Abstract
Breast cancer is one of the most common cancers worldwide, which makes it a very impactful malignancy in the society. Breast cancers can be classified through different systems based on the main tumor features and gene, protein, and cell receptors expression, which will determine the most advisable therapeutic course and expected outcomes. Multiple therapeutic options have already been proposed and implemented for breast cancer treatment. Nonetheless, their use and efficacy still greatly depend on the tumor classification, and treatments are commonly associated with invasiveness, pain, discomfort, severe side effects, and poor specificity. This has demanded an investment in the research of the mechanisms behind the disease progression, evolution, and associated risk factors, and on novel diagnostic and therapeutic techniques. However, advances in the understanding and assessment of breast cancer are dependent on the ability to mimic the properties and microenvironment of tumors in vivo, which can be achieved through experimentation on animal models. This review covers an overview of the main animal models used in breast cancer research, namely in vitro models, in vivo models, in silico models, and other models. For each model, the main characteristics, advantages, and challenges associated to their use are highlighted.
1. Introduction
Over the last few decades, breast cancer has been the target of several studies regarding its molecular status, epidemiology, diagnosis, and treatment. Breast cancer is a major concern in women’ health due to its high incidence and mortality rate. However, it must be noted that it also affects men, although with much less frequency. After analyzing breast cancer’s trends and predictions for 2020 [1], the overall decline in breast cancer mortality rate over the last decades stands out, and it appears that it will continue decreasing. In the European Union, breast cancer mortality rate declined 15.3% between 2002 and 2012, and it is predicted to fall around 10% until 2020. The same tendency applies for the USA. It is also important to mention that although Western lifestyle appears to be a major risk factor for breast cancer, the increased awareness of the population and the development of novel improved diagnostic and treatment strategies are major contributors to this decline. In addition, it is impossible to ignore the importance of the evolution in the research field when analyzing those numbers.
Breast cancers are differentiated in several types based on their features, allowing the best treatment course selection and better prediction of the expected outcomes [2]. Breast cancers can be distinguished based on histopathological types, tumor grade, tumor stage, and molecular subtypes, as schematically represented on Figure 1.
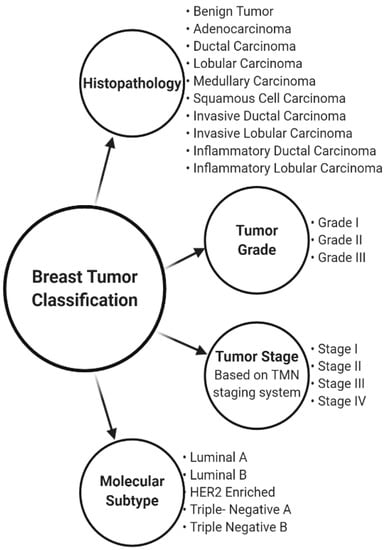
Figure 1.
Schematic representation of the breast cancer classification depending on histopathological characteristics, tumor grade, tumor stage, and molecular subtype.
Histopathology characterizes the tumors regarding its site and morphology based on a microscopic evaluation of the cells. It gives information about tumor size, description (presence or absence of masses or margins), texture, and histological and architectural type. Based on the location of the tumors, they can be classified as lobular or ductal if they begin in the lobules or in the ducts, respectively. Moreover, depending on the morphology of the cells, tumors can be divided into non-invasive or benign, and invasive or malign. Non-invasive tumors, also known as carcinomas in situ, are limited to the area where they are original set and have typically better prognosis, although they can also evolve to invasive cancers.
The tumor grade classification, also known as the Elston and Ellis modified Scarff–Bloom–Richardson (SBR) is based on a numeric score attributed to a tumor [3]. It assesses how closely cancer cells resemble normal cells depending on three parameters: the tubule formation, nuclear pleomorphism, and mitotic rate.
The most used tumor stage classification system is based on the TNM staging system developed by the American Joint Committee on Cancer [4,5]. TNM is a two-step procedure, and it stands for Tumor, Nodes, and Metastasis [4,5]. Firstly, the cancer is classified based on its size and extension, on the spreading or not to lymphatic nodes, and on the presence or absence of metastasis. Then, the combination of the previous factors will be translated in an overall TMN score.
The molecular subtyping of breast cancer is based on the expression of specific genes, proteins, and cell receptors [6,7,8,9,10], such as Estrogen Receptor (ER), Progesterone Receptor (PR), Human Epidermal Growth Factor Receptor 2 (HER2), Tyrosine Kinase Receptor, and Ki-67 protein. Additionally, over the last few years, Androgen Receptors (AR) have drawn attention as possible new markers for breast cancer classification, prognosis predictors, and therapeutic targets [11,12,13]; however, their use is still controversial. Based on the ER, PR, and HER2 receptors status and on other proteins, breast cancers can be classified as Luminal A (LA), Luminal B (LB), HER2-Enriched (H), Triple-Negative A or basal-like (TNA), or as Triple-Negative B or normal-like (TNB), as represented on Table 1. This subtyping reflects not only differences in the molecular expression of the cancers, but also on their incidence, prognosis, and sensitivity to specific treatments [6,8,14].

Table 1.
Molecular classification of breast cancer based on Estrogen Receptor (ER), Progesterone Receptor (PR) and Human Epidermal Growth Factor Receptor 2 (HER2) status [10,15,16,17,18,19].
Nowadays, there are several therapeutic options available for breast cancer, which are possibly divided into two main categories: local treatments and systemic treatments. Local treatments allow minimizing the effect over healthy tissues, and they include [20,21,22,23] tumor surgery (which is considered the primary treatment for early stage breast cancer) and radiation therapy [24,25,26]. Systemic treatments are those in which the administered drugs will travel through the blood stream, reaching cells all over the body [21]. Regarding breast cancer systemic therapeutic options, four major groups can be considered: chemotherapy [20,21,22], hormone therapy [21,22], targeted therapy [27,28,29,30], and immunotherapy [31,32,33,34].
Despite the multiple therapies available for breast cancer treatment, their use and efficacy greatly depend on the patient, tumor classification and analysis of the clinician. Moreover, there are still some concerns that need to be addressed, including extensive side effects, the need of using higher doses of treatments to surpass the lack of specific targeting, and the few therapeutics available for certain cancers, in particular triple-negative breast cancer. In addition, breast cancer resistance to treatments and recurrence deserves special attention and further research. In this line, Breast Cancer Stem Cells (BCSC) are considered to play an important role [35,36], and thus, several studies have already hypothesized the importance of studying these cells to improve breast cancer prognosis and treatment efficacy [35,36,37]. For these reasons, it is imperative to continue the search for novel therapeutic options, which not only requires the search for new drugs, formulations, and therapies, but also the search and development of reliable breast models, whether in vitro, in vivo, or in silico. Thus, advances in breast cancer therapeutic approaches greatly depend on the availability of experimental models accurately mimicking human breast cancer’ features and microenvironments, which by their turn, depend on the ability of accurately detecting and classifying breast cancers.
2. Animal Models in Breast Cancer
As aforementioned, breast cancer is still one of the most common cancers in humans, which has boosted multifield efforts either to understand the mechanisms behind the disease progression, evolution, and associated risk factors, both to discover and develop innovative diagnostic and therapeutic tools. Nonetheless, advances in the breast cancer field depend on the ability to mimic tumors’ properties and microenvironment, which can be achieved using animal models experimentation.
Great advances in animal models experimentation have been done over the last years to meet the demands of biomedical, medical, and pharmaceutical research [38,39]. Currently, animal models can be gathered into different groups depending on their biological complexity and if there is a coupling to non-biological entities or not, as represented in Figure 2 and explored further in this article. The simpler models in terms of biological complexity are in vitro models [19,40,41], which consist in the controlled growth, maintenance, and testing of single isolated cell lines or a synergy of different cell lines. Then, with higher complexity, there are in vivo models [42,43], which correspond to animal experimentation, representing a closer proximity to what is seen in humans, since they embody a better mimicking of biological microenvironments, as well as immunological responses. Furthermore, there are two other classes of models, the in silico models [44,45] and other models. In silico models are computational models that use mathematical equations and high computational power to represent and simulate biological properties and mechanisms. Other models include Phantoms and Microfluidic three-dimensional models, which use non-biological entities alone or coupled with cells, respectively, to physically mimic biological features and actions.
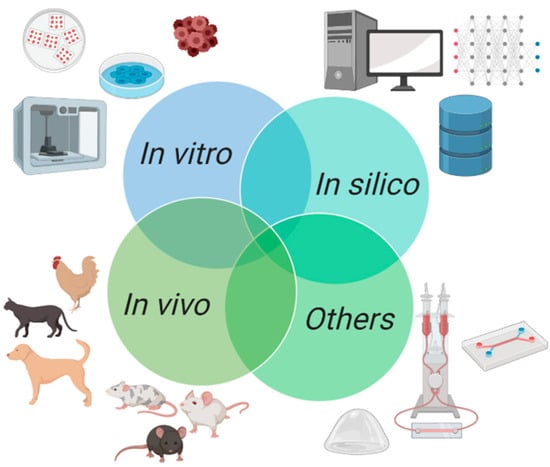
Figure 2.
Schematic representation of experimental models currently available for breast cancer research, which are classified as in vitro models, in vivo models, in silico models, and others, including phantoms and 3D microfluidic chips.
The choice of the animal model that is most suitable for each project will greatly depend on several aspects, namely ethical concerns, the purpose and the severity of the procedures, the restrictions imposed on the models to use (genetic variability, immunity…), financial and logistic resources, and the time available for the development of the project.
Currently, ethical aspects play an increasingly important role at the time of the decision of which animal model to use, because the well-being of the animals is becoming more and more valued [46,47,48]. In order to respect the animals’ welfare and ensure the use of the strictly necessary number of animals, guidelines [49,50], included in the legislation, for the use of animals in scientific research were created. In Europe, the Federation of European Laboratory Animal Science Associations (FELASA) guidelines apply [50], which include the 3Rs Principles, which were firstly proposed by Russel and Burch in 1959 [51,52]. The use of animals for research purposes in Europe is regulated by the European Convention (ETS 123) for the Protection of Vertebrate Animals used for Experimental and Other Scientific Purposes [53], which was adopted on 18th March 1986 and by the Directive 2013/63/EU of the European Parliament and of the Council on the Protection of Animals Used for Scientific Purposes [54], adopted on 22n September 2010, both including the 3Rs Principles. Furthermore, in Europe, there is also the European Animal Research Association (EARA) [55], which is an organization whose main goals are to inform the community about animal experimentation in biomedical research and healthcare and to ensure transparency regarding the use of animals in research. Moreover, most countries, as well as each animal research center, create their own Animal Welfare and Ethics Body (ORBEA) to regulate and evaluate animal experimentation. The 3Rs stand for Replacement, Reduction, and Refinement, which compile a group of measures to encourage, among others, the reduction of the number of animals used by selecting other models. They also prompt the care of the animals to minimize their pain and distress as much as possible. Currently, a fourth R is included for responsibility, appealing to the sense of responsibility of the researchers to respect animals’ welfare and to follow the animal experimentation guidelines. The application of the 4Rs implies not only ethical considerations, but also economical and common-sense considerations, because it is important to share the results of research projects, no matter good or bad, in order to avoid multiple experiments on the same subject leading to the sacrifice of more animals.
2.1. In Vitro Models in Breast Cancer
In vitro experimentation is based on the growth of single or multiple isolated cell lines in culture media. The cells can be grown on flat surfaces, on scaffold-based structures, or on an extracellular matrix. Cell culture can here be interpreted as the maintenance of animal or human cells outside of the donor in a viable state for long periods of time, leading to the creation of new generations of cells [56]. The in vitro models can use the culture of cellular adherent monolayers or suspensions of cells, resulting in two-dimensional models (2D models) or three-dimensional models (3D models).
In vitro models are a good alternative to implement the 3Rs Principles, since they may reduce the extension of the animal experimentation used, although in vitro experimentation also uses components from animal sources.
In vitro experiments allow studying several intracellular and intercellular mechanisms under controllable environmental conditions, such as cellular signaling pathways, metabolism, the absorption and excretion of substances, metastasis, and cellular proliferation. This type of experiments is frequently used because it allows the study of various cellular mechanisms and also because there is a vast list of cell lines available. Moreover, this method is considered to be easy and cheap to maintain compared to others. Nonetheless, it also presents some disadvantages such as the difficult mimicking of the conditions surrounding the cells, the lack of contextualization regarding the microenvironment and immunological response, and the different genetic expression and cellular markers of the cells, since those are the specific features of each individual.
2.1.1. 2D Models
Cell line cultures are one of the tools that researchers use the most in laboratory research in in vitro models. Apart from their advantages of unlimited self-replication and their ease of being handled and preserved, there are already multiple cell lines catalogued into different breast cancer subtypes, as summarized on Table 2, which make them meaningful models. Nonetheless, cell lines are known to accumulate genetic changes and mutations over time, which presents a big disadvantage of cell culture models. This phenomenon led researchers to debate whether or not cell lines are capable of maintaining their characteristics [40], because DNA alterations appear to be two times more common in breast cancer cell lines than in original tumors [15], which can cause phenotypical changes in the cells. Nonetheless, Comparative Genomic Hybridization (CGH) analysis hinted that cell lines are still representative of tumors, and that the same mutations happen in both the tumor and cell lines [57]. Historically, the first breast cancer cell model that was discovered, established, and used was BT-20, in 1958 [58]. This cell line was derived from a 74-year-old female with a triple-negative invasive ductal carcinoma, and it was mainly used for pre-clinical studies. Later, in 1973, the Michigan Cancer Foundation discovered the MCF-7 cells [59], which is the most commonly used cell line in breast cancer research [41]. The popularity of MCF-7 is largely due to its exquisite hormone sensitivity through the expression of ER. This fact makes it an ideal model to study hormone response in breast cancer. Afterwards, other cell lines were discovered and categorized in tumor subtypes. The second most used breast cancer cell line is MDA-MB-231, followed by the T47D cell line [41].

Table 2.
Characterization of breast cancer cell lines based on the receptors they express: Estrogen Receptor (ER), Progesterone Receptor (PR), Androgen Receptor (AR), and Human Epidermal Growth Factor Receptor 2 (HER2); and classification of the tumor they origin, into breast cancer subtypes depending on the receptors they express: Luminal A (LA), Luminal B (LB), HER2 Enriched (H), Triple-Negative A or basal-like (TNA), and Triple-Negative B or normal-like (TNB); and on their histopathological features: benign tumor (B), adenocarcinoma (AC), ductal carcinoma (DC), invasive ductal carcinoma (IDC), invasive lobular carcinoma (ILC), inflammatory carcinoma (InfC), inflammatory ductal carcinoma (InfDC), medullary carcinomas (MC), or squamous carcinoma (SqC). Additional information on the culture medias more commonly used for each cell line: Ham’s F12 liquid medium (HAM’s F12), Dulbecco’s Modified Eagle’s Medium (DMEM), Roswell Park Memorial Institute (RPMI) medium and Minimal Essential Medium Eagle-alpha modified with nucleosides and DFC1 medium (α-MEM/DFC1) [11,15,16,18,66,67,68,69,70,71,72,73].
The cell line nomenclature [15,60] is based not on the phenotypical characteristics of the cells, but on their origin: if they were isolated at the same laboratory, if they were from the same patient, or if they were isolated by serial subculture from the same initial population. For instance, the MCF series means that the cells were isolated at the Michigan Cancer Foundation [59]; the MDA series of cells were established at the M. D. Anderson Hospital and Tumor Institute [61]; 21 series were established from a patient diagnosed as having an infiltrating and intra-ductal mammary carcinoma (regardless their phenotypes or genotypes) [62]; and the SUM series of cells were isolated from different tumor species but established using the same selective media [63].
Molecular classification of the cell lines is very important, since it will indicate which cell line to choose, preferably depending on the end goal and the characteristics of the study [15,19,41]. Based on gene expression profiling, the cells in Table 2 are characterized by the status of four receptors normally used in breast cancer subtyping: ER, PR, AR and HER2. The nomenclature adopted to characterize the tumors depends on the receptors they express—Luminal A, Luminal B, HER2-Enriched and Triple-Negative—and the tumor type: adenocarcinoma (AC), benign tumor (B), carcinoma (C), ductal carcinoma (DC), invasive ductal carcinoma (IDC), invasive lobular carcinoma (ILC), inflammatory ductal carcinoma (InfDC), medullary carcinoma (MC), squamous carcinoma (SqC) or non-available information (NA).
The culture medium, or basic medium, is also an important factor to take into consideration regarding cells’ needs, once each cell line has specific conditions to maintain its physiological and molecular features. In order to make the culture medium as similar to tumor microenvironments as possible, some refinement methods can be adopted, which can include the addition of elements such as glucose, amino acids, vitamins, inorganic salts, and even serums to provide attachment factors, hormones, and growth factors [64,65]. The main basic media utilized are [65] Ham’s F12 liquid medium, Dulbecco’s Modified Eagle’s Medium (DMEM), and the Roswell Park Memorial Institute (RPMI) medium. The most commonly used culture media for each cell line are also presented in Table 2.
To conclude, despite all the advantages that cell lines present, they are still viewed as basic models that are unable to capture all the features and heterogeneity of cancer, so more additional advanced models must be addressed.
2.1.2. 3D Models
Biological systems are characterized by constant dynamic interactions between multiple elements, which complicate the understanding, mimicking, and knowledge of biological mechanisms, either in normal physiological conditions or altered states. By altered states, it is meant in the presence of pathology or in response to different environmental conditions, such as contact with chemicals, exposure to radiation, lack of nutrients, etc. Tumors present highly differentiated and heterogeneous structures [78], whose growth and proliferation are based on complex interactions between multiple cell types and the surrounding environment. This fact hinders the understanding of biological mechanisms and the discovery of more efficient treatments, since it requires the creation and use of more complex models that are able to successfully mimic complex tumor microenvironments.
It was in response to the need of developing more complex models that 3D in vitro models appeared. 3D in vitro models can be developed from previously well-known, isolated, and commercialized cell lines or from patient-derived samples collected through biopsies. They allow studying cell–cell interactions and cell–extracellular matrix (ECM) interactions. Moreover, 3D in vitro models make it possible to analyze tumor formation, progression, and metastasis. This class of models includes tissue slice models, organoids, spheroids, and scaffold-based models.
Tissue Slice Models
Tissue slice models consist of thin slices of tissues collected from patients through biopsies, retaining their native state and preserving cell–cell and cell–ECM connections [79]. These models are very delicate to maintain and are subjected to higher ethical concerns once they use patient samples.
The preservation of the interactions between cells and cells and ECM makes the diffusion of the medium through the tissues possible [79], allowing the application of these models to study drug response [80] and the use of adenoviral vector strategies to develop novel gene therapies and virotherapies [81,82,83].
Organoids
Organoids are 3D assemblies of cells of one or more types with histologic arrangements at the micron scale similar to in vivo tumors [79,84]. More commonly, organoids are composed of malignant cells and supporting cells from the tumor microenvironment, such as fibroblasts, leucocytes, and endothelial cells, among others. Frequently, the cells used to create organoids are derived from patient samples obtained through biopsies, which leads to the formation of 3D structures that are able to mimic the tissue from where they were derived, representing a more realistic model. When patient samples are used, one of the techniques proposed to produce organoids is based on the disaggregation of the sample tissue followed by the reaggregation of the cells, allowing analysis of the ability of the cells to self-organize.
The use of organoids in breast cancer research has been proposed to study the interaction between fibroblasts and epithelial breast tumor cells [85], to study tumor progression [86], and to predict a drug response [87]. Nevertheless, these structures also present some drawbacks, namely the lack of blood flow. Some organoids are able to develop a vascular system, although it requires very complex production techniques, and the results are very variable [84].
Spheroids
Three-dimensional 3D spheroid models include sophisticated structures composed by one or more types of cells which, when grown enough, present a necrotic core surrounded by actively proliferating layers of cells [79,88]. The complex formed also includes an extracellular matrix (ECM) formed of proteins produced by the cells. These models are very similar to in vivo tumors, since they present similar genetic expression, growth kinetics, and cellular heterogeneity [88]. Moreover, the presence of a natural ECM allows mimicking a natural barrier to study the drug penetration, while the internal structure of the spheroids represents a great mimicking of in vivo tumors, where the internal region appears necrotic due to the deprivation of nutrients and oxygen.
Several methodologies were already proposed to produce 3D spheroids [79,88], such as the spinner flask technique, in which a suspension of cells is continuously stirred, encouraging the formation of cell aggregates; the liquid overlay technique [89,90], where the cells are cultured in non-adherent surfaces, supporting the formation of aggregates rather than adherence to the flask surface; the hanging drop technique [91], in which cells are cultured as droplets of medium on the lid of a petri dish, and the lid is turned over and placed on the bottom petri dish, followed by the culture of the cells until the spheroids reach the desired size; and magnetic levitation [92,93], where the cells aggregate in response to the application of magnetic fields. The methodology used will determine the production cost, since some techniques imply the use of specific materials and/or equipment. Moreover, the chosen production technique will influence the homogeneity of the spheroids obtained, which despite the efforts made so far is still a major concern.
Due to their potential as similar models to in vivo tumors, 3D spheroids have been increasingly explored [88,94]. They have already been used to study breast cancer cells invasiveness [91] and in drug delivery systems efficacy and distribution studies [89,90].
Scaffold-Based Models
Three-dimensional scaffold-based models rely on the growth of cells in 3D structures that will, in a certain way, mimic the extracellular matrix. The structures used in the models can be made of natural (e.g., collagen), semi-synthetic (e.g., chitosan), or synthetic (e.g., polycaprolactone) biomaterials [88]. Moreover, the cells can be cultured on previously fabricated/synthesized structures [95], or they can be 3D bio-printed [96]. The use of structural entities allows modeling small complex shapes [79], while simultaneously, when simulating the ECM, it creates some resistance to drug penetration, which is similar to what happens in in vivo tumors. Moreover, these models are usually characterized by spontaneous cell organization, possible heterogeneity, gene expression, and cellular phenotype, similar to in vivo tumors [88]. Despite the undeniable advantages of these models, they present some hindrances as well, such as the use of artificial ECM structures. Furthermore, it is required to use structural materials with high biocompatibility that are able to support the cells, which in the case of bio-printed structures must also be efficiently printable.
These models have been reported in projects involving the study of breast cancer’ initiation, its hypoxia-driven cancer progression [97], and its response to treatments [95,98]. Additionally, they have been studied to replicate the tumor microenvironment and to be used in drug screening assays [99].
2.2. In Vivo Models in Breast Cancer
As mentioned above, the understanding of the mechanisms underlying breast cancer pathology and the development of new diagnostic and therapeutic options depend on the ability of animal models to mimic the tumors’ properties and environment. When considering in vivo models, there are different animal species already reported, such as cats, dogs, rats, and mice [100,101].
2.2.1. Canine and Feline Models
Cats and dogs are natural models to study mammary tumors because they develop spontaneous malignant tumors with similar characteristics to human tumors, namely histopathological features [102,103]. They have genetic variability comparable to humans and might be exposed to several carcinogens, similar to the ones to which humans [104] are prone.
To study the basis of age onset, incidence, and pattern of metastasis, cats are good models [103,105], presenting histological characteristics more similar to human breast cancer than murine and canine models [103,106]. Some feline mammary carcinomas present particular characteristics, such as overexpression of the HER2 receptor [107], tyrosine kinase receptor [105], and cyclin A [103], and the nuclear accumulation of the p53 [103] gene, making them suitable natural models of hormone-independent, HER2 overexpressing, and aggressive human breast cancer [38].
Canine models of mammary tumors have also similar characteristics to human breast cancer, namely the biological behavior and histological type of tumors [108]. Moreover, canine mammary tumors have comparable epidemiological factors, such as age-related incidence rate and the protective effect of early pregnancy [108,109]. HER2 overexpression and the deregulation of BRCA1 and BRCA2 genes, which are related to development and progression of breast tumors in humans, are also found in dogs’ mammary tumors [109,110,111]. Furthermore, genetic alterations in the p53 gene in dogs can be associated with the development of canine mammary carcinomas and their increased malignancy [107,112].
2.2.2. Murine Models
In the European Union, the species more frequently used in experimental research are rats, Mus musculus, and mice, Rattus norvergicus [101,113]. Rats and mice present multiple advantages when compared with other animal species, such as they are small and relatively cheap animals; they are easy to accommodate and manipulate; and they have many similarities with humans, namely a 98% genetic homology [114], which is one of the most important characteristics [100,101,113,115,116,117,118]. Two of the main differences between these two rodents species are size and weight, with mice possibly weighting 10 times less than rats [119,120].
In breast cancer-induced models, rat models, similar to what is seen in human breast cancer, present a high frequency of hormone dependence and show a tumor progression from ductal hyperplasia and ductal carcinoma in situ [121,122,123]. Those characteristics make them good models to study the development of breast cancer. Contrary to rats, mice models when compared with humans have a low frequency of hormone dependence and show a tumor progression from alveolar hyperplasia [121,122,123]. Regardless, they are good models to study the effect of genetic manipulations.
Within each species, the choice of the strain is a very important factor, since some features, such as the genetic variability, might affect the experimental design and the reproducibility of the models [124]. Depending on the genetic uniformity of laboratory rodents’ colonies, they can be divided into inbred and outbred. Inbred animals are characterized by genetic uniformity, as a result of more than 20 generations of brother–sister mating or the equivalent. This class of animals is very useful to reduce the number of animals used in studies, to increase statistical power, and to improve experimental reproducibility, which represents major advantages when compared to the use of outbred colonies. There are different inbred colonies of rats—namely Fischer, F-344, and Lewis—and of mice, such as BALB/c and C57BL/6 [124,125,126,127]. In contrast, outbred colonies have no genetic uniformity, which is important to maintain the individual diversity, a characteristic that seems to be more similar to what happens in the human population. Moreover, outbred colonies allow increasing the fecundity and reducing costs [126,128]. There are different outbred colonies of rats, such as Sprague–Dawley, Wistar, and Long-Evans; and of mice, such as Black Swiss and CD-1 [124,125,126,127].
Researchers look for animal models that focus on particular features that better replicate the human breast cancer. However, this is a multifactorial and polygenic disease with significant morphological, histopathological, and molecular variation, as mentioned above. For this reason, it’s difficult to find a single model representative of all human breast cancers. Nonetheless, a set of models can be used, with each one of them representing a particular subtype of tumor or characteristics of the disease [129,130].
There are numerous animal models reported to simulate different human cancers [131], specially breast cancer-induced models, which can include environmental models with chemical induction, transplanted tumors, and genetically engineered models [131].
Chemically-Induced Models
Environmental models consist in the induction of tumors through contact with chemical compounds, namely, compounds to which people can be exposed daily, through different pathways. Currently, there are only two chemical carcinogenic agents (7,12-dimethylbenzantracene (DMBA) and N-methyl-N-nitrosourea (NMU)) that are able to induce mammary tumors in rats that exhibit histopathological characteristics and genetic alterations similar to those described in humans [101,132]. Both compounds are complete carcinogens that are able to induce cancer without the additional functions of tumor-promoting agents, and, in rats, they are capable of inducing hormone-dependent tumors [131,133,134,135]. In contrast, in mice, the tumors induced by the mentioned carcinogens tend to be hormone-independent [136]. DMBA is a compound that is present in cigarette smoke, coal, petrol, diesel engines, meat grilling, and roasting, that can be absorbed by the skin, respiratory, and gastrointestinal tracts. The inhalation of cigarette smoke can be one of the main examples of the human exposure to DMBA [131,137,138,139,140,141,142]. DMBA is a classical polycyclic aromatic hydrocarbon (PAH), an indirect-action carcinogen, that needs to be previously bioactivated by cytochrome P-450 located in the liver, and that can form adducts with DNA [143,144,145,146]. On the other hand, MNU is a direct alkylating agent by the methylation of guanine nucleosides that does not need previous bioactivation [101,147,148]. A single dose of both compounds can induce mammary tumors in Sprague–Dawley rats [149,150,151,152]. DMBA, administered at 50–56 days of age by gavage, in a single dose from 50 to 100 mg/kg can induce a high number of mammary tumors with 100% incidence in Sprague–Dawley rats [144,153]. Wistar rats can also be used, but recent studies showed that the number of mammary tumors induced in Wistar rats is lower when comparing with Sprague–Dawley rats [101,154]. A single intraperitoneal injection of MNU, at 50 days of age, with a dose of 50 mg/kg, is also capable of inducing mammary tumors with 100% incidence in Sprague–Dawley rats [101,117,148,155,156]. MNU can also be administered by other routes such as subcutaneously, intravenously, and by gavage, but the number of tumors induced is higher by intraperitoneal administration [101,157]. Comparing the mammary tumors induced with MNU and DMBA, MNU-induced mammary tumors tend to be locally more aggressive and spontaneously metastasized [158], and they can be used to induce other types of cancer models such as gastric, colorectal, and esophageal cancers [159,160,161]. Meanwhile, DMBA-induced tumors tend to be less aggressive, lacking local invasion and metastasis to distant organs [158,162].
Tumor latency is inversely related to carcinogen dose, whereas tumor incidence and the number of tumors per animal is related to the age of animals at the time of the carcinogen administration [163,164]. The age of the animals at the carcinogen administration time is considered to be the crucial point for carcinogenesis initiation [132,163,165]. The optimal age for carcinogen administration occurs between 45 and 60 days age, corresponding to the age of rat puberty, which is the stage where a faster expansion of the mammary gland epithelium occurs. At this age, the terminal end buds (TEBs) of mammary glands are numerous and differentiating into alveolar buds and terminal ducts, which maximizes the number of foci of tumoral initiation [132,163]. After this time, there is a decrease in the number of undifferentiated structures and the susceptibility of mammary glands to carcinogenesis decreases [166]. Similarly to what happens in rats, in human breast cancer during early adulthood, the cell replication rate is maximal, which is a phenomenon that tends to decrease considerably with age. This indicates that at the puberty age, the cellular turnover is faster, and consequently, there are increased chances for neoplastic processes to be initiated [166]. All this highlights the importance of the carcinogenesis initiation at the early adulthood stage.
In summary, chemically induced models have been widely studied and allow the investigation of certain compounds’ effects on the initiation, promotion, and progression of breast cancer. Thus, they represent very useful tools to understand the impact of environmental factors. However, these models are time-consuming, and the tumors generated are still variable regarding their size and location.
Transplanted Tumors Models
Transplanted tumors models consist in the transplantation of living cancer cells in suspension or solid tumors obtained from a donor. They can be classified in cell-derived xenografts (CDX), patient-derived xenografts (PDX), or syngeneic models (allograft models) [167]. When the tumor donor and host are from the same species, the models are classified as allograft. Contrarily, when the tumor donor and host are from different species, they are classified as xenograft models. In the case of xenografts, the host animals must be immunocompromised to achieve greater tolerance to tumor transplantation [131,168]. In both models, if the tumor cells are implanted in the tumor site of origin, they are classified as orthotopic models, the transplantation of mammary tumor cells to the mammary fat pad as an example. On the other hand, when the tumor cells are implanted in a different place from their origin, they are classified as heterotopic models, the transplantation of tumor cells intraperitoneally or intramuscular being examples [101,133,169]. There are different tumor transplantation models, as described in Table 3.

Table 3.
Summary of breast cancer cell-derived xenograft (CDX), patient-derived xenograft (PDX), and syngeneic mouse models [167].
Transplanted tumors models are well-established models that were mainly used to study new drugs’ toxicity and therapeutic value [167]. CDX models are mostly useful to analyze tumor initiation and growth. They can also be used to study the metastatic process, although their clinical predictability is still limited. PDX models are based on the implementation of tumor fragments in murine animals, which leads to the development of tumors with the same features as the original tumor. Those features make these models especially advantageous to explore personalized treatments. Both CDX and PDX models might require the use of immunocompromised animals, which does not mimic what really happens in humans and increases the cost of the models. Syngeneic models present metastatic responses more alike to what is seen in human tumors, making them profitable resources to study the metastatic process and to test new anti-cancer and anti-metastatic drugs. Moreover, they do not require the use of immunocompromised animals, representing a better mimicking of real situations, although expressing genetic and histological properties possibly different from the humans. Additionally, they potentiate the study of novel immunotherapies.
Genetically Engineered Models
Genetically engineered models (GEM) correspond to animal strains with manipulated genetic alterations that can be classified as transgenic, knock-in, or knock-out, depending on if the addition, modification, or removal of DNA sequences occurred, respectively [101,170]. The main alterations involve the overexpression of oncogenes or loss of tumor-suppressor genes [171]. Comparing the number of genetically modified rats and mice, there are more genetically modified mice [172]. However, genetically modified rats can be good models to evaluate the role of Ras, BRCA1, and BRCA2 genes in the progression of malignant mammary tumors [151,152,173,174]. Genetically modified mice models of breast tumors can include the application of growth factors and receptors (TGF-alpha, Erbb2, and IGF-II), nuclear oncogenes (c-myc), viral oncogenes (polyoma middle T and SV40 large T), Ras genes (Ha-Ras and N-Ras), INT genes (Wnt family, INT family, FGF family), growth suppressor genes (p53, TGF-beta, BRCA1, and BRCA2), and genes affecting the cell cycle (cyclin-D1) [43].
Radiation Models
Ionizing radiation can induce tumors in the case of radiation overdose and overtime exposure, and mammary tissue is one of the most sensitive tissues to this kind of radiation [175]. The risk of developing breast cancer in cases of radiation exposure is related to the age at which exposure occurs, the risk being augmented in women under 20 years of age and more than 50 years old [176]. There are different types of radiations with a potential carcinogenic effect, such as atomic radiation (alpha, beta, gamma, and neutrons), X-rays, and ultraviolet radiation [177]. The first report of X-ray irradiation and the induction of mammary cancer was in Sprague–Dawley rats [178]. More recent studies also used other rat strains such as F-344 [179,180], Wistar Furth, WAG, WM, and Lewis [181,182,183,184], but the Sprague–Dawley strain is considered the most likely to develop mammary cancer through radiation exposure [180,185,186]. Mice models can also be used to study radiation carcinogenesis, such as BALB/c [187]. Moreover, different mutant strains such as BALB/c with mutation in tumor suppressor gene p53 [188,189] and mice with double mutation in BRCA1 and p53 (strain unspecified) [190] can also be used. This type of models shows potential for studying the effect of the radiation exposure over the mammary tissue regarding tumor induction. Nonetheless, the data published so far on these models is still controversial, and consequently, more studies are required to prove their value and usefulness.
Despite the variety of in vivo models described in the literature, the choice of the most suitable model for each project is not obvious, so to make this task easier, Table 4 presents a summarized comparison of the main advantages and disadvantages identified by the authors (based on personal opinion) for each type of in vivo model here enumerated.

Table 4.
Summary table exposing the main advantages and disadvantages identified by the authors for each breast cancer in vivo model mentioned in this review article.
2.3. In Silico Models in Breast Cancer
Some of the main challenges and disadvantages of using in vitro and in vivo models are related to the difficulties intrinsic to the maintenance of the models, the high biological variability, and the ethical concerns inherent to the use of biological species. To surpass the mentioned disadvantages, in silico models were created to perform biological studies without using biological entities, allowing higher control over the experiments, to increase the number of parameter/variabilities to test and eliminating all ethical apprehensions [44,45].
In silico models combine complex mathematical equations with potent computational resources to simulate and investigate biological mechanisms. These models can be designed by gathering and analyzing the data obtained from previous in vitro and in vivo experiments [45,191], and/or by applying predictive and modeling techniques. They allow obtaining important information regarding chemical, physiological, and pathophysiological features, but at the moment, their validation is still dependent on in vitro and in vivo experiments.
The applications extent of in silico models is far-reaching. There are examples of models participating in different research fronts, going from the identification and understanding of the disease’s cancer-driver factors [192,193,194,195,196], to its assessment regarding metabolism [197,198] and progression [199,200], to its diagnostic [201,202,203,204] and up to its treatment [205,206,207,208,209,210].
As aforementioned, the majority of the data analyzed for the design and development of in silico models comes from previous in vitro and in vivo experiments, which can be obtained from people belonging to the same group, from already published works [201], or from open sources such as libraries and databases. There are already well-organized and big databases gathering information about several cancers, especially regarding the cancer genome [211,212] and cancer proteome [213]. Moreover, researchers created a more advanced database with a million profiles, collecting information on genes, drugs, and disease states, which led to the creation of the so-called connectivity maps [214], potentiating the development of novel cancer-targeted therapies. Furthermore, some of the data used for creating in silico models can also be obtained from patients’ medical exams [209,215] acquired prior their diagnostic, during the course of their treatments, or afterwards.
Genomic and proteomic data allow the development of computational methods not only to identify cancer-driver mutations and pathways, but also to discover novel drugs and more targeted therapies. The methods used to identify cancer-driver mutations and pathways can rely on [44,192]:
- using known pathways from public databases [194,216,217], which consists of the comparison of genetic modifications related to cancer with pathways already reported in the literature through the application of statistical and machine learning methods;
- network-based methods [193,195,218], identifying cancer-associated genes and pathways related with interactions at the cellular and molecular level in biological networks;
- learning cancer pathways de novo [196,219], making deductions on cancer genes and pathways based on the identification of co-occurrence patterns or mutual exclusivity between genetic aberrations, without replicating state-of-the-art knowledge.
The data extracted from databases and from work already published allow creating simulations of breast cancer fundamental aspects such as tumor growth [199,200], metabolism [197], and solute transport in tumor tissues [198]. Moreover, researchers developed computational methods to evaluate the efficacy of new targeted drugs. The use of machine learning to predict potential targeting sites was also described based on multiple data types, foreseeing, simultaneously, new drug repositioning opportunities and identifying similarities between drug classes [206]. It allowed making assumptions regarding previously unexplained clinical observations. There are also reports of the use of in silico methods combined with in vitro methods to assess the potential of new drugs as anti-cancer agents [207]. Tumors response to a single drug or a combination of drugs was already simulated [205,220], providing a very useful tool for developing more targeted and efficient drugs and for comparing the efficacy of different medicines.
Another sector in which in silico methods present high potentiality is in computer-aided decision making [215] which, likely, allows validating medical assessments regarding diagnosis and treatment options, leading to a reduction of the probability of human error occurrence. The majority of the computer-aided decision methods are related with the classification of the tumor types based on medical exams’ data, such as mammograms [201,202] and infrared spectroscopy [203,204].
Additionally, in silico models can be applied to create virtual tissue models, also known as virtual phantoms, for assessing and predicting different tissues responses to different stimuli. This kind of model can be designed based on medical data or can result from mathematical simulations. When considering the virtual models constructed based on medical data, there are projects reporting the use of computerized tomography data [221,222] and others reporting the use of 3D histopathological reconstructions [223,224]. Other types of models are designed by applying mathematical models, which are used for instance to optimize the acquisition of medical images [225] or by using finite element simulations to estimate the viscoelastic profile of breast tissues [226,227], as an example.
As discussed in this section, in silico models are valuable resources for doing a deeper and more complete analysis of the data obtained from in vitro and in vivo experiments. Moreover, computational simulations of biological systems, structures, and environments offer more controllable and flexible tools with enhanced easiness to evaluate multiple parameters in the same study than in vitro and in vivo systems. Nonetheless, it must be kept in mind that the greater the complexity of the system to study, the more computational power needed, and the extra technical and computational skills required.
2.4. Other Models Helpful in Breast Cancer
In addition to the models already mentioned, there are two different ones worth mentioning: the physical phantoms and 3D microfluidic models.
Physical phantoms are physical structures designed and produced to mimic biological structures’ properties. Phantoms are frequently used to calibrate and optimize diagnostic techniques, such as X-ray-based imaging systems [228,229], microwave breast imaging systems [230,231], and terahertz spectroscopy systems [232]. Another common application of phantoms is to assess radiation doses reaching the tissues upon exposure to a radiation source. They are particularly important to determine the radiation dose reaching the tissues surrounding the irradiated area at radiotherapy sessions [233,234], since it will allow better treatment planning by adjusting the intensity, exposure time, and directionality of the radiation needed. Furthermore, the use of phantoms was already reported to mimic the dielectric properties of healthy and tumor tissues for further applications in novel diagnostic tools using millimeter-wave imaging [235].
3D Microfluidic models, also called on-chip 3D models, are particular models that combine in the same system micron-sized fluidic channels with tubing pumping peripherals, fluids, and cells. Once these models have cells, they could be considered as in vitro models, but since they use a microfluidic system, they will be here considered as a different class of models. The combination of cell culture and microfluidic systems bring multiple advantages for a closer approach to in vivo systems. The fact of using microchannels limits the volume of reagents needed, which makes cost reduction possible. Moreover, the use of circulating fluid allows maintaining a continuous perfusion of the cells and removing the degradation products resultant from the cellular metabolism, which is similar to what happens in vivo, making it possible to keep longer and more stable cell cultures. Furthermore, the fluid is pumped with a laminar flow, which is highly studied, allowing a controllable and predictable behavior, applied not only to the fluid but also to the entities flowing in the fluid, such as particles or cells. In summary, these systems demonstrate a terrific potential to better mimic in vivo tumor microenvironments, which make them ideal for studies of drug screening and disease modeling. On-chip 3D models are frequently used for drug screening assays, existing reports on the creation and characterization of these models [236], and reports already demonstrating the application of the models for drug screening tests [237], such as Carnosic Acid and Doxorubicin [238]. These models are also commonly used for therapy assessment, including the mimicking of magnetic particles guidance through narrow mammary ducts models [239] for developing possible future treatments for the evaluation of photodynamic [240] and photothermal [241] therapy systems. Moreover, these models are ideal candidates for modeling tumors’ invasion mechanisms [242,243,244], tumors’ microenvironments [245] and tumors’ metastatic behavior [246,247]. Despite the undeniable potential and advantages of these models, they also present their weaknesses, such as the complexity to design, produce, and handle the systems’ hardware. Moreover, the use of cells, although presenting less ethical concerns than the use of animals, still implies the use of resources extracted from biological sources.
Overall, a brief comparison of the main advantages and disadvantages identified by the authors (based on personal opinion) for each type of model is summarized in Table 5.

Table 5.
Summary table exposing the main advantages and disadvantages identified by the authors for each breast cancer model mentioned in this review article.
3. Conclusions
Breast cancer was always considered a complex disease, but with the evolution of the research, its complexity was proven and there is still a lot to discover. Research evolution and society pressure has demanded more and more improvements in what is known about cancer fundamentals, diagnostics, and treatments, which boosts novel and innovative projects on breast cancer. These current demands have already led to important and impactful discoveries, which while providing useful answers also demand the development of more advanced tools to fulfill the research needs. Some of those demands include the development of more sophisticated models to allow performing cutting edge and more accurate studies.
This work presents an extensive review on the main models used in breast cancer research, highlighting their main advantages, challenges, disadvantages, and potential depending on the applications. In the authors’ opinion, in vivo models and 3D microfluidic models are overall the most informative and complete models when considering that they do a better job of mimicking live organisms systems. Nonetheless, they present strong disadvantages such as the ethical concerns inherent to the use of animals and the complexity of the microfluidic devices, respectively.
Among the in vivo models, there are several types of models already being used but there is no an ideal one, since it will greatly depend on the focus of the study. In CDX, PDX, and Syngeneic models, the development of tumors is more controlled, and their characteristics are more homogeneous and better defined. However, they use immunocompromised animals, which does not mimic what happens in humans and results in a lack of immune system response. In contrast, chemically induced models in addition to developing more heterogeneous tumors with longer latency do a better job of mimicking what may be underlined in the development of breast cancer in humans. These models might allow the analysis of different types of tumors, their development (carcinogenesis), and the impact of constant exposure to environmental toxic agents. Moreover, this type of model offers higher cost-effectiveness and greater ease of application.
To conclude, the authors believe that there is no single optimal model that is ideal for all the applications or studies, but a combination of multiple models may lead to exquisite results and discoveries and will surpass the main disadvantages inherent to individual models. The choice of the most suitable model will certainly depend on the research topic, the resources available, the preferences of the researcher, and on ethical concerns related to the use of biological resources, so there is no standard model or group of models to be used; instead, it will be tailored to each project.
Author Contributions
Conceptualization, A.S.C., I.V.F. and C.P.R.; methodology and formal analysis, E.C., T.F.-G. and G.C.; writing—original draft preparation, E.C., T.F.-G. and G.C.; writing—review and editing, E.C., T.F.-G., A.S.C., I.V.F. and C.P.R.; supervision, A.S.C., I.V.F. and C.P.R.; funding acquisition, Fundação para a Ciência e Tecnologia (FCT), Portugal. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Fundação para a Ciência e Tecnologia (FCT), Portugal under the Project Reference UID/DTP/04138/2019 and UIDB/00313/2020. T.F.-G. was supported by FCT, Portugal under the Reference SFRH/BD/147306/2019.
Acknowledgments
The authors would like to thank TripleBlue-Pharma Consulting, Lda. (info@tripleblue.pt) for revising and editing the manuscript in order to meet English standards.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Carioli, G.; Malvezzi, M.; Rodriguez, T.; Bertuccio, P.; Negri, E.; La Vecchia, C. Trends and predictions to 2020 in breast cancer mortality in Europe. Breast 2017, 36, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, G.K.; Zhao, X.; Band, H.; Band, V. Histological, molecular and functional subtypes of breast cancers. Cancer Biol. Ther. 2010, 10, 955–960. [Google Scholar] [CrossRef] [PubMed]
- Bansal, C.; Singh, U.S.; Misra, S.; Sharma, K.L.; Tiwari, V.; Srivastava, A.N. Comparative evaluation of the modified Scarff-Bloom-Richardson grading system on breast carcinoma aspirates and histopathology. Cytojournal 2012, 9, 4. [Google Scholar] [CrossRef] [PubMed]
- Singletary, S.E.; Allred, C.; Ashley, P.; Bassett, L.W.; Berry, D.; Bland, K.I.; Borgen, P.I.; Clark, G.; Edge, S.B.; Hayes, D.F.; et al. Revision of the American Joint Committee on Cancer staging system for breast cancer. J. Clin. Oncol. 2002, 20, 3628–3636. [Google Scholar] [CrossRef]
- American Joint Committee on Cancer. AJCC Cancer Staging Manual, 7th ed.; Edge, S.B., Byrd, D.R., Compton, C.C., Fritz, A.G., Greene, F.L., Trotti, A., Eds.; Springer: Chicago, IL, USA, 2010; ISBN 978-0-387-88440-0. [Google Scholar]
- Sotiriou, C.; Pusztai, L. Gene-expression signatures in breast cancer. N. Engl. J. Med. 2009, 360, 790–800. [Google Scholar] [CrossRef]
- Li, J.; Chen, Z.; Su, K.; Zeng, J. Clinicopathological classification and traditional prognostic indicators of breast cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 8500–8505. [Google Scholar]
- Rakha, E.A.; Reis-Filho, J.S.; Ellis, I.O. Combinatorial biomarker expression in breast cancer. Breast Cancer Res. Treat. 2010, 120, 293–308. [Google Scholar] [CrossRef]
- Prat, A.; Karginova, O.; Parker, J.S.; Fan, C.; He, X.; Bixby, L.; Harrell, J.C.; Roman, E.; Adamo, B.; Troester, M.; et al. Characterization of cell lines derived from breast cancers and normal mammary tissues for the study of the intrinsic molecular subtypes. Breast Cancer Res. Treat. 2013, 142, 237–255. [Google Scholar] [CrossRef]
- Eroles, P.; Bosch, A.; Perez-Fidalgo, J.A.; Lluch, A. Molecular biology in breast cancer: Intrinsic subtypes and signaling pathways. Cancer Treat. Rev. 2012, 38, 698–707. [Google Scholar] [CrossRef]
- Giovannelli, P.; Di Donato, M.; Galasso, G.; Di Zazzo, E.; Bilancio, A.; Migliaccio, A. The androgen receptor in breast cancer. Front. Endocrinol. (Lausanne) 2018, 9, 1–8. [Google Scholar] [CrossRef]
- Sunar, V.; Dogan, H.T.; Sarici, F.; Ates, O.; Akin, S.; Baspinar, B.; Aksoy, S.; Altundag, K. Association between androgen receptor status and prognosis in triple negative breast cancer. JBUON 2018, 23, 1325–1330. [Google Scholar]
- Gerratana, L.; Basile, D.; Buono, G.; De Placido, S.; Giuliano, M.; Minichillo, S.; Coinu, A.; Martorana, F.; De Santo, I.; Del Mastro, L.; et al. Androgen receptor in triple negative breast cancer: A potential target for the targetless subtype. Cancer Treat. Rev. 2018, 68, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Perou, C.M.; Borresen-Dale, A.-L. Systems biology and genomics of breast cancer. Cold Spring Harb. Perspect. Biol. 2011, 3. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Cheng, H.; Bai, Z.; Li, J. Breast cancer cell line classification and Its relevance with breast tumor subtyping. J. Cancer 2017, 8, 3131–3141. [Google Scholar] [CrossRef] [PubMed]
- Subik, K.; Lee, J.-F.; Baxter, L.; Strzepek, T.; Costello, D.; Crowley, P.; Xing, L.; Hung, M.-C.; Bonfiglio, T.; Hicks, D.G.; et al. The Expression Patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67 and AR by Immunohistochemical Analysis in Breast Cancer Cell Lines. Breast Cancer (Auckl.) 2010, 4, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Goldhirsch, A.; Winer, E.P.; Coates, A.S.; Gelber, R.D.; Piccart-Gebhart, M.; Thurlimann, B.; Senn, H.-J. Personalizing the treatment of women with early breast cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2013, 24, 2206–2223. [Google Scholar] [CrossRef]
- Vuong, D.; Simpson, P.T.; Green, B.; Cummings, M.C.; Lakhani, S.R. Molecular classification of breast cancer. Virchows Arch. 2014, 465, 1–14. [Google Scholar] [CrossRef]
- Holliday, D.L.; Speirs, V. Choosing the right cell line for breast cancer research. Breast Cancer Res. 2011, 13, 215. [Google Scholar] [CrossRef]
- American Cancer Society Breast Cancer: Treatment and Side Effects. Available online: https://www.cancer.org/cancer/breast-cancer.html (accessed on 7 December 2019).
- American Cancer Society. Breast Cancer: Treatment Guideline for Patients, Version VIII. J. Natl. Compr. Cancer Netw. 2006. [Google Scholar]
- Waks, A.G.; Winer, E.P. Breast Cancer Treatment: A Review. JAMA 2019, 321, 288–300. [Google Scholar] [CrossRef]
- Gradishar, W.J.; Anderson, B.O.; Balassanian, R.; Blair, S.L.; Burstein, H.J.; Cyr, A.; Elias, A.D.; Farrar, W.B.; Forero, A.; Giordano, S.H.; et al. Clinical practice guidelines in oncology. J. Natl. Compr. Cancer Netw. 2018, 16, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Castaneda, S.A.; Strasser, J. Updates in the Treatment of Breast Cancer with Radiotherapy. Surg. Oncol. Clin. N. Am. 2017, 26, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Wu, H.; Gao, F.; Su, Y.; Li, Q.; Liu, S.; Cai, J. Brachytherapy in the treatment of breast cancer. Int. J. Clin. Oncol. 2017, 22, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Giugliano, F.M.; Falivene, S.; Esposito, E.; Di Franco, R.; Muto, M.; D’Aiuto, M.; Muto, P. External radiotherapy for breast cancer in the elderly. Aging Clin. Exp. Res. 2017, 29, 149–157. [Google Scholar] [CrossRef]
- Meisel, J.L.; Venur, V.A.; Gnant, M.; Carey, L. Evolution of Targeted Therapy in Breast Cancer: Where Precision Medicine Began. In American Society of Clinical Oncology Educational Book; American Society of Clinical Oncology: Alexandria, VA, USA, 2018; Volume 38, pp. 78–86. [Google Scholar] [CrossRef]
- Munagala, R.; Aqil, F.; Gupta, R. Promising molecular targeted therapies in breast cancer. Indian J. Pharmacol. 2011, 43, 236–245. [Google Scholar] [CrossRef]
- Liyanage, P.Y.; Hettiarachchi, S.D.; Zhou, Y.; Ouhtit, A.; Seven, E.S.; Oztan, C.Y.; Celik, E.; Leblanc, R.M. Nanoparticle-mediated targeted drug delivery for breast cancer treatment. Biochim. Biophys. Acta Rev. Cancer 2019, 1871, 419–433. [Google Scholar] [CrossRef]
- Tray, N.; Adams, S.; Esteva, F.J. Antibody-drug conjugates in triple negative breast cancer. Future Oncol. 2018, 14, 2651–2661. [Google Scholar] [CrossRef]
- Perez-Garcia, J.; Munoz-Couselo, E.; Cortes, J.; Scaltriti, M. Therapeutic antibodies in breast cancer. Semin. Oncol. 2014, 41, 576–588. [Google Scholar] [CrossRef]
- Ernst, B.; Anderson, K.S. Immunotherapy for the treatment of breast cancer. Curr. Oncol. Rep. 2015, 17, 5. [Google Scholar] [CrossRef]
- Heimes, A.-S.; Schmidt, M. Atezolizumab for the treatment of triple-negative breast cancer. Expert Opin. Investig. Drugs 2019, 28, 1–5. [Google Scholar] [CrossRef]
- Schmid, P.; Adams, S.; Rugo, H.S.; Schneeweiss, A.; Barrios, C.H.; Iwata, H.; Dieras, V.; Hegg, R.; Im, S.-A.; Shaw Wright, G.; et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N. Engl. J. Med. 2018, 379, 2108–2121. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Xu, J.; Tang, L.; Guan, X. Breast cancer stem cell: The roles and therapeutic implications. Cell. Mol. Life Sci. 2017, 74, 951–966. [Google Scholar] [CrossRef] [PubMed]
- Giovannelli, P.; Di Donato, M.; Galasso, G.; Di Zazzo, E.; Medici, N.; Bilancio, A.; Migliaccio, A.; Castoria, G. Breast cancer stem cells: The role of sex steroid receptors. World J. Stem Cells 2019, 11, 594–603. [Google Scholar] [CrossRef] [PubMed]
- O’Conor, C.J.; Chen, T.; González, I.; Cao, D.; Peng, Y. Cancer stem cells in triple-negative breast cancer: A potential target and prognostic marker. Biomark. Med. 2018, 12, 813–820. [Google Scholar] [CrossRef] [PubMed]
- El-Abd, E.; Shalaby, E.; Matalkah, F. Animal Models of Breast Cancer. In Omics Approaches in Breast Cancer: Towards Next-Generation Diagnosis, Prognosis and Therapy; Springer: Berlin, Germany, 2014; pp. 297–314. [Google Scholar]
- Barré-Sinoussi, F.; Montagutelli, X. Animal models are essential to biological research: Issues and perspectives. Future Sci. OA 2015, 1, 4–6. [Google Scholar] [CrossRef] [PubMed]
- Burdall, S.E.; Hanby, A.M.; Lansdown, M.R.J.; Speirs, V. Breast cancer cell lines: Friend or foe? Breast Cancer Res. 2003, 5, 89–95. [Google Scholar] [CrossRef]
- Liu, K.; Newbury, P.A.; Glicksberg, B.S.; Zeng, W.Z.D.; Paithankar, S.; Andrechek, E.R.; Chen, B. Evaluating cell lines as models for metastatic breast cancer through integrative analysis of genomic data. Nat. Commun. 2019, 10, 2138. [Google Scholar] [CrossRef]
- Holen, I.; Speirs, V.; Morrissey, B.; Blyth, K. In vivo models in breast cancer research: Progress, challenges and future directions. Dis. Model. Mech. 2017, 10, 359–371. [Google Scholar] [CrossRef]
- Akbari Bazm, M.; Naseri, L.; Khazaei, M. Methods of inducing breast cancer in animal models: A systemic review. World Cancer Res. J. 2018, 5, 1182. [Google Scholar]
- Jean-Quartier, C.; Jeanquartier, F.; Jurisica, I.; Holzinger, A. In silico cancer research towards 3R. BMC Cancer 2018, 18, 1–12. [Google Scholar] [CrossRef]
- Colquitt, R.B.; Colquhoun, D.A.; Thiele, R.H. In silico modelling of physiologic systems. Best Pract. Res. Clin. Anaesthesiol. 2011, 25, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Sneddon, L.U.; Halsey, L.G.; Bury, N.R. Considering aspects of the 3Rs principles within experimental animal biology. J. Exp. Biol. 2017, 220, 3007–3016. [Google Scholar] [CrossRef] [PubMed]
- Parker, R.M.A.; Browne, W.J. The place of experimental design and statistics in the 3Rs. Inst. Lab. Anim. Res. J. 2014, 55, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Franco, N.H.; Olsson, I.A.S. Scientists and the 3Rs: Attitudes to animal use in biomedical research and the effect of mandatory training in laboratory animal science. Lab. Anim. 2014, 48, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Workman, P.; Aboagye, E.O.; Balkwill, F.; Balmain, A.; Bruder, G.; Chaplin, D.J.; Double, J.A.; Everitt, J.; Farningham, D.A.H.; Glennie, M.J.; et al. Guidelines for the welfare and use of animals in cancer research. Br. J. Cancer 2010, 102, 1555–1577. [Google Scholar] [CrossRef] [PubMed]
- Guillen, J. FELASA guidelines and recommendations. J. Am. Assoc. Lab. Anim. Sci. 2012, 51, 311–321. [Google Scholar]
- Russel, W.M.S.; Burch, R.L. The Principles of Humane Experimental Technique; Methuen & Co. Ltd.: London, UK, 1959. [Google Scholar]
- Tannenbaum, J.; Bennett, B.T. Russell and Burch’s 3Rs then and now: The need for clarity in definition and purpose. J. Am. Assoc. Lab. Anim. Sci. 2015, 54, 120–132. [Google Scholar]
- Council of Europe. European Convention for the Protection of Vertebrate Animals used for Experimental and Other Scientific Purposes; European Treaty Series No. 123; Council of Europe: Strasbourg, France, 1986. [Google Scholar]
- The European Parliament and the Council of the European Union. Directive 2010/63/EU of the European Parliament and of the Council of 22 September 2010 on the protection of animals used for scientific purposes Text with EEA relevance. Off. J. Eur. Union 2010, 53, 33–79. [Google Scholar] [CrossRef]
- European Animal Research Association. European Animal Research Association: Goals and Milestones. Available online: http://eara.eu/en/about-us/info-brochure/ (accessed on 8 December 2019).
- Cavanaugh, P.; Haier, J. Basic Tissue and Cell Culture in Cancer Research. In The Cancer Handbook; American Cancer Society: Atlanta, GA, USA, 2007; ISBN 9780470025079. [Google Scholar]
- Forozan, F.; Mahlamaki, E.H.; Monni, O.; Chen, Y.; Veldman, R.; Jiang, Y.; Gooden, G.C.; Ethier, S.P.; Kallioniemi, A.; Kallioniemi, O.P. Comparative genomic hybridization analysis of 38 breast cancer cell lines: A basis for interpreting complementary DNA microarray data. Cancer Res. 2000, 60, 4519–4525. [Google Scholar]
- Lasfargues, E.Y.; Ozzello, L. Cultivation of Human Breast Carcinomas. JNCI J. Natl. Cancer Inst. 1958, 21, 1131–1147. [Google Scholar] [CrossRef]
- Soule, H.D.; Vazquez, J.; Long, A.; Albert, S.; Brennan, M. A Human Cell Line From a Pleural Effusion Derived From a Breast Carcinoma2. JNCI J. Natl. Cancer Inst. 1973, 51, 1409–1416. [Google Scholar] [CrossRef] [PubMed]
- International Cell Line Authentication Committee Naming a Cell Line. Available online: https://iclac.org/resources/cell-line-names/ (accessed on 9 December 2019).
- Cailleau, R.; Olive, M.; Cruciger, Q.V. Long-term human breast carcinoma cell lines of metastatic origin: Preliminary characterization. In Vitro 1978, 14, 911–915. [Google Scholar] [CrossRef] [PubMed]
- Band, V.; Zajchowski, D.; Swisshelm, K.; Trask, D.; Kulesa, V.; Cohen, C.; Connolly, J.; Sager, R. Tumor progression in four mammary epithelial cell lines derived from the same patient. Cancer Res. 1990, 50, 7351–7357. [Google Scholar] [PubMed]
- Ethier, S.P.; Mahacek, M.L.; Gullick, W.J.; Frank, T.S.; Weber, B.L. Differential isolation of normal luminal mammary epithelial cells and breast cancer cells from primary and metastatic sites using selective media. Cancer Res. 1993, 53, 627–635. [Google Scholar]
- Selenius, L.A.; Wallenberg Lundgren, M.; Jawad, R.; Danielsson, O.; Bjornstedt, M. The Cell Culture Medium Affects Growth, Phenotype Expression and the Response to Selenium Cytotoxicity in A549 and HepG2 Cells. Antioxidants 2019, 8, 130. [Google Scholar] [CrossRef]
- Arora, M. Cell Culture Media: A Review. Mater. Methods 2013, 3. [Google Scholar] [CrossRef]
- Garcia-Carpizo, V.; Ruiz-Llorente, S.; Sarmentero, J.; Gonzalez-Corpas, A.; Barrero, M.J. CREBBP/EP300 bromodomain inhibition affects the proliferation of AR-positive breast cancer cell lines. Mol. Cancer Res. 2019, 17, 720–730. [Google Scholar] [CrossRef]
- Barton, V.N.; D’Amato, N.C.; Gordon, M.A.; Lind, H.T.; Spoelstra, N.S.; Babbs, B.L.; Heinz, R.E.; Elias, A.; Jedlicka, P.; Jacobsen, B.M.; et al. Multiple Molecular Subtypes of Triple-Negative Breast Cancer Critically Rely on Androgen Receptor and Respond to Enzalutamide In Vivo. Mol. Cancer Ther. 2015, 14, 769–778. [Google Scholar] [CrossRef]
- Riaz, M.; van Jaarsveld, M.T.M.; Hollestelle, A.; Prager-van der Smissen, W.J.C.; Heine, A.A.J.; Boersma, A.W.M.; Liu, J.; Helmijr, J.; Ozturk, B.; Smid, M.; et al. miRNA expression profiling of 51 human breast cancer cell lines reveals subtype and driver mutation-specific miRNAs. Breast Cancer Res. 2013, 15, R33. [Google Scholar] [CrossRef]
- Neve, R.M.; Chin, K.; Fridlyand, J.; Yeh, J.; Baehner, F.L.; Fevr, T.; Clark, L.; Bayani, N.; Coppe, J.-P.; Tong, F.; et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 2006, 10, 515–527. [Google Scholar] [CrossRef]
- Hollestelle, A.; Nagel, J.H.A.; Smid, M.; Lam, S.; Elstrodt, F.; Wasielewski, M.; Ng, S.S.; French, P.J.; Peeters, J.K.; Rozendaal, M.J.; et al. Distinct gene mutation profiles among luminal-type and basal-type breast cancer cell lines. Breast Cancer Res. Treat. 2010, 121, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Kao, J.; Salari, K.; Bocanegra, M.; Choi, Y.-L.; Girard, L.; Gandhi, J.; Kwei, K.A.; Hernandez-Boussard, T.; Wang, P.; Gazdar, A.F.; et al. Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PLoS ONE 2009, 4, e6146. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, M.; Leclercq, G. Relevance of breast cancer cell lines as models for breast tumours: An update. Breast Cancer Res. Treat. 2004, 83, 249–289. [Google Scholar] [CrossRef] [PubMed]
- Speers, C.; Zhao, S.G.; Chandler, B.; Liu, M.; Wilder-Romans, K.; Olsen, E.; Nyati, S.; Ritter, C.; Alluri, P.G.; Kothari, V.; et al. Androgen receptor as a mediator and biomarker of radioresistance in triple-negative breast cancer. NPJ Breast Cancer 2017, 3, 1–10. [Google Scholar] [CrossRef]
- Kurebayashi, J.; Kurosumi, M.; Sonoo, H. A new human breast cancer cell line, KPL-1 secretes tumour-associated antigens and grows rapidly in female athymic nude mice. Br. J. Cancer 1995, 71, 845–853. [Google Scholar] [CrossRef]
- Briand, P.; Lykkesfeldt, A.E. An in vitro model of human breast carcinogenesis: Epigenetic aspects. Breast Cancer Res. Treat. 2001, 65, 179–187. [Google Scholar] [CrossRef]
- Micci, F.; Teixeira, M.R.; Heim, S. Complete cytogenetic characterization of the human breast cancer cell line MA11 combining G-banding, comparative genomic hybridization, multicolor fluorescence in situ hybridization, RxFISH, and chromosome-specific painting. Cancer Genet. Cytogenet. 2001, 131, 25–30. [Google Scholar] [CrossRef]
- Rye, P.D.; Norum, L.; Olsen, D.R.; Garman-Vik, S.; Kaul, S.; Fodstad, Ø. Brain metastasis model in athymic nude mice using a novel MUC1-secreting human breast-cancer cell line, MA11. Int. J. Cancer 1996, 68, 682–687. [Google Scholar] [CrossRef]
- Stadler, M.; Walter, S.; Walzl, A.; Kramer, N.; Unger, C.; Scherzer, M.; Unterleuthner, D.; Hengstschläger, M.; Krupitza, G.; Dolznig, H. Increased complexity in carcinomas: Analyzing and modeling the interaction of human cancer cells with their microenvironment. Semin. Cancer Biol. 2015, 35, 107–124. [Google Scholar] [CrossRef]
- Roberts, S.; Peyman, S.; Speirs, V. Current and Emerging 3D Models to Study Breast Cancer. In Breast Cancer Metastasis and Drug Resistance: Challenges and Progress; Ahmad, A., Ed.; Springer: Cham, Switzerland, 2019; pp. 413–427. ISBN 978-3-030-20301-6. [Google Scholar]
- Grosso, S.H.G.; Katayama, M.L.H.; Roela, R.A.; Nonogaki, S.; Soares, F.A.; Brentani, H.; Lima, L.; Folgueira, M.A.A.K.; Waitzberg, A.F.L.; Pasini, F.S.; et al. Breast cancer tissue slices as a model for evaluation of response to rapamycin. Cell Tissue Res. 2013, 352, 671–684. [Google Scholar] [CrossRef]
- Pennington, K.; Chu, Q.D.; Curiel, D.T.; Li, B.D.L.; Mathis, J.M. The utility of a tissue slice model system to determine breast cancer infectivity by oncolytic adenoviruses. J. Surg. Res. 2010, 163, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Stoff-Khalili, M.A.; Stoff, A.; Rivera, A.A.; Banerjee, N.S.; Everts, M.; Young, S.; Siegal, G.P.; Richter, D.F.; Wang, M.; Dall, P.; et al. Preclinical evaluation of transcriptional targeting strategies for carcinoma of the breast in a tissue slice model system. Breast Cancer Res. 2005, 7, R1141–R1152. [Google Scholar] [CrossRef] [PubMed]
- Stoff-Khalili, M.A.; Stoff, A.; Rivera, A.A.; Mathis, J.M.; Everts, M.; Wang, M.; Kawakami, Y.; Waehler, R.; Mathews, Q.L.; Yamamoto, M.; et al. Gene transfer to carcinoma of the breast with fiber-modified adenoviral vectors in a tissue slice model system. Cancer Biol. Ther. 2005, 4, 1203–1220. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.A. Chapter 1—Organoids and mini-organs: Introduction, history, and potential. In Organs and Organoids; Davies, J.A., Lawrence, M.L., Eds.; Academic Press: Amsterdam, the Netherlands, 2018; pp. 3–23. ISBN 978-0-12-812636-3. [Google Scholar]
- Koledova, Z. 3D Coculture of Mammary Organoids with Fibrospheres: A Model for Studying Epithelial--Stromal Interactions During Mammary Branching Morphogenesis. In 3D Cell Culture: Methods and Protocols; Koledova, Z., Ed.; Springer: New York, NY, USA, 2017; pp. 107–124. ISBN 978-1-4939-7021-6. [Google Scholar]
- Djomehri, S.I.; Burman, B.; Gonzalez, M.E.; Takayama, S.; Kleer, C.G. A reproducible scaffold-free 3D organoid model to study neoplastic progression in breast cancer. J. Cell Commun. Signal. 2019, 13, 129–143. [Google Scholar] [CrossRef]
- Walsh, A.J.; Cook, R.S.; Sanders, M.E.; Aurisicchio, L.; Ciliberto, G.; Arteaga, C.L.; Skala, M.C. Quantitative Optical Imaging of Primary Tumor Organoid Metabolism Predicts Drug Response in Breast Cancer. Cancer Res. 2014. [Google Scholar] [CrossRef]
- Costa, E.C.; Moreira, A.F.; de Melo-Diogo, D.; Gaspar, V.M.; Carvalho, M.P.; Correia, I.J. 3D tumor spheroids: An overview on the tools and techniques used for their analysis. Biotechnol. Adv. 2016, 34, 1427–1441. [Google Scholar] [CrossRef]
- Rodallec, A.; Sicard, G.; Giacometti, S.; Carré, M.; Pourroy, B.; Bouquet, F.; Savina, A.; Lacarelle, B.; Ciccolini, J.; Fanciullino, R. From 3D spheroids to tumor bearing mice: Efficacy and distribution studies of trastuzumab-docetaxel immunoliposome in breast cancer. Int. J. Nanomed. 2018, 13, 6677–6688. [Google Scholar] [CrossRef]
- Ji, Y.; Li, J.; Zhao, J.; Shan, S.; Chu, C.C. A light-facilitated drug delivery system from a pseudo-protein/hyaluronic acid nanocomplex with improved anti-tumor effects. Nanoscale 2019, 11, 9987–10003. [Google Scholar] [CrossRef]
- Brown, M.J.; Bahsoun, S.; Morris, M.A.; Akam, E.C. Determining conditions for successful culture of multi-cellular 3D tumour spheroids to investigate the effect of mesenchymal stem cells on breast cancer cell invasiveness. Bioengineering 2019, 6, 101. [Google Scholar] [CrossRef]
- Jaganathan, H.; Gage, J.; Leonard, F.; Srinivasan, S.; Souza, G.R.; Dave, B.; Godin, B. Three-dimensional in vitro co-culture model of breast tumor using magnetic levitation. Sci. Rep. 2014, 4, 6468. [Google Scholar] [CrossRef]
- Leonard, F.; Godin, B. 3D In Vitro Model for Breast Cancer Research Using Magnetic Levitation and Bioprinting Method. Methods Mol. Biol. 2016, 1406, 239–251. [Google Scholar] [CrossRef]
- Almarshad, H.A.; Madhavan, M.; Hoshino, K. Focused Ion Beam-Based Milling, Imaging and Analysis of 3D Tumor Spheroids. In Proceedings of the 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 17–21 July 2018; pp. 4480–4483. [Google Scholar] [CrossRef]
- Ranamukhaarachchi, S.K.; Modi, R.N.; Han, A.; Velez, D.O.; Kumar, A.; Engler, A.J.; Fraley, S.I. Macromolecular crowding tunes 3D collagen architecture and cell morphogenesis. Biomater. Sci. 2019, 7, 618–633. [Google Scholar] [CrossRef] [PubMed]
- Reid, J.A.; Mollica, P.A.; Bruno, R.D.; Sachs, P.C. Consistent and reproducible cultures of large-scale 3D mammary epithelial structures using an accessible bioprinting platform. Breast Cancer Res. 2018, 20, 122. [Google Scholar] [CrossRef] [PubMed]
- Liverani, C.; De Vita, A.; Minardi, S.; Kang, Y.; Mercatali, L.; Amadori, D.; Bongiovanni, A.; La Manna, F.; Ibrahim, T.; Tasciotti, E. A biomimetic 3D model of hypoxia-driven cancer progression. Sci. Rep. 2019, 9, 12263. [Google Scholar] [CrossRef] [PubMed]
- Mollica, P.A.; Booth-Creech, E.N.; Reid, J.A.; Zamponi, M.; Sullivan, S.M.; Palmer, X.L.; Sachs, P.C.; Bruno, R.D. 3D bioprinted mammary organoids and tumoroids in human mammary derived ECM hydrogels. Acta Biomater. 2019, 95, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, S.; Hamid, Q.; Sun, W.; Clyne, A.M. Bioprinting of 3D breast epithelial spheroids for human cancer models. Biofabrication 2019, 11, 25003. [Google Scholar] [CrossRef]
- Conn, P.M. Animal Models for the Study of Human Disease; Elsevier: Amsterdam, the Netherlands, 2013. [Google Scholar]
- Alvarado, A.; Faustino-Rocha, A.I.; Colaço, B.; Oliveira, P.A. Experimental mammary carcinogenesis—Rat models. Life Sci. 2017, 173, 116–134. [Google Scholar] [CrossRef]
- de las Mulas, J.M.; Reymundo, C. Animal models of human breast carcinoma: Canine and feline neoplasms. Rev. Oncol. 2000, 2, 274–281. [Google Scholar] [CrossRef]
- De Maria, R.; Olivero, M.; Iussich, S.; Nakaichi, M.; Murata, T.; Biolatti, B.; Di Renzo, M.F. Spontaneous Feline Mammary Carcinoma Is a Model of HER2 Overexpressing Poor Prognosis Human Breast Cancer. Cancer Res. 2005, 65, 907–912. [Google Scholar]
- Antuofermo, E.; Miller, M.A.; Pirino, S.; Xie, J.; Badve, S.; Mohammed, S.I. Spontaneous Mammary Intraepithelial Lesions in Dogs—A Model of Breast Cancer. Cancer Epidemiol. Biomark. Prev. 2007, 16, 2247–2256. [Google Scholar] [CrossRef]
- De Maria, R.; Maggiora, P.; Biolatti, B.; Prat, M.; Comoglio, P.M.; Castagnaro, M.; Di Renzo, M.F. Feline STK gene expression in mammary carcinomas. Oncogene 2002, 21, 1785–1790. [Google Scholar] [CrossRef] [PubMed]
- Uva, P.; Aurisicchio, L.; Watters, J.; Loboda, A.; Kulkarni, A.; Castle, J.; Palombo, F.; Viti, V.; Mesiti, G.; Zappulli, V.; et al. Comparative expression pathway analysis of human and canine mammary tumors. BMC Genom. 2009, 10, 135. [Google Scholar] [CrossRef]
- Lee, C.H.; Kim, W.H.; Lim, J.H.; Kang, M.S.; Kim, D.Y.K.O. Mutation and overexpression of p53 as a prognostic factor in canine mammary tumors. J. Vet. Sci. 2004, 5, 63–69. [Google Scholar] [CrossRef] [PubMed]
- MacEwen, E.G.; Patnaik, A.K.; Harvey, H.J.; Panko, W.B. Estrogen Receptors in Canine Mammary Tumors. Cancer Res. 1982, 42, 2255–2259. [Google Scholar] [PubMed]
- Pinho, S.S.; Carvalho, S.; Cabral, J.; Reis, C.A.; Gärtner, F. Canine tumors: A spontaneous animal model of human carcinogenesis. Transl. Res. 2012, 165–172. [Google Scholar] [CrossRef]
- Nieto, A.; Pérez-Alenza, M.D.; Del Castillo, N.; Tabanera, E.; Castaño, M.; Peña, L. BRCA1 Expression in Canine Mammary Dysplasias and Tumours: Relationship with Prognostic Variables. J. Comp. Pathol. 2003, 128, 260–268. [Google Scholar] [CrossRef]
- Klopfleisch, R.; Gruber, A.D. Increased Expression of BRCA2 and RAD51 in Lymph Node Metastases of Canine Mammary Adenocarcinomas. Vet. Pathol. 2009, 46, 416–422. [Google Scholar] [CrossRef]
- Klopfleisch, R.; Gruber, A.D. Differential expression of cell cycle regulators p21, p27 and p53 in metastasizing canine mammary adenocarcinomas versus normal mammary glands. Res. Vet. Sci. 2009, 87, 91–96. [Google Scholar] [CrossRef]
- European Commission. Seventh Report on the Statistics on the Number of Animals Used for Experimental and Other Scientific Purposes in the Member States of the European Union; European Commission: Brussels, Belgium.
- Understanding Animal Research What Is Animal Research? 10 Facts-Mouse. Available online: http://www.understandinganimalresearch.org.uk/animals/10-facts/mouse/ (accessed on 14 January 2020).
- Iannaccone, P.M.; Jacob, H.J. Rats! Dis. Model. Mech. 2009, 2, 206–210. [Google Scholar] [CrossRef]
- Clarke, R. Animal models of breast cancer: Their diversity and role in biomedical research. Breast Cancer Res. Treat. 1996, 39, 1–6. [Google Scholar] [CrossRef]
- Faustino-Rocha, A.I.; Ferreira, R.; Oliveira, P.A.; Gama, A.; Ginja, M. N-Methyl-N-nitrosourea as a mammary carcinogenic agent. Tumor Biol. 2015, 36, 9095–9117. [Google Scholar] [CrossRef] [PubMed]
- Fagundes, D.J.; Taha, M.O. Animal disease model: Choice’s criteria and current animals specimens. Acta Cir. Bras. 2004, 19, 59–65. [Google Scholar] [CrossRef]
- Kjell, J.; Olson, L. Rat models of spinal cord injury: From pathology to potential therapies. Dis. Model. Mech. 2016, 9, 1125–1137. [Google Scholar] [CrossRef] [PubMed]
- Ellenbroek, B.; Youn, J. Rodent models in neuroscience research: Is it a rat race? Dis. Model. Mech. 2016, 9, 1079–1087. [Google Scholar] [CrossRef]
- Thompson, H.J.; Singh, M. Rat Models of Premalignant Breast Disease. J. Mamm. Gland Biol. Neop. 2000, 5, 409–420. [Google Scholar] [CrossRef]
- Nandi, S.; Guzman, R.C.; Yang, J. Hormones and mammary carcinogenesis in mice, rats, and humans: A unifying hypothesis. Proc. Natl. Acad. Sci. USA 1995, 92, 3650–3657. [Google Scholar] [CrossRef]
- Gould, M.N. Rodent models for the study of etiology, prevention and treatment of breast cancer. Semin. Cancer Biol. 1995, 6, 147–152. [Google Scholar] [CrossRef]
- Brekke, T.D.; Steele, K.A.; Mulley, J.F. Inbred or outbred? Genetic diversity in laboratory rodent colonies. G3 Genes Genomes Genet. 2018, 8, 679–686. [Google Scholar] [CrossRef]
- Eppig, J.T. Chapter 5—Mouse Strain and Genetic Nomenclature: An Abbreviated Guide. In The Mouse in Biomedical Research, 2nd ed.; Fox, J.G., Davisson, M.T., Quimby, F.W., Barthold, S.W., Newcomer, C.E., Smith, A.L., Eds.; Academic Press: Burlington, VT, USA, 2007; pp. 79–98. ISBN 978-0-12-369454-6. [Google Scholar]
- Jensen, V.S.; Porsgaard, T.; Lykkesfeldt, J.; Hvid, H. Rodent model choice has major impact on variability of standard preclinical readouts associated with diabetes and obesity research. Am. J. Transl. Res. 2016, 8, 3574–3584. [Google Scholar]
- Festing, M.F.W. Inbred strains should replace outbred stocks in toxicology, safety testing, and drug development. Toxicol. Pathol. 2010, 38, 681–690. [Google Scholar] [CrossRef]
- Gill, T.J. The use of randomly bred and genetically defined animals in biomedical research. Am. J. Pathol. 1980, 101, S21–S32. [Google Scholar] [PubMed]
- Herschkowitz, J.I.; Simin, K.; Weigman, V.J.; Mikaelian, I.; Usary, J.; Hu, Z.; Rasmussen, K.E.; Jones, L.P.; Assefnia, S.; Chandrasekharan, S.; et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007, 8, R76. [Google Scholar] [CrossRef] [PubMed]
- Matulka, L.A.; Wagner, K.U. Models of breast cancer. Drug Discov. Today Dis. Model. 2005, 2, 1–6. [Google Scholar] [CrossRef]
- Liu, Y.; Yin, T.; Feng, Y.; Cona, M.M.; Huang, G.; Liu, J.; Song, S.; Jiang, Y.; Xia, Q.; Swinnen, J.V.; et al. Mammalian models of chemically induced primary malignancies exploitable for imaging-based preclinical theragnostic research. Quant. Imaging Med. Surg. 2015, 5, 708–729. [Google Scholar] [CrossRef]
- Medina, D. Chemical carcinogenesis of rat and mouse mammary glands. Breast Dis. 2007, 28, 63–68. [Google Scholar] [CrossRef]
- Vargo-Gogola, T.; Rosen, J.M. Modelling breast cancer: One size does not fit all. Nat. Rev. Cancer 2007, 7, 659–672. [Google Scholar] [CrossRef]
- Trosko, J.E. Commentary: Is the concept of “tumor promotion” a useful paradigm? Mol. Carcinog. 2001, 30, 131–137. [Google Scholar] [CrossRef]
- Luch, A. Nature and nurture—lessons from chemical carcinogenesis. Nat. Rev. Cancer 2005, 5, 113–125. [Google Scholar] [CrossRef]
- Zarbl, H. Toxicogenomic analyses of genetic susceptibility to mammary gland carcinogenesis in rodents: Implications for human breast cancer. Breast Dis. 2007, 28, 87–105. [Google Scholar] [CrossRef]
- Santarelli, R.L.; Pierre, F.; Corpet, D.E. Processed meat and colorectal cancer: A review of epidemiologic and experimental evidence. Nutr. Cancer 2008, 60, 131–144. [Google Scholar] [CrossRef]
- Ward, M.H.; Cross, A.J.; Divan, H.; Kulldorff, M.; Nowell-Kadlubar, S.; Kadlubar, F.F.; Sinha, R. Processed meat intake, CYP2A6 activity and risk of colorectal adenoma. Carcinogenesis 2007, 28, 1210–1216. [Google Scholar] [CrossRef]
- Pohanish, R.P. Sittig’s Handbook of Toxic and Hazardous Chemicals and Carcinogens; Elsevier Science: Amsterdam, the Netherlands, 2011; ISBN 9781437778694. [Google Scholar]
- Agrawal, A.; Verma, P.; Goyal, P.K. Chemomodulatory Effects of Aegle Marmelos Against DMBA-Induced Skin Tumorigenesis in Swiss Albino Mice. Asian Pac. J. Cancer Prev. J Cancer Prev. 2010, 11, 1311–1314. [Google Scholar]
- Dimitrova-Shumkovska, J.; Veenman, L.; Ristoski, T.; Leschiner, S.; Gavish, M. Decreases in Binding Capacity of the Mitochondrial 18 kDa Translocator Protein Accompany Oxidative Stress and Pathological Signs in Rat Liver After DMBA Exposure. Toxicol. Pathol. 2010, 38, 957–968. [Google Scholar] [CrossRef]
- Loomis, T.A.; Hayes, A.W. Loomis’s Essentials of Toxicology; Academic Press: Amsterdam, the Netherlands, 1996; ISBN 9780080535630. [Google Scholar]
- Russo, J.; Russo, I.H. Atlas and Histologic Classification of Tumors of the Rat Mammary Gland. J. Mamm. Gland Biol. Neop. 2000, 5, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Al-Dhaheri, W.S.; Hassouna, I.; Al-Salam, S.; Karam, S.M. Characterization of Breast Cancer Progression in the Rat. Ann. N. Y. Acad. Sci. 2008, 1138, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Currier, N.; Solomon, S.E.; Demicco, E.G.; Chang, D.L.F.; Farago, M.; Ying, H.; Dominguez, I.; Sonenshein, G.E.; Cardiff, R.D.; Jim Xiao, Z.X.; et al. Oncogenic Signaling Pathways Activated in DMBA-Induced Mouse Mammary Tumors. Toxicol. Pathol. 2005, 33, 726–737. [Google Scholar] [CrossRef] [PubMed]
- Russo, J.; Russo, I.H. Differentiation and breast cancer development. In Breast Cancer; Bittar, E.E., Heppner, G., Peters, W.P., Visscher, D.W., Eds.; Elsevier: Amsterdam, the Netherlands, 1999; Volume 2, pp. 1–10. ISBN 1569-254X. [Google Scholar]
- Martinez, A.; Merchan, J.; Sala, M.L.; Renedo, G.; Fernandez-Pascual, J.; Bullón, A.; Blanes, A.; Renedo, G.; Bullón, A., Jr.; Bullón, F.; et al. Carcinogenesis y Nitrosoamidas. Patologia (Mex.) 1974, VII, 225–230. [Google Scholar]
- Murray, T.J.; Ucci, A.A.; Maffini, M.V.; Sonnenschein, C.; Soto, A.M. Histological analysis of low dose NMU effects in the rat mammary gland. BMC Cancer 2009, 9, 267. [Google Scholar] [CrossRef]
- Lyng, H.; Olsen, D.R.; Southon, T.E.; Rofstad, E.K. 31P-nuclear magnetic resonance spectroscopy in vivo of four human melanoma xenograft lines: Spin-lattice relaxation times. Br. J. Cancer 1993, 68, 1061–1070. [Google Scholar] [CrossRef][Green Version]
- Wu, I.; Wang, H.; Huso, D.; Wahl, R.L. Optimal definition of biological tumor volume using positron emission tomography in an animal model. EJNMMI Res. 2015, 5, 58. [Google Scholar] [CrossRef]
- Cotroneo, M.S.; Haag, J.D.; Zan, Y.; Lopez, C.C.; Thuwajit, P.; Petukhova, G.V.; Camerini-Otero, R.D.; Gendron-Fitzpatrick, A.; Griep, A.E.; Murphy, C.J.; et al. Characterizing a rat Brca2 knockout model. Oncogene 2007, 26, 1626–1635. [Google Scholar] [CrossRef] [PubMed]
- Zan, Y.; Haag, J.D.; Chen, K.-S.; Shepel, L.A.; Wigington, D.; Wang, Y.-R.; Hu, R.; Lopez-Guajardo, C.C.; Brose, H.L.; Porter, K.I.; et al. Production of knockout rats using ENU mutagenesis and a yeast-based screening assay. Nat. Biotechnol. 2003, 21, 645–651. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, V.; Ankola, D.D.; Gupta, S.C.; Schneider, M.; Lehr, C.M.; Kumar, M.N.V.R. PLGA nanoparticles stabilized with cationic surfactant: Safety studies and application in oral delivery of paclitaxel to treat chemical-induced breast cancer in rat. Pharm. Res. 2009, 26, 2495–2503. [Google Scholar] [CrossRef] [PubMed]
- Gal, A.; Baba, A.; Miclaus, V.; Bouari, C.; Bolfă, P.; Borza, G.; Catoi, C. Comparative aspects regarding MNU-induced mammary carcinogenesis in immature Sprague-Dowley and Whistar rats. Bull. Univ. Agric. Sci. Vet. Med. Cluj-Napoca Vet. Med. 2011, 68, 159–163. [Google Scholar] [CrossRef]
- Alvarado, A.; Faustino-Rocha, A.I.; Ferreira, R.; Mendes, R.; Duarte, J.A.; Pires, M.J.; Colaço, B.; Oliveira, P.A. Prognostic factors in an exercised model of chemically-induced mammary cancer. Anticancer Res. 2016, 36, 2181–2188. [Google Scholar]
- Faustino-Rocha, A.I.; Gama, A.; Oliveira, P.A.; Alvarado, A.; Neuparth, M.J.; Ferreira, R.; Ginja, M. Effects of lifelong exercise training on mammary tumorigenesis induced by MNU in female Sprague–Dawley rats. Clin. Exp. Med. 2017, 17, 151–160. [Google Scholar] [CrossRef]
- Jiang, C.; Mitrenga, T.; Cutter, G.; Thompson, H. Pathogenic characterization of 1-methyl-1-nitrosourea-induced mammary carcinomas in the rat. Carcinogenesis 1998, 19, 223–227. [Google Scholar]
- Perše, M.; Cerar, A.; Injac, R.; Štrukelj, B. N-methylnitrosourea Induced Breast Cancer in Rat, the Histopathology of the Resulting Tumours and its Drawbacks as a Model. Pathol. Oncol. Res. 2009, 15, 115–121. [Google Scholar] [CrossRef]
- Takayama, S.; Thorgeirsson, U.P.; Adamson, R.H. Chemical carcinogenesis studies in nonhuman primates. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2008, 84, 176–188. [Google Scholar] [CrossRef]
- Yang, J.; Shikata, N.; Mizuoka, H.; Tsubura, A. Colon carcinogenesis in shrews by intrarectal infusion of N-methyl-N-nitrosourea. Cancer Lett. 1996, 110, 105–112. [Google Scholar] [CrossRef]
- Leung, W.K.; Wu, K.; Wong, C.Y.P.; Cheng, A.S.L.; Ching, A.K.K.; Chan, A.W.H.; Chong, W.W.S.; Go, M.Y.Y.; Yu, J.; To, K.-F.; et al. Transgenic cyclooxygenase-2 expression and high salt enhanced susceptibility to chemical-induced gastric cancer development in mice. Carcinogenesis 2008, 29, 1648–1654. [Google Scholar] [CrossRef] [PubMed]
- McCormick, D.L.; Adamowski, C.B.; Fiks, A.; Moon, R.C. Lifetime dose-response relationships for mammary tumor induction by a single administration of N-methyl-N-nitrosourea. Cancer Res. 1981, 41, 1690–1694. [Google Scholar]
- Russo, I.H.; Russo, J. Mammary gland neoplasia in long-term rodent studies. Environ. Health Perspect. 1996, 104, 938–967. [Google Scholar] [CrossRef] [PubMed]
- Russo, J.; Russo, I.H. Experimentally induced mammary tumors in rats. Breast Cancer Res. Treat. 1996, 39, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Sternlicht, M.D. Key stages in mammary gland development: The cues that regulate ductal branching morphogenesis. Breast Cancer Res. 2006, 8, 201. [Google Scholar] [CrossRef]
- Russo, J. Significance of Rat mammary tumors for human risk assessment. Toxicol. Pathol. 2015, 43, 145–170. [Google Scholar] [CrossRef]
- Park, M.K.; Lee, C.H.; Lee, H. Mouse models of breast cancer in preclinical research. Lab. Anim. Res. 2018, 34, 160. [Google Scholar] [CrossRef]
- Ni, Y.; Wang, H.; Chen, F.; Li, J.; DeKeyzer, F.; Feng, Y.; Yu, J.; Bosmans, H.; Marchal, G. Tumor models and specific contrast agents for small animal imaging in oncology. Methods 2009, 48, 125–138. [Google Scholar] [CrossRef]
- Sano, D.; Myers, J.N. Xenograft models of head and neck cancers. Head Neck Oncol. 2009, 1, 32. [Google Scholar] [CrossRef]
- Forabosco, F.; Löhmus, M.; Rydhmer, L.; Sundström, L.F. Genetically modified farm animals and fish in agriculture: A review. Livest. Sci. 2013, 153, 1–9. [Google Scholar] [CrossRef]
- Doetschman, T. GI GEMs: Genetically engineered mouse models of gastrointestinal disease. Gastroenterology 2011, 140, 380–385.e2. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Workshop, E.C. Of mice and men—are mice relevant models for human disease? In Proceedings of the Outcomes of the European Commission Workshop ‘Are Mice Relevant Models for Human Disease?’, London, UK, 21 May 2010; Health Directorate, DG Research, European Commission: Brussels, Belgium, 2010; p. 10. [Google Scholar]
- Hoenerhoff, M.J.; Shibata, M.A.; Bode, A.; Green, J.E. Pathologic progression of mammary carcinomas in a C3(1)/SV40 T/t-antigen transgenic rat model of human triple-negative and Her2-positive breast cancer. Transgenic Res. 2011, 20, 247–259. [Google Scholar] [CrossRef]
- Mullins, L.J.; Brooker, G.; Mullins, J.J. Transgenesis in the Rat BT—Transgenesis Techniques: Principles and Protocols; Clarke, A.R., Ed.; Springer: Totowa, NJ, USA, 2002; pp. 255–270. ISBN 978-1-59259-178-7. [Google Scholar]
- Ito, N.; Hasegawa, R.; Sano, M.; Tamano, S.; Esumi, H.; Takayama, S.; Sugimura, T. A new colon and mammary carcinogen in cooked food, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP). Carcinogenesis 1991, 12, 1503–1506. [Google Scholar] [CrossRef] [PubMed]
- Ronckers, C.M.; Erdmann, C.A.; Land, C.E. Radiation and breast cancer: A review of current evidence. Breast Cancer Res. 2005, 7, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Imaoka, T.; Nishimura, M.; Iizuka, D.; Daino, K.; Takabatake, T.; Okamoto, M.; Kakinuma, S.; Shimada, Y. Radiation-Induced Mammary Carcinogenesis in Rodent Models: What’s Different from Chemical Carcinogenesis? J. Radiat. Res. 2009, 50, 281–293. [Google Scholar] [CrossRef]
- Finerty, J.C.; Binhammer, R.T.; Schneider, M.; Cunningham, A.W.B. Neoplasms in Rats Exposed to Single-Dose Total-Body X Radiation2. J. Natl. Cancer Inst. 1953, 14, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Holtzman, S.; Stone, J.P.; Shellabarger, C.J. Synergism of diethylstilbestrol and radiation in mammary carcinogenesis in female F344 rats. J. Natl. Cancer Inst. 1979, 63, 1071–1074. [Google Scholar]
- Vogel, H.H.; Turner, J.E. Genetic Component in Rat Mammary Carcinogenesis. Radiat. Res. 1982, 89, 264–273. [Google Scholar] [CrossRef]
- Haag, J.D.; Hsu, L.-C.; Newton, M.A.; Gould, M.N. Allelic imbalance in mammary carcinomas induced by either 7,12-dimethylbenz[a]anthracene or ionizing radiation in rats carrying genes conferring differential susceptibilities to mammary carcinogenesis. Mol. Carcinog. 1996, 17, 134–143. [Google Scholar] [CrossRef]
- Bartstra, R.W.; Bentvelzen, P.A.J.; Zoetelief, J.; Mulder, A.H.; Broerse, J.J.; van Bekkum, D.W. Induction of Mammary Tumors in Rats by Single-Dose Gamma Irradiation at Different Ages. Radiat. Res. 1998, 150, 442–450. [Google Scholar] [CrossRef]
- Inano, H.; Suzuki, K.; Ishii-Ohba, H.; Ikeda, K.; Wakabayashi, K. Pregnancy-dependent initiation in tumorigenesis of Wistar rat mammary glands by 60Co-irradiation. Carcinogenesis 1991, 12, 1085–1090. [Google Scholar] [CrossRef] [PubMed]
- Shellabarger, C.J. Mammary Neoplastic Response of Lewis and Sprague-Dawley Female Rats to 7,12-Dimethylbenz(a)anthracene or X-ray. Cancer Res. 1972, 32, 883–885. [Google Scholar] [PubMed]
- Shellabarger, C.J.; Stone, J.P.; Holtzman, S. Rat Differences in Mammary Tumor Induction With Estrogen and Neutron Radiation. J. Natl. Cancer Inst. 1978, 61, 1505–1508. [Google Scholar] [CrossRef] [PubMed]
- Imaoka, T.; Nishimura, M.; Kakinuma, S.; Hatano, Y.; Ohmachi, Y.; Yoshinaga, S.; Kawano, A.; Maekawa, A.; Shimada, Y. High Relative Biologic Effectiveness of Carbon Ion Radiation on Induction of Rat Mammary Carcinoma and its Lack of H-ras and Tp53 Mutations. Int. J. Radiat. Oncol. 2007, 69, 194–203. [Google Scholar] [CrossRef]
- Ullrich, R.L.; Preston, R.J. Radiation Induced Mammary Cancer. J. Radiat. Res. 1991, 32, 104–109. [Google Scholar] [CrossRef]
- Mori, N.; Yamate, J.; Umesako, S.-I.; Hong, D.-P.; Okumoto, M.; Nakao, R. Preferential Induction of Mammary Tumors in p53 Hemizygous BALB/c Mice by Fractionated Irradiation of a Sub-lethal Dose of X-rays. J. Radiat. Res. 2003, 44, 249–254. [Google Scholar] [CrossRef][Green Version]
- Backlund, M.G.; Trasti, S.L.; Backlund, D.C.; Cressman, V.L.; Godfrey, V.; Koller, B.H. Impact of Ionizing Radiation and Genetic Background on Mammary Tumorigenesis in p53-deficient Mice. Cancer Res. 2001, 61, 6577–6582. [Google Scholar]
- Cressman, V.L.; Backlund, D.C.; Hicks, E.M.; Gowen, L.C.; Godfrey, V.; Koller, B.H. Mammary Tumor Formation in p53- and BRCA1-deficient Mice. Cell Growth Differ. 1999, 10, 1–10. [Google Scholar]
- Bassaganya-Riera, J.; Hontecillas, R.; Abedi, V.; Carbo, A.; Philipson, C.; Hoops, S. Computational Modeling; Elsevier Inc.: Amsterdam, the Netherlands, 2016; ISBN 9780128037157. [Google Scholar]
- Dimitrakopoulos, C.M.; Beerenwinkel, N. Computational approaches for the identification of cancer genes and pathways. Wiley Interdiscip. Rev. Syst. Biol. Med. 2017, 9, 1–18. [Google Scholar] [CrossRef]
- Fang, G.; Wang, W.; Paunic, V.; Heydari, H.; Costanzo, M.; Liu, X.; Liu, X.; VanderSluis, B.; Oately, B.; Steinbach, M.; et al. Discovering genetic interactions bridging pathways in genome-wide association studies. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef]
- Dunning, A.M.; Michailidou, K.; Kuchenbaecker, K.B.; Thompson, D.; French, J.D.; Beesley, J.; Healey, C.S.; Kar, S.; Pooley, K.A.; Lopez-Knowles, E.; et al. Breast cancer risk variants at 6q25 display different phenotype associations and regulate ESR1, RMND1 and CCDC170. Nat. Genet. 2016, 48, 374–386. [Google Scholar] [CrossRef] [PubMed]
- Gitter, A.; Braunstein, A.; Pagnani, A.; Baldassi, C.; Borgs, C.; Chayes, J.; Zecchina, R.; Fraenkel, E. Sharing information to reconstruct patient-specific pathways in heterogeneous diseases. In Proceedings of the Pacific Symposium on Biocomputing 2014, Fairmont Orchid, HI, USA, 3–7 January 2014; pp. 39–50. [Google Scholar]
- Bao, S.; Zhao, H.; Yuan, J.; Fan, D.; Zhang, Z.; Su, J.; Zhou, M. Computational identification of mutator-derived lncRNA signatures of genome instability for improving the clinical outcome of cancers: A case study in breast cancer. Brief. Bioinform. 2019, 00, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, A.; Nielsen, J. Genome scale metabolic modeling of cancer. Metab. Eng. 2017, 43, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Ozkan, A.; Rylander, M.N.; Woodward, W.A.; Vlachos, P. Mixture theory modeling for characterizing solute transport in breast tumor tissues. J. Biol. Eng. 2019, 13, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Benzekry, S.; Lamont, C.; Beheshti, A.; Tracz, A.; Ebos, J.M.L.; Hlatky, L.; Hahnfeldt, P. Classical Mathematical Models for Description and Prediction of Experimental Tumor Growth. PLoS Comput. Biol. 2014, 10. [Google Scholar] [CrossRef]
- Norton, L. A Gompertzian Model of Human Breast Cancer Growth. Cancer Res. 1988, 48, 7067–7071. [Google Scholar]
- Gardezi, S.J.S.; Elazab, A.; Lei, B.; Wang, T. Breast cancer detection and diagnosis using mammographic data: Systematic review. J. Med. Internet Res. 2019, 21, 1–22. [Google Scholar] [CrossRef]
- Bornefalk, H.; Hermansson, A.B. On the comparison of FROC curve in mammography CAD system. Med. Phys. 2005, 32, 412–417. [Google Scholar] [CrossRef]
- Tiwari, S.; Bhargava, R. Extracting knowledge from chemical imaging data using computational algorithms for digital cancer diagnosis. Yale J. Biol. Med. 2015, 88, 131–143. [Google Scholar]
- Fabian, H.; Thi, N.A.N.; Eiden, M.; Lasch, P.; Schmitt, J.; Naumann, D. Diagnosing benign and malignant lesions in breast tissue sections by using IR-microspectroscopy. Biochim. Biophys. Acta Biomembr. 2006, 1758, 874–882. [Google Scholar] [CrossRef][Green Version]
- Kirouac, D.C.; Du, J.Y.; Lahdenranta, J.; Overland, R.; Yarar, D.; Paragas, V.; Pace, E.; McDonagh, C.F.; Nielsen, U.B.; Onsum, M.D. Computational modeling of ERBB2-amplified breast cancer identifies combined ErbB2/3 blockade as superior to the combination of MEK and AKT inhibitors (Science Signaling 6: 288 (ra68)). Sci. Signal. 2014, 7, er5. [Google Scholar] [CrossRef]
- Madhukar, N.S.; Khade, P.K.; Huang, L.; Gayvert, K.; Galletti, G.; Stogniew, M.; Allen, J.E.; Giannakakou, P.; Elemento, O. A Bayesian machine learning approach for drug target identification using diverse data types. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, Z.; Mahmood, T.; Hassan, M.; Afzal, S.; Rafique, H.; Afzal, K.; Latip, J. Dexibuprofen amide derivatives as potential anticancer agents: Synthesis, in silico docking, bioevaluation, and molecular dynamic simulation. Drug Des. Dev. Ther. 2019, 13, 1643–1657. [Google Scholar] [CrossRef] [PubMed]
- Spinello, A.; Ritacco, I.; Magistrato, A. Recent advances in computational design of potent aromatase inhibitors: Open-eye on endocrine-resistant breast cancers. Expert Opin. Drug Discov. 2019, 14, 1065–1076. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Abramson, R.G.; Arlinghaus, L.R.; Chakravarthy, A.B.; Abramson, V.; Mayer, I.; Farley, J.; Delbeke, D.; Yankeelov, T.E. An algorithm for longitudinal registration of PET/CT images acquired during neoadjuvant chemotherapy in breast cancer: Preliminary results. EJNMMI Res. 2012, 2, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Wang, J.; Zhang, H.H. Computerized PET/CT image analysis in the evaluation of tumour response to therapy. Br. J. Radiol. 2015, 88, 10–12. [Google Scholar] [CrossRef]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Uhlén, M.; Björling, E.; Agaton, C.; Szigyarto, C.A.K.; Amini, B.; Andersen, E.; Andersson, A.C.; Angelidou, P.; Asplund, A.; Asplund, C.; et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol. Cell. Proteom. 2005, 4, 1920–1932. [Google Scholar] [CrossRef]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K.; et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017, 171, 1437–1452.e17. [Google Scholar] [CrossRef]
- Castellino, R.A. Computer aided detection (CAD): An overview. Cancer Imaging 2005, 5, 17–19. [Google Scholar] [CrossRef] [PubMed]
- Badgujar, N.V.; Tarapara, B.V.; Shah, F.D. Computational analysis of high-risk SNPs in human CHK2 gene responsible for hereditary breast cancer: A functional and structural impact. PLoS ONE 2019, 14, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Seo, J.-H.; Stranger, B.; McKenna, A.; Pe’er, I.; Laframboise, T.; Brown, M.; Tyekucheva, S.; Freedman, M.L. Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell 2013, 152, 633–641. [Google Scholar] [CrossRef] [PubMed]
- Chuang, H.Y.; Lee, E.; Liu, Y.T.; Lee, D.; Ideker, T. Network-based classification of breast cancer metastasis. Mol. Syst. Biol. 2007, 3, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, J.; Liu, E.; Kinnebrew, G.; Zhang, X.; Stover, D.; Huo, Y.; Zeng, Z.; Jiang, W.; Cheng, L.; et al. Identification of alternatively-activated pathways between primary breast cancer and liver metastatic cancer using microarray data. Genes (Basel) 2019, 10, 753. [Google Scholar] [CrossRef]
- Lai, X.; Geier, O.M.; Fleischer, T.; Garred, Y.; Borgen, E.; Funke, S.W.; Kumar, S.; Rognes, M.E.; Seierstad, T.; Børresen-Dale, A.L.; et al. Toward personalized computer simulation of breast cancer treatment: A multiscale pharmacokinetic and pharmacodynamic model informed by multitype patient data. Cancer Res. 2019, 79, 4293–4304. [Google Scholar] [CrossRef]
- Li, C.M.; Segars, W.P.; Tourassi, G.D.; Boone, J.M.; Dobbins, J.T. Methodology for generating a 3D computerized breast phantom from empirical data. Med. Phys. 2009, 36, 3122–3131. [Google Scholar] [CrossRef]
- Hsu, C.M.L.; Palmeri, M.L.; Segars, W.P.; Veress, A.I.; Dobbins, J.T. Generation of a suite of 3D computer-generated breast phantoms from a limited set of human subject data. Med. Phys. 2013, 40, 1–11. [Google Scholar] [CrossRef]
- Booth, M.E.; Nash, C.E.; Roberts, N.P.; Magee, D.R.; Treanor, D.; Hanby, A.M.; Speirs, V. 3-D tissue modelling and virtual pathology as new approaches to study ductal carcinoma in Situ. ATLA Altern. Lab. Anim. 2015, 43, 377–383. [Google Scholar] [CrossRef]
- Booth, M.E.; Treanor, D.; Roberts, N.; Magee, D.R.; Speirs, V.; Hanby, A.M. Three-dimensional reconstruction of ductal carcinoma in situ with virtual slides. Histopathology 2015, 66, 966–973. [Google Scholar] [CrossRef]
- Graff, C.G. A new, open-source, multi-modality digital breast phantom. In Medical Imaging 2016: Physics of Medical Imaging; San Diego, CA, USA, 2016; Volume 9783, p. 978309. Available online: https://www.spiedigitallibrary.org/conference-proceedings-of-spie/9783/978309/A-new-open-source-multi-modality-digital-breast-phantom/10.1117/12.2216312.short?SSO=1 (accessed on 7 December 2019). [CrossRef]
- Widmer, A.; Hu, Y. A viscoelastic model of a breast phantom for real-time palpation. In Proceedings of the 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Boston, MA, USA, 30 August–3 September 2011; pp. 4546–4549. [Google Scholar] [CrossRef]
- Celi, S.; Di Puccio, F.; Forte, P. Advances in finite element simulations of elastosonography for breast lesion detection. J. Biomech. Eng. 2011, 133, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ikejimba, L.C.; Graff, C.G.; Rosenthal, S.; Badal, A.; Ghammraoui, B.; Lo, J.Y.; Glick, S.J. A novel physical anthropomorphic breast phantom for 2D and 3D X-ray imaging. Med. Phys. 2017, 44, 407–416. [Google Scholar] [CrossRef] [PubMed]
- Kiarashi, N.; Nolte, A.C.; Sturgeon, G.M.; Segars, W.P.; Ghate, S.V.; Nolte, L.W.; Samei, E.; Lo, J.Y. Development of realistic physical breast phantoms matched to virtual breast phantoms based on human subject data. Med. Phys. 2015, 42, 4116–4126. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.T.; Samsuzzaman, M.; Kibria, S.; Islam, M.T. Experimental breast phantoms for estimation of breast tumor using microwave imaging systems. IEEE Access 2018, 6, 78587–78597. [Google Scholar] [CrossRef]
- Joachimowicz, N.; Duchêne, B.; Conessa, C.; Meyer, O. Easy-to-produce adjustable realistic breast phantoms for microwave imaging. In Proceedings of the 2016 10th European Conference Antennas Propagation (EuCAP 2016), Davos, Switzerland, 10–15 April 2016; pp. 3–6. [Google Scholar] [CrossRef]
- Truong, B.C.Q.; Fitzgerald, A.J.; Fan, S.; Wallace, V.P. Concentration analysis of breast tissue phantoms with terahertz spectroscopy. Biomed. Opt. Express 2018, 9, 1334. [Google Scholar] [CrossRef] [PubMed]
- Behmadi, M.; Gholamhosseinian, H.; Mohammadi, M.; Naseri, S.H.; Momennezhad, M.; Bayani, S.H.; Bahreyni Tossi, M.T. Evaluation of breast cancer radiation therapy techniques in outfield organs of rando phantom with thermoluminescence dosimeter. J. Biomed. Phys. Eng. 2019, 9, 179–188. [Google Scholar] [CrossRef]
- Quinn, A.; Holloway, L.; Metcalfe, P. Image guidance during breast radiotherapy: A phantom dosimetry and radiation-induced second cancer risk study. J. Phys. Conf. Ser. 2013, 444. [Google Scholar] [CrossRef]
- Di Meo, S.; Pasotti, L.; Iliopoulos, I.; Pasian, M.; Ettorre, M.; Zhadobov, M.; Matrone, G. Tissue-mimicking materials for breast phantoms up to 50 GHz. Phys. Med. Biol. 2019, 64. [Google Scholar] [CrossRef]
- Sabhachandani, P.; Motwani, V.; Cohen, N.; Sarkar, S.; Torchilin, V.; Konry, T.; Fenway, T.; Avenue, H. Fluorescent Aliphatic Hyperbranched Polyether: Chromophores-free and without any N and P Atoms. Phys. Chem. Chem. Phys. 2016, 16, 497–505. [Google Scholar] [CrossRef]
- Hwang, H.; Park, J.; Shin, C.; Do, Y.; Cho, Y.K. Three dimensional multicellular co-cultures and anti-cancer drug assays in rapid prototyped multilevel microfluidic devices. Biomed. Microdevices 2013, 15, 627–634. [Google Scholar] [CrossRef]
- Yildiz-Ozturk, E.; Gulce-Iz, S.; Anil, M.; Yesil-Celiktas, O. Cytotoxic responses of carnosic acid and doxorubicin on breast cancer cells in butterfly-shaped microchips in comparison to 2D and 3D culture. Cytotechnology 2017, 69, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Grafton, M.M.G.; Wang, L.; Vidi, P.A.; Leary, J.; Lelièvre, S.A. Breast on-a-chip: Mimicry of the channeling system of the breast for development of theranostics. Integr. Biol. 2011, 3, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yang, X.; Zou, J.; Jia, C.; Hu, Y.; Du, H.; Wang, H. Evaluation of photodynamic therapy efficiency using an in vitro three-dimensional microfluidic breast cancer tissue model. Lab Chip 2015, 15, 735–744. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.M.; Seo, H.I.; Bae, J.H.; Chung, B.G. Hydrogel microfluidic co-culture device for photothermal therapy and cancer migration. Electrophoresis 2017, 38, 1318–1324. [Google Scholar] [CrossRef]
- Sung, K.E.; Yang, N.; Pehlke, C.; Keely, P.J.; Eliceiri, K.W.; Friedl, A.; Beebe, D.J. Transition to invasion in breast cancer: A microfluidic in vitro model enables examination of spatial and temporal effects. Integr. Biol. (Camb.) 2011, 3, 439–450. [Google Scholar] [CrossRef]
- Bischel, L.L.; Beebe, D.J.; Sung, K.E. Microfluidic model of ductal carcinoma in situ with 3D, organotypic structure. BMC Cancer 2015, 15, 1–10. [Google Scholar] [CrossRef]
- Truong, D.; Puleo, J.; Llave, A.; Mouneimne, G.; Kamm, R.D.; Nikkhah, M. Breast cancer cell invasion into a three dimensional tumor-stroma microenvironment. Sci. Rep. 2016, 6, 34094. [Google Scholar] [CrossRef]
- Kwak, B.; Ozcelikkale, A.; Shin, C.S.; Park, K.; Han, B. Simulation of complex transport of nanoparticles around a tumor using tumor-microenvironment-on-chip. J. Control. Release 2014, 194, 157–167. [Google Scholar] [CrossRef]
- Bersini, S.; Jeon, J.S.; Dubini, G.; Arrigoni, C.; Chung, S.; Charest, J.L.; Moretti, M.; Kamm, R.D. A microfluidic 3D in vitro model for specificity of breast cancer metastasis to bone. Biomaterials 2014, 35, 2454–2461. [Google Scholar] [CrossRef]
- Song, J.W.; Cavnar, S.P.; Walker, A.C.; Luker, K.E.; Gupta, M.; Tung, Y.C.; Luker, G.D.; Takayama, S. Microfluidic endothelium for studying the intravascular adhesion of metastatic breast cancer cells. PLoS ONE 2009, 4. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).