accurate Enough to Study Proteins in Crowded Environments? PLoS Comput. Biol. 2014, 5, e1003638. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.W.S.; Vranken, W.F. ACPYPE—AnteChamber PYthon Parser interfacE. BMC Res. Notes 2012, 5, 1–8. [Google Scholar] [CrossRef]
- Wang, H.; Gao, X.; Fang, J. Multiple Staggered Mesh Ewald: Boosting the Accuracy of the Smooth Particle Mesh Ewald Method. J. Chem. Theory Comput. 2016, 11, 5596–5608. [Google Scholar] [CrossRef]
- Kumari, R.; Kumar, R. g_mmpbsa—A GROMACS Tool for High-Throughput MM-PBSA Calculations. J. Chem. Inf. Model. 2014, 7, 1951–1962. [Google Scholar] [CrossRef]
- Tabrizi, A.M.; Goossens, S.; Rahimi, A.M.; Knepley, M.; Bardhan, J.P. Predicting solvation free energies and thermodynamics in polar solvents and mixtures using a solvation-layer interface condition. J. Chem Phys. 2017, 9, 094103. [Google Scholar] [CrossRef]
- Yamagishi, J.; Okimoto, N.; Morimoto, G.; Taiji, M. A new set of atomic radii for accurate estimation of solvation free energy by Poisson–Boltzmann solvent model. J. Comput. Chem. 2014, 29, 2132. [Google Scholar] [CrossRef]
- Shoichet, B.K.; McGovern, S.L.; Wei, B.; Irwin, J.J. Lead discovery using molecular docking. Curr. Opin. Chem. Biol. 2002, 4, 439–446. [Google Scholar] [CrossRef]
- Rozwarski, D.A.; Vilchèze, C.; Sugantino, M.; Bittman, R.; Sacchettini, J.C. Crystal structure of the Mycobacterium tuberculosis enoyl-ACP reductase, InhA, in complex with NAD+ and a C16 fatty acyl substrate. J. Biol. Chem. 1999, 22, 15582–15589. [Google Scholar] [CrossRef] [PubMed]
- Suhasini, M.; Sailatha, E.; Gunasekaran, S.; Ramkumaar, G.R. Vibrational and electronic investigations, thermodynamic parameters, HOMO and LUMO analysis on Lornoxicam by density functional theory. J. Mol. Struct. 2015, 1100, 116–128. [Google Scholar] [CrossRef]
- Chen, F.; Liu, H.; Sun, H.; Pan, P.; Li, Y.; Li, D. Assessing the performance of the MM/PBSA and MM/GBSA methods. 6. Capability to predict protein-protein binding free energies and re-rank binding poses generated by protein-protein docking. Phys. Chem. Chem. Phys. 2016, 32, 22129–22139. [Google Scholar] [CrossRef] [PubMed]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 3, 727–748. [Google Scholar] [CrossRef] [PubMed]
- Seifert, M.; Catanzaro, D.; Catanzaro, A.; Rodwell, T.C. Genetic mutations associated with isoniazid resistance in Mycobacterium tuberculosis: A systematic review. PLoS ONE 2015, 3, e0119628. [Google Scholar] [CrossRef] [PubMed]
- Dessen, A.; Quemard, A.; Blanchard, J.S.; Jacobs, W.R., Jr.; Sacchettini, J.C. Cyrstal structure and function of the isoniazid target of Mycobacterium tuberculosis. Science 1995, 5204, 1638–1641. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).