Promising Anti-MRSA Activity of Brevibacillus sp. Isolated from Soil and Strain Improvement by UV Mutagenesis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Bacterial Isolation
2.2. Determination of Antibacterial Activity
2.2.1. Cross-Streak Method
2.2.2. Agar Well Diffusion Assay
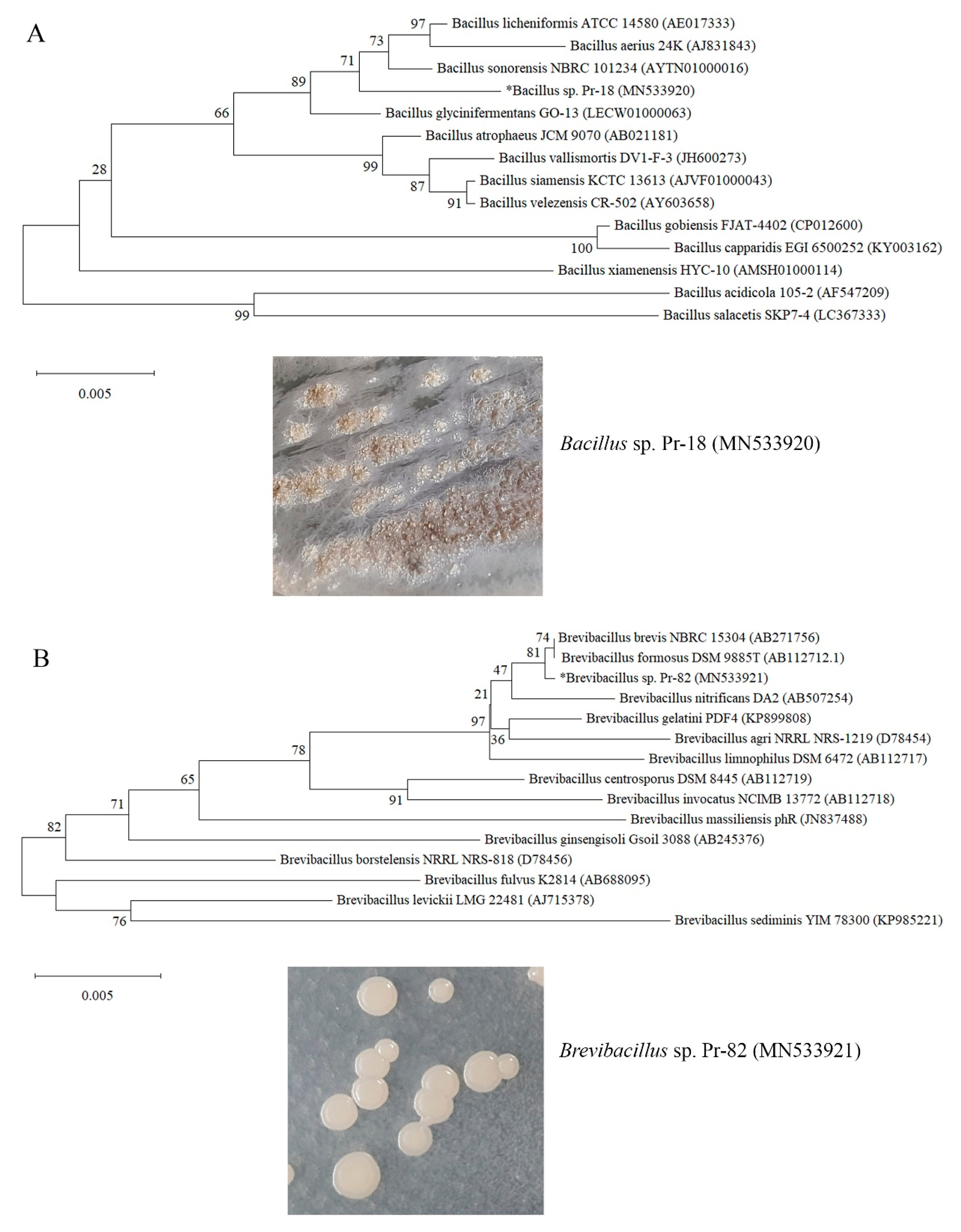
2.3. Bacterial Identification by 16S rRNA Sequencing
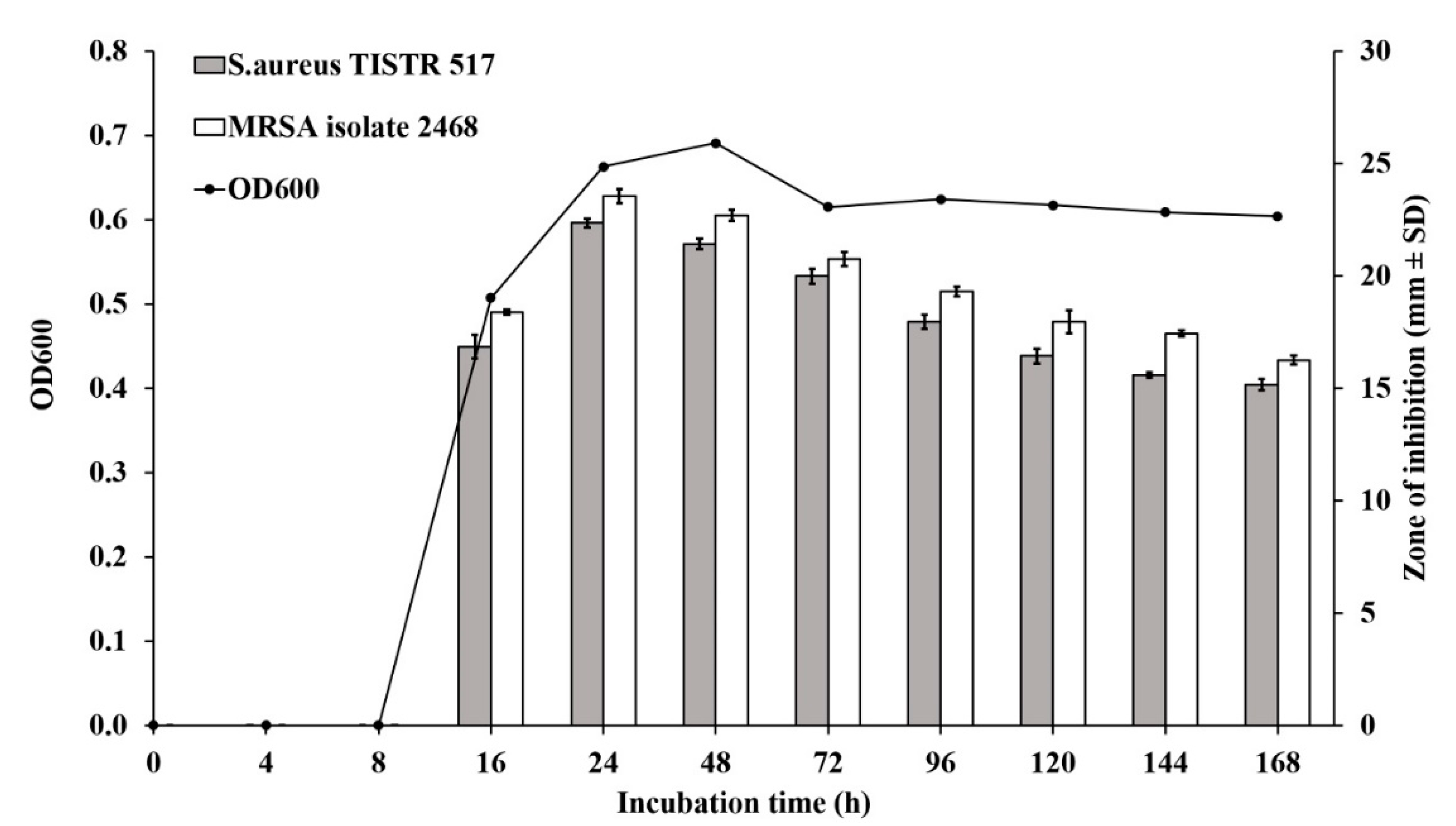
2.4. Production Kinetics of Anti-MRSA Substances
2.5. Stability Study of Antibacterial Substances
2.5.1. Thermal Stability
2.5.2. pH Sensitivity
2.5.3. Catalytic Enzyme and Surfactant Stability
2.6. Strain Improvement by UV Mutagenesis
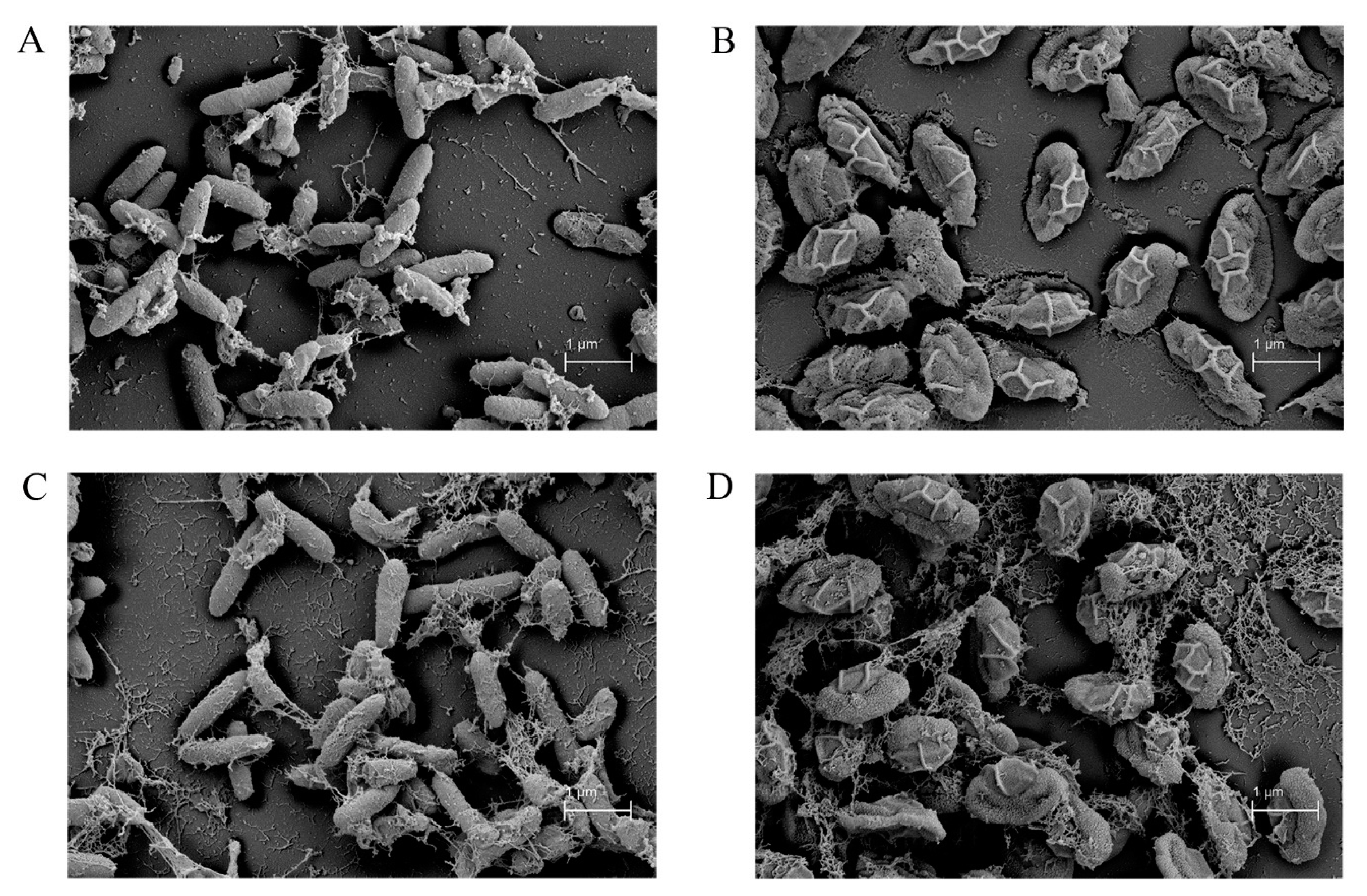
2.7. Scanning Electron Microscope (SEM)
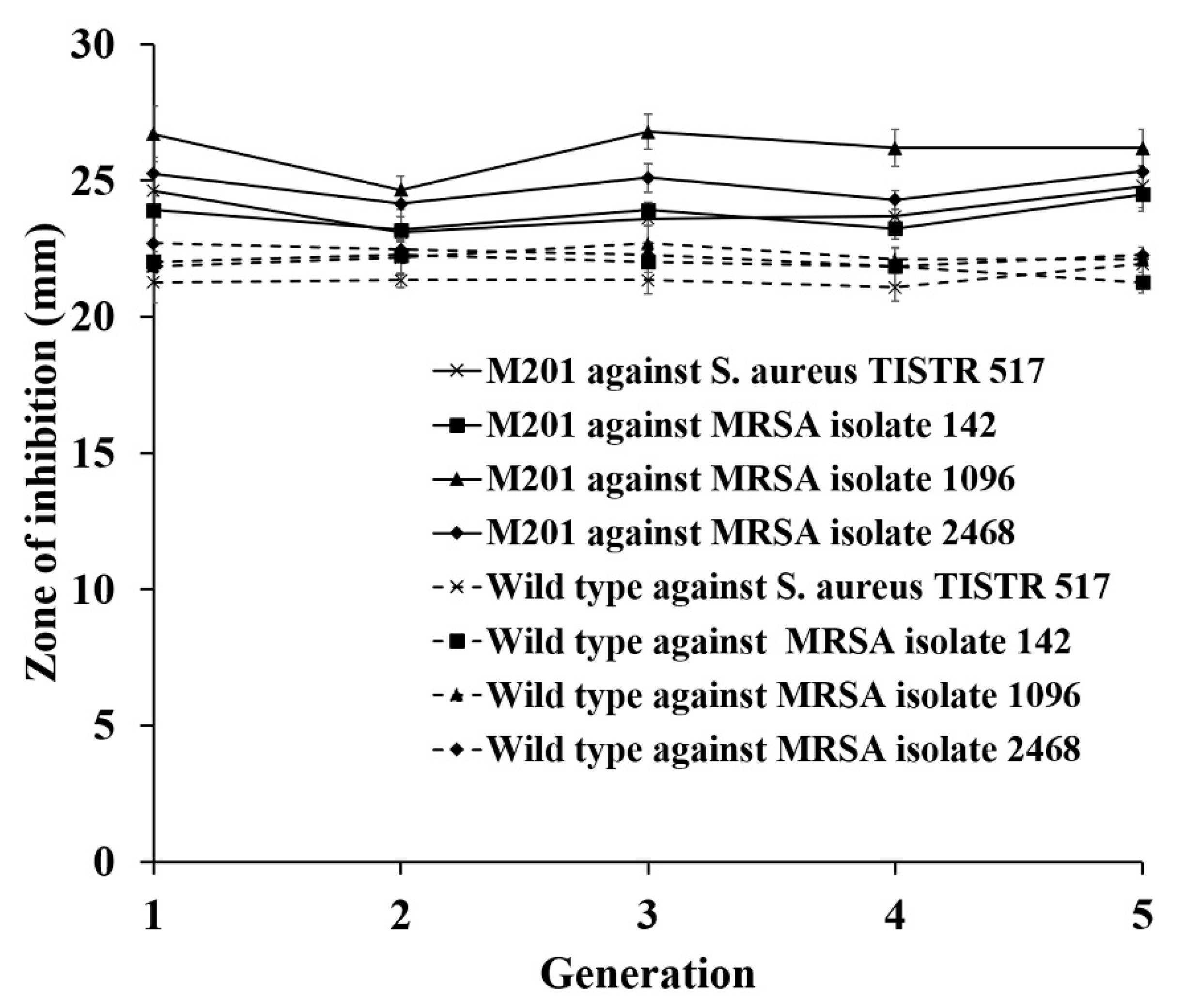
2.8. Strain Stability of the Mutant on the Antibacterial Activity
2.9. Susceptibility Study of the Mutant Strain
3. Results
3.1. Bacterial Isolation and Antibacterial Activity
3.2. Bacterial Identification and Phylogenetic Tree Construction
3.3. Production Kinetics of Anti-MRSA Substances from Brevibacillus sp. SPR-20
3.4. Stability Study of Antibacterial Substances
3.5. Strain Improvement of SPR-20 by UV Mutagenesis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- O’Neill, J. Review on Antimicrobial Resistance, Tackling Drug-resistant Infections Globally: Final Report and Recommendations; Wellcome Trust and HM Government: London, UK, 2016; pp. 10–16.
- Antimicrobial Resistance Division, World Health Organization. 2019 Antibacterial Agents in Clinical Development: An Analysis of the Antibacterial Clinical Development Pipeline; Weiss, G., Ed.; World Health Organization: Geneva, Switzerland, 2019; pp. 1–24. [Google Scholar]
- Hiramatsu, K.; Katayama, Y.; Matsuo, M.; Sasaki, T.; Morimoto, Y.; Sekiguchi, A.; Baba, T. Multi-drug-resistant Staphylococcus aureus and future chemotherapy. J. Infect. Chemother. 2014, 20, 593–601. [Google Scholar] [CrossRef] [Green Version]
- Kapoor, G.; Saigal, S.; Elongavan, A. Action and resistance mechanisms of antibiotics: A guide for clinicians. J. Anaesthesiol. Clin. Pharmacol. 2017, 33, 300–305. [Google Scholar] [CrossRef]
- Bin Zaman, S.; Hussain, M.A.; Nye, R.; Mehta, V.; Mamun, K.T.; Hossain, N. A Review on antibiotic resistance: Alarm Bells are ringing. Cureus 2017, 9, e1403. [Google Scholar] [CrossRef] [Green Version]
- Zulkeflle, S.N.M.; Yusaimi, Y.A.; Sugiura, N.; Iwamoto, K.; Goto, M.; Utsumi, M.; Othman, N.B.; Zakaria, Z.; Hara, H. Phenotypic and genetic characterization of multidrug-resistant Staphylococcus aureus in the tropics of Southeast Asia. Microbiology 2016, 162, 2064–2074. [Google Scholar] [CrossRef]
- Hug, J.J.; Bader, C.D.; Remškar, M.; Cirnski, K.; Müller, R. Concepts and methods to access novel antibiotics from actinomycetes. Antibiot 2018, 7, 44. [Google Scholar] [CrossRef] [Green Version]
- Andayani, D.G.S.; Sukandar, U.; Sukandar, E.Y.; Adnyana, I.K. Antibacterial, antifungal and anticancer activity of five strains of soil microorganisms isolated from Tangkuban Perahu mountain by fermentation. HAYATI J. Biosci. 2015, 22, 186–190. [Google Scholar] [CrossRef] [Green Version]
- Sumi, C.D.; Yang, B.W.; Yeo, I.-C.; Hahm, Y.T. Antimicrobial peptides of the genus Bacillus: A new era for antibiotics. Can. J. Microbiol. 2015, 61, 93–103. [Google Scholar] [CrossRef]
- Ahmad, M.S.; El-Gendy, A.O.; Ahmed, R.R.; Hassan, H.M.; El-Kabbany, H.M.; Merdash, A.G. Exploring the antimicrobial and antitumor potentials of Streptomyces sp. AGM12-1 isolated from egyptian soil. Front. Microbiol. 2017, 8, 438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saxena, S. Strategies of Strain Improvement of Industrial Microbes. In Applied Microbiology; Springer: New Delhi, India, 2015; pp. 155–171. [Google Scholar] [CrossRef]
- Ikehata, H.; Ono, T. The mechanisms of UV mutagenesis. J. Radiat. Res. 2011, 52, 115–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nasfi, Z.; Busch, H.; Kehraus, S.; Linares-Otoya, L.; König, G.M.; Schäberle, T.F.; Bachoual, R. Soil bacteria isolated from tunisian arid areas show promising antimicrobial activities against gram-negatives. Front. Microbiol. 2018, 9, 2742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selvin, J.; Shanmughapriya, S.; Gandhimathi, R.; Kiran, G.S.; Ravji, T.R.; Natarajaseenivasan, K.; Hema, T.A. Optimization and production of novel antimicrobial agents from sponge associated marine actinomycetes Nocardiopsis dassonvillei MAD08. Appl. Microbiol. Biotechnol. 2009, 83, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Valgas, C.; De Souza, S.M.; Smânia, E.F.A.; Smânian, A.S., Jr. Screening methods to determine antibacterial activity of natural products. Braz. J. Microbiol. 2007, 38, 369–380. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Goh, H.F.; Philip, K. Purification and characterization of bacteriocin produced by Weissella confusa A3 of dairy origin. PLoS ONE 2015, 10, e0140434. [Google Scholar] [CrossRef]
- Chalasani, A.G.; Dhanarajan, G.; Nema, S.; Sen, R.; Roy, U. An antimicrobial metabolite from Bacillus sp.: Significant activity against pathogenic bacteria including multidrug-resistant clinical strains. Front. Microbiol. 2015, 6, 1335. [Google Scholar] [CrossRef] [Green Version]
- Hmani, M.; Boukedi, H.; Ben Khedher, S.; Elleuch, A.; Tounsi, S.; Abdelkefi-Mesrati, L. Improvement of Vip3Aa16 toxin production and efficiency through nitrous acid and UV mutagenesis of Bacillus thuringiensis (Bacillales: Bacillaceae). J. Econ. Entomol. 2018, 111, 108–111. [Google Scholar] [CrossRef]
- Chatterjee, S.; Shekhawat, K.; Gupta, N. Bioreduction of toxic hexavalent chromium by novel indigenous microbe Brevibacillus agri isolated from tannery wastewater. Int. J. Environ. Sci. Technol. 2019, 16, 3549–3556. [Google Scholar] [CrossRef]
- CLSI. M100 Performance Standards for Antimicrobial Susceptibility Testing, 13th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020; pp. 58–66. [Google Scholar]
- Ma, X.-J.; Zhang, H.-M.; Lu, X.-F.; Han, J.; Zhu, H.-X.; Wang, H.; Yao, R.-S. Mutant breeding of Starmerella bombicola by atmospheric and room-temperature plasma (ARTP) for improved production of specific or total sophorolipids. Bioprocess Biosyst. Eng. 2020, 43, 1869–1883. [Google Scholar] [CrossRef]
- Bérdy, J. Bioactive microbial metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Yousef, A.E. Antimicrobial peptides produced by Brevibacillus spp.: Structure, classification and bioactivity: A mini review. World J. Microbiol. Biotechnol. 2018, 34, 57. [Google Scholar] [CrossRef] [PubMed]
- Ghadbane, M.; Harzallah, D.; Ibn Laribi, A.; Jaouadi, B.; Belhadj, H. Purification and biochemical characterization of a highly thermostable bacteriocin isolated from Brevibacillus brevis strain GM100. Biosci. Biotechnol. Biochem. 2013, 77, 151–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Huang, E.; Yuan, C.; Zhang, L.; Yousef, A.E. Isolation and structural elucidation of brevibacillin, an antimicrobial lipopeptide from Brevibacillus laterosporus that combats drug-resistant gram-positive bacteria. Appl. Environ. Microbiol. 2016, 82, 2763–2772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chawawisit, K.; Lertcanawanichakul, M. Minimum inhibitory concentration (MIC) of crude preparations of Brevibacillus laterosporus SA14 bioactive material compared to vancomycin and oxacillin, against clinical isolates of methicillin-resistant Staphylococcus aureus. World J. Microbiol. Biotechnol. 2008, 24, 2199–2204. [Google Scholar] [CrossRef]
- Song, J.; Wang, Y.; Song, Y.; Zhao, B.; Wang, H.; Zhou, S.; Kong, D.; Guo, X.; Li, Y.; He, M.; et al. Brevibacillus halotolerans sp. nov., isolated from saline soil of a paddy field. Int. J. Syst. Evol. Microbiol. 2017, 67, 772–777. [Google Scholar] [CrossRef]
- Glare, T.; Durrant, A.; Berry, C.; Palma, L.; Ormskirk, M.M.; Cox, M.P. Phylogenetic determinants of toxin gene distribution in genomes of Brevibacillus laterosporus. Genomics 2019, 112, 1042–1053. [Google Scholar] [CrossRef]
- Chopra, L.; Singh, G.; Choudhary, V.; Sahoo, D.K. Sonorensin: An antimicrobial peptide, belonging to the heterocycloanthracin subfamily of bacteriocins, from a new marine isolate, Bacillus sonorensis MT93. Appl. Environ. Microbiol. 2014, 80, 2981–2990. [Google Scholar] [CrossRef] [Green Version]
- Song, Z.; Liu, Q.; Guo, H.; Ju, R.; Zhao, Y.; Li, J.; Liu, X. Tostadin, a novel antibacterial peptide from an antagonistic microorganism Brevibacillus brevis XDH. Bioresour. Technol. 2012, 111, 504–506. [Google Scholar] [CrossRef]
- Holcapkova, P.; Raskova, Z.K.; Hrabalikova, M.; Salakova, A.; Drbohlav, J.; Sedlarik, V. Isolation and thermal stabilization of bacteriocin nisin derived from whey for antimicrobial modifications of polymers. Int. J. Polym. Sci. 2017, 2017, 3072582. [Google Scholar] [CrossRef] [Green Version]
- Moreno, I.; Lerayer, A.L.S.; Baldini, V.L.S.; Leitão, M.F. Characterization of bacteriocins produced by Lactococcus lactis strains. Braz. J. Microbiol. 2000, 31, 183–191. [Google Scholar] [CrossRef] [Green Version]
- Slootweg, J.C.; Liskamp, R.M.J.; Rijkers, D.T.S. Scalable purification of the lantibiotic nisin and isolation of chemical/enzymatic cleavage fragments suitable for semi-synthesis. J. Pept. Sci. 2013, 19, 692–699. [Google Scholar] [CrossRef]
- Liu, K.; Yang, L.; Peng, X.; Gong, H.; Wang, J.; Lu, J.R.; Xu, H. Effects of conventional surfactants on the activity of designed antimicrobial peptide. Langmuir 2020, 36, 3531–3539. [Google Scholar] [CrossRef]
- Saleem, F.; Ahmad, S.; Yaqoob, Z.; Rasool, S.A. Comparative study of two bacteriocins produced by representative indigenous soil bacteria. Pak. J. Pharm. Sci. 2009, 22, 252–258. [Google Scholar]
- Faheem, F.; Saeed, S.; Rasool, S.A. Studies on Brevicin AF01: A bacteriocin like inhibitory substance active against methicillin resistant Staphylococcus aureus. Pak. J. Bot. 2007, 39, 1293–1302. [Google Scholar]
- Cherif, A.J.A.A.; Chehimi, S.; Limem, F.; Hansen, B.; Hendriksen, N.; Daffonchio, D.; Boudabous, A. Detection and characterization of the novel bacteriocin entomocin 9, and safety evaluation of its producer, Bacillus thuringiensis ssp. entomocidus HD9. J. Appl. Microbiol. 2003, 95, 990–1000. [Google Scholar] [CrossRef] [PubMed]
- Aunpad, R.; Na-Bangchang, K. Pumilicin 4, A Novel Bacteriocin with anti-MRSA and Anti-VRE activity produced by newly isolated bacteria Bacillus pumilus strain WAPB4. Curr. Microbiol. 2007, 55, 308–313. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Huang, E.; Yuan, C.; Zhang, L.; Yousef, A.E. Isolation of a Paenibacillus sp. strain and structural elucidation of its broad-spectrum lipopeptide antibiotic. Appl. Environ. Microbiol. 2012, 78, 3156–3165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parekh, S.; Vinci, V.A.; Strobel, R.J. Improvement of microbial strains and fermentation processes. Appl. Microbiol. Biotechnol. 2000, 54, 287–301. [Google Scholar] [CrossRef]
- Mukherjee, A.K.; Rai, S.K.; Bordoloi, N.K. Biodegradation of waste chicken-feathers by an alkaline β-keratinase (Mukartinase) purified from a mutant Brevibacillus sp. strain AS-S10-II. Int. Biodeterior. Biodegrad. 2011, 65, 1229–1237. [Google Scholar] [CrossRef]
- Suribabu, K.; Lalitha Govardhan, T.; Hemalatha, K.P.J. Strain improvement of Brevibacillus borostelensis R1 for optimization of α-amylase production by mutagens. J. Microb. Biochem. Technol. 2014, 6, 123–127. [Google Scholar] [CrossRef] [Green Version]
- Ali, S.; Nelofer, R.; Andleeb, S.; Baig, S.; Shakir, H.A.; Qazi, J.I. Biosynthesis and optimization of bacitracin by mutant Bacillus licheniformis using submerged fermentation. Indian J. Biotechnol. 2018, 17, 251–260. [Google Scholar]
- Che, J.; Liu, B.; Liu, G.; Chen, Q.; Huang, D. Induced mutation breeding of Brevibacillus brevis FJAT-0809-GLX for improving ethylparaben production and its application in the biocontrol of Lasiodiplodia theobromae. Postharvest Biol. Technol. 2018, 146, 60–67. [Google Scholar] [CrossRef]
- Gamalier, J.P.; Silva, T.P.; Zarantonello, V.; Dias, F.F.; Melo, R.C. Increased production of outer membrane vesicles by cultured freshwater bacteria in response to ultraviolet radiation. Microbiol. Res. 2017, 194, 38–46. [Google Scholar] [CrossRef] [PubMed]


| Sampling Location | S. aureus TISTR 517 | E. coli TISTR 887 |
|---|---|---|
| 8°33′55.4″ N, 99°31′12.1″ E | 1 | 0 |
| 8°46′03.3″ N, 99°48′09.3″ E | 5 | 2 |
| 12°25′07.3″ N, 99°58′58.3″ E | 29 | 20 |
| 9°05′22.6″ N, 99°53′39.9″ E | 4 | 5 |
| 9°00′50.3″ N, 99°46′18.0″ E | 7 | 1 |
| 8°33′33.4″ N, 99°46′55.6″ E | 7 | 6 |
| 8°38′31.7″ N, 99°53′38.7″ E | 13 | 8 |
| 8°38′55.6″ N, 99°52′51.4″ E | 6 | 0 |
| 8°38′49.7″ N, 54°45′50″ E | 5 | 8 |
| 8°38′18.3″ N, 54°45′48.5″ E | 17 | 17 |
| Total | 94 | 67 |
| Isolates | S. aureus TISTR 517 (mm) | MRSA Isolate 142 (mm) | MRSA Isolate 1096 (mm) | MRSA Isolate 2468 (mm) | E. coli TISTR 887 (mm) | P. aeruginosa TISTR 357 (mm) | C. albicans TISTR 5554 (mm) |
|---|---|---|---|---|---|---|---|
| AK-74 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 10.89 ± 0.16 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| B6-28 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 11.95 ± 0.27 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Play-88 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 11.26 ± 0.53 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Pr-18 | 15.41 ± 0.61 | 16.01 ± 0.42 | 16.12 ± 0.35 | 16.07 ± 0.24 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Pr-82 | 15.75 ± 0.33 | 16.38 ± 0.13 | 17.14 ± 0.32 | 17.09 ± 0.27 | 12.65 ± 0.39 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Pr-83 | 13.12 ± 0.92 | 10.99 ± 0.26 | 13.09 ± 0.35 | 11.82 ± 0.57 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Pr-117 | 14.02 ± 0.11 | 13.70 ± 0.55 | 14.60 ± 0.27 | 14.02 ± 0.47 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Pr-122 | 15.08 ± 0.05 | 15.46 ± 0.16 | 14.00 ± 0.44 | 15.97 ± 0.26 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| SPR-02 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 13.34 ± 0.34 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| SPR-14 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 10.42 ± 0.50 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| SPR-20 | 17.52 ± 0.60 | 18.28 ± 0.58 | 18.03 ± 0.36 | 19.05 ± 0.84 | 13.29 ± 0.12 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| S-STD-1 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 11.18 ± 0.44 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Su-7 | 11.80 ± 0.76 | 13.62 ± 0.19 | 14.25 ± 0.57 | 13.60 ± 1.08 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| WUL-02 | 11.49 ± 0.81 | 12.10 ± 0.05 | 0.00 ± 0.00 | 12.06 ± 0.11 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| WUL-10 | 14.59 ± 0.06 | 15.91 ± 0.58 | 15.11 ± 0.43 | 14.66 ± 0.06 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Cefoxitin (30 µg) | 35.98 ± 0.53 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 34.71 ± 0.73 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Oxacillin (1 µg) | 30.82 ± 1.40 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Vancomycin (30 µg) | 25.48 ± 0.15 | 25.74 ± 0.15 | 25.74 ± 0.29 | 25.99 ± 0.29 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Conditions | % Activity | |||
|---|---|---|---|---|
| S. aureus TISTR 517 | MRSA Isolate 2468 | |||
| SPR-20 | Nisin | SPR-20 | Nisin | |
| Untreated Sample | 100.00 ± 4.46 | 100.00 ± 3.78 | 100.00 ± 4.93 | 100.00 ± 1.95 |
| Thermal Stability | ||||
| Sample 60 °C, 1 h | 100.00 ± 1.37 | 89.13 ± 3.77 * | 99.31 ± 3.17 | 92.43 ± 2.49 * |
| Sample 80 °C, 1 h | 96.44 ± 1.81 * | 73.91 ± 2.49 * | 97.92 ± 1.59 | 88.05 ± 1.38 * |
| Sample 100 °C, 1 h | 95.65 ± 1.81 * | 0.00 ± 0.00 * | 96.54 ± 1.04 * | 69.72 ± 1.38 * |
| Sample 121 °C, 15 psi, 15 min (autoclave) | 91.30 ± 3.14 * | 72.28 ± 3.39 * | 92.39 ± 1.80 * | 92.43 ± 0.69 * |
| Enzyme Stability | ||||
| Sample + Proteinase K (1 mg/mL) | 96.08 ± 0.68 * | 0.00 ± 0.00 * | 95.90 ± 2.33 | 0.00 ± 0.00 * |
| Sample + Trypsin (1 mg/mL) | 98.00 ± 1.83 | 0.00 ± 0.00 * | 96.72 ± 0.63 * | 0.00 ± 0.00 * |
| Sample + α-Chymotrypsin (1 mg/mL) | 95.20 ± 1.83 * | 0.00 ± 0.00 * | 94.89 ± 3.34 | 68.95 ± 3.20 * |
| Proteinase K (1 mg/mL) | 0.00 ± 0.00 * | 0.00 ± 0.00 * | 0.00 ± 0.00 * | 0.00 ± 0.00 * |
| Trypsin (1 mg/mL) | 0.00 ± 0.00 * | 0.00 ± 0.00 * | 0.00 ± 0.00 * | 0.00 ± 0.00 * |
| α-Chymotrypsin (1 mg/mL) | 0.00 ± 0.00 * | 0.00 ± 0.00 * | 0.00 ± 0.00 * | 0.00 ± 0.00 * |
| Surfactant Stability | ||||
| Sample + 1% SDS | 108.81 ± 2.76 * | 130.86 ± 0.99 * | 102.95 ± 1.28 * | 95.14 ± 1.40 * |
| Sample + 1% Triton X-100 | 109.24 ± 0.75 * | 148.00 ± 1.98 * | 106.36 ± 1.94 * | 121.05 ± 4.27 * |
| 1% SDS | 105.29 ± 0.75 * | 137.71 ± 1.98 * | 102.95 ± 2.53 | 99.60 ± 1.21 |
| 1% Triton X-100 | 0.00 ± 0.00 * | 0.00 ± 0.00 * | 0.00 ± 0.00 * | 0.00 ± 0.00 * |
| pH Stability | ||||
| pH 2 | 102.51 ± 1.92 | 98.86 ± 0.99 | 102.33 ± 1.16 | 97.98 ± 2.42 |
| pH 4 | 100.84 ± 2.90 | 100.57 ± 0.99 | 102.33 ± 0.00 | 97.18 ± 1.85 |
| pH 5 | 100.84 ± 3.16 | 97.71 ± 1.71 | 102.33 ± 2.01 | 98.39 ± 1.85 |
| pH 6 | 100.00 ± 2.90 | 96.57 ± 1.98 | 101.55 ± 2.93 | 100.00 ± 1.85 |
| pH 7 | 100.42 ± 2.17 | 97.71 ± 1.71 | 100.39 ± 0.67 | 97.98 ± 1.21 |
| pH 8 | 100.00 ± 1.45 | 101.14 ± 2.97 | 100.00 ± 1.16 | 99.19 ± 1.21 |
| pH 9 | 101.67 ± 1.26 | 98.29 ± 1.98 | 101.57 ± 0.00 | 99.18 ± 0.71 |
| pH 11 | 101.26 ± 0.72 | 96.00 ± 1.71 * | 100.00 ± 0.68 | 97.95 ± 1.42 |
| pH 12 | 102.93 ± 1.26 | 0.00 ± 0.00 * | 100.39 ± 1.18 | 78.69 ± 2.13 * |
| pH 13 | 102.51 ± 1.45 | 0.00 ± 0.00 * | 101.97 ± 1.36 | 0.00 ± 0.00 * |
| pH 14 | 99.16 ± 2.17 | 0.00 ± 0.00 * | 101.97 ± 0.63 | 0.00 ± 0.00 * |
| Isolates | S. aureus TISTR 517 (mm) | MRSA Isolate 142 (mm) | MRSA Isolate 1096 (mm) | MRSA Isolate 2468 (mm) |
|---|---|---|---|---|
| M5 | 20.56 ± 0.25 * | 20.85 ± 0.73 | 22.02 ± 1.21 | 21.93 ± 0.26 |
| M6 | 21.41 ± 0.40 | 21.76 ± 0.27 | 21.70 ± 0.64 | 23.26 ± 0.56 |
| M7 | 21.32 ± 0.25 | 21.67 ± 0.37 | 22.46 ± 0.30 | 22.53 ± 0.39 |
| M8 | 22.27 ± 0.52 * | 22.20 ± 0.41 | 22.46 ± 0.29 | 23.53 ± 1.21 |
| M10 | 20.99 ± 0.62 | 20.70 ± 0.93 | 19.89 ± 0.97 | 22.02 ± 1.07 |
| M11 | 21.32 ± 0.53 | 21.00 ± 0.14 * | 21.54 ± 0.38 | 21.68 ± 0.53 |
| M19 | 21.57 ± 0.50 | 22.01 ± 0.18 | 22.22 ± 0.42 | 22.70 ± 0.26 |
| M56 | 20.90 ± 0.39 | 21.59 ± 0.43 | 21.96 ± 0.52 | 22.10 ± 0.44 |
| M79 | 21.57 ± 0.29 | 21.76 ± 0.14 | 22.21 ± 0.29 | 22.70 ± 0.73 |
| M83 | 20.73 ± 0.67 | 21.34 ± 0.00 * | 21.21 ± 0.52 | 21.34 ± 0.51 |
| M142 | 21.92 ± 0.40 | 21.93 ± 0.77 | 21.61 ± 0.25 * | 22.99 ± 0.31 * |
| M149 | 21.57 ± 1.09 | 21.88 ± 0.73 | 23.17 ± 1.36 | 23.04 ± 1.03 |
| M151 | 20.87 ± 0.42 | 21.47 ± 0.58 | 22.67 ± 0.16 * | 23.03 ± 0.16 * |
| M159 | 21.82 ± 0.50 | 22.13 ± 0.18 | 22.53 ± 0.24 | 22.45 ± 0.15 |
| M178 | 21.42 ± 0.16 | 22.41 ± 0.28 * | 22.67 ± 0.16 * | 23.40 ± 0.75 |
| M201 | 24.62 ± 0.67 * | 23.91 ± 0.52 * | 26.70 ± 1.02 * | 25.24 ± 0.61 * |
| M203 | 21.49 ± 0.25 | 21.50 ± 0.52 | 22.12 ± 0.66 | 22.01 ± 0.52 |
| M212 | 21.82 ± 0.64 | 21.67 ± 0.29 | 22.87 ± 0.66 | 22.79 ± 0.52 |
| M214 | 21.51 ± 0.28 | 21.38 ± 0.16 * | 22.21 ± 0.16 * | 22.27 ± 0.00 |
| M220 | 21.15 ± 0.58 | 22.09 ± 0.87 | 22.62 ± 0.43 | 21.93 ± 0.40 |
| M228 | 21.57 ± 0.66 | 21.63 ± 0.54 | 21.80 ± 0.24 | 22.53 ± 0.15 |
| M231 | 21.40 ± 0.29 | 22.00 ± 0.43 | 21.56 ± 0.37 | 23.14 ± 0.45 |
| Wild-type | 21.23 ± 0.69 | 21.76 ± 0.82 | 22.21 ± 0.83 | 22.27 ± 0.83 |
| Antibiotics | Wild-Type Strain (mm) | Mutant 201 Strain (mm) |
|---|---|---|
| Cefoxitin (30 µg) | 36.24 ± 0.96 | 36.58 ± 1.10 |
| Ceftriaxone (30 µg) | 32.34 ± 0.24 | 36.91 ± 0.96 * |
| Ciprofloxacin (5 µg) | 34.71 ± 0.63 | 35.90 ± 0.86 |
| Erythromycin (15 µg) | 37.25 ± 0.86 | 38.78 ± 1.27 |
| Gentamicin (10 µg) | 26.42 ± 1.10 | 26.25 ± 1.46 |
| Imipenem (10 µg) | 36.58 ± 0.83 | 38.10 ± 0.41 |
| Tetracycline (30 µg) | 28.79 ± 0.24 | 30.31 ± 1.04 |
| Vancomycin (30 µg) | 19.64 ± 0.24 | 21.67 ± 0.86 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Songnaka, N.; Lertcanawanichakul, M.; Atipairin, A. Promising Anti-MRSA Activity of Brevibacillus sp. Isolated from Soil and Strain Improvement by UV Mutagenesis. Sci. Pharm. 2021, 89, 1. https://doi.org/10.3390/scipharm89010001
Songnaka N, Lertcanawanichakul M, Atipairin A. Promising Anti-MRSA Activity of Brevibacillus sp. Isolated from Soil and Strain Improvement by UV Mutagenesis. Scientia Pharmaceutica. 2021; 89(1):1. https://doi.org/10.3390/scipharm89010001
Chicago/Turabian StyleSongnaka, Nuttapon, Monthon Lertcanawanichakul, and Apichart Atipairin. 2021. "Promising Anti-MRSA Activity of Brevibacillus sp. Isolated from Soil and Strain Improvement by UV Mutagenesis" Scientia Pharmaceutica 89, no. 1: 1. https://doi.org/10.3390/scipharm89010001
APA StyleSongnaka, N., Lertcanawanichakul, M., & Atipairin, A. (2021). Promising Anti-MRSA Activity of Brevibacillus sp. Isolated from Soil and Strain Improvement by UV Mutagenesis. Scientia Pharmaceutica, 89(1), 1. https://doi.org/10.3390/scipharm89010001







