Machine Learning-Based Identifications of COVID-19 Fake News Using Biomedical Information Extraction
Abstract
1. Introduction
2. Materials and Methods
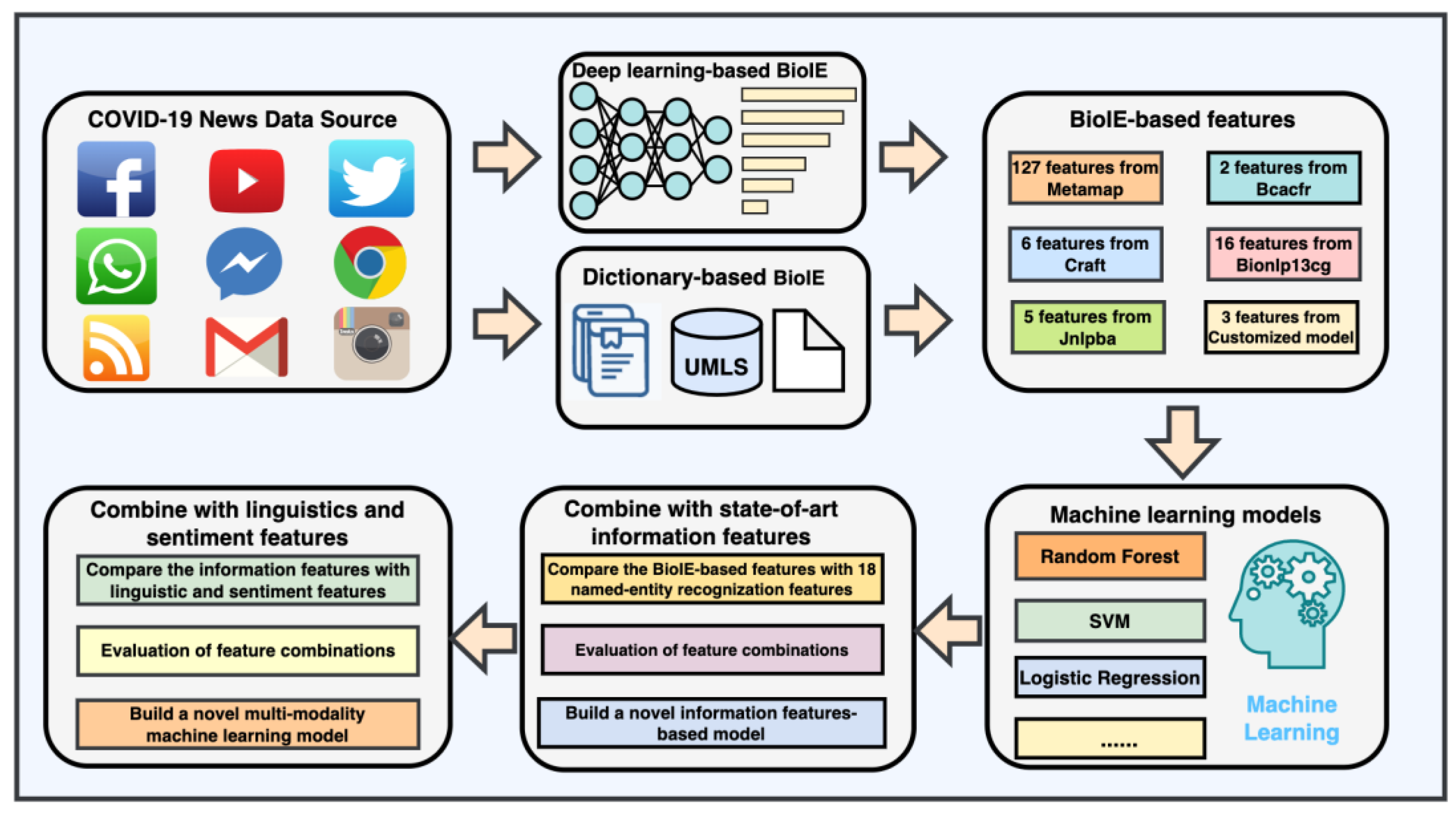
2.1. Data
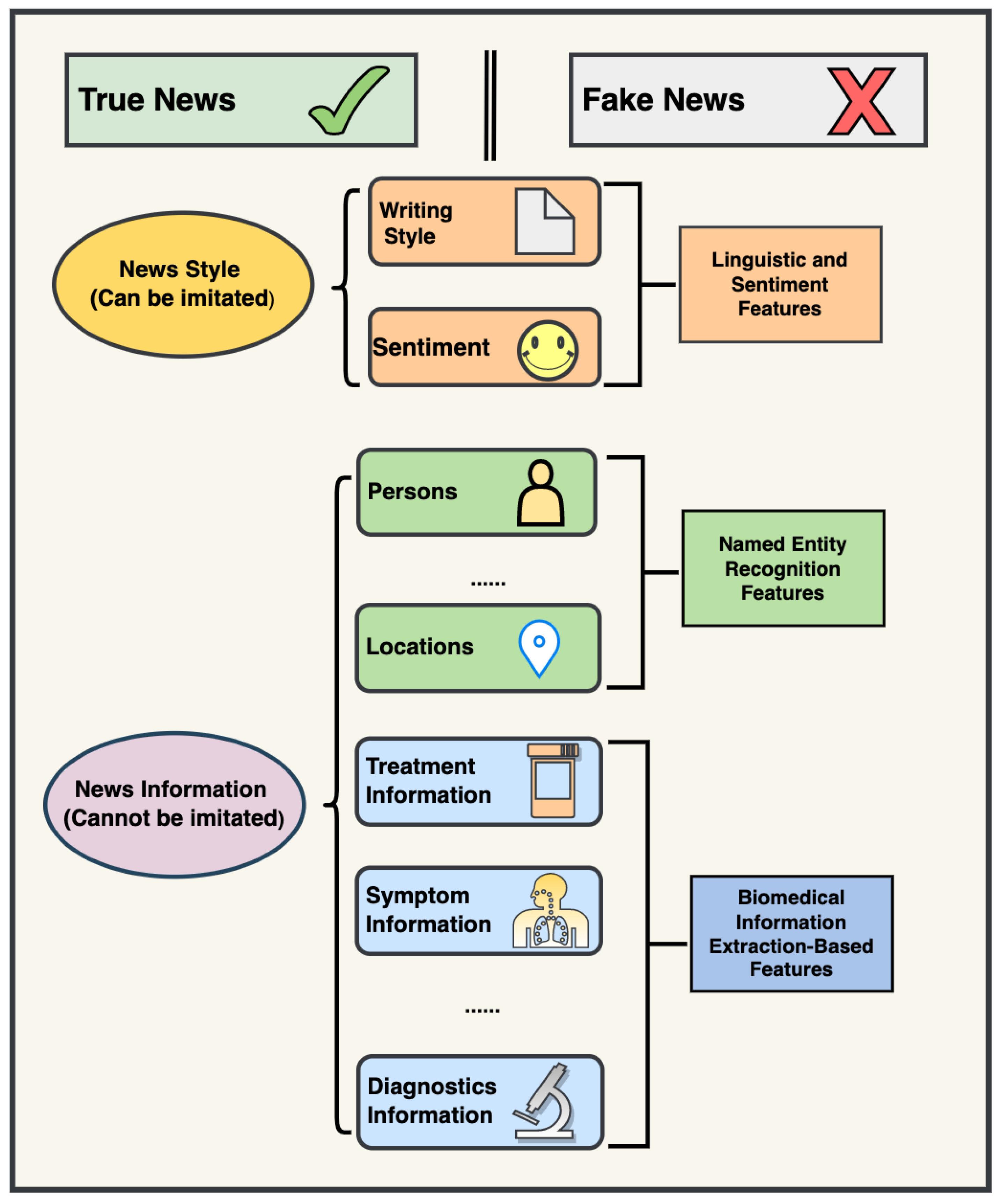
2.2. Constructing Novel Features with Biomedical Information Extraction
2.3. Training and Evaluation with Biomedical Information Extraction-Based Feature
2.4. Training and Evaluation through a Combination of State-of-the-Art Information Features and Biomedical Information Extraction-Based Features
2.5. A Novel Biomedical Information Extraction-Driven Multi-Modality Machine Learning Model
3. Results
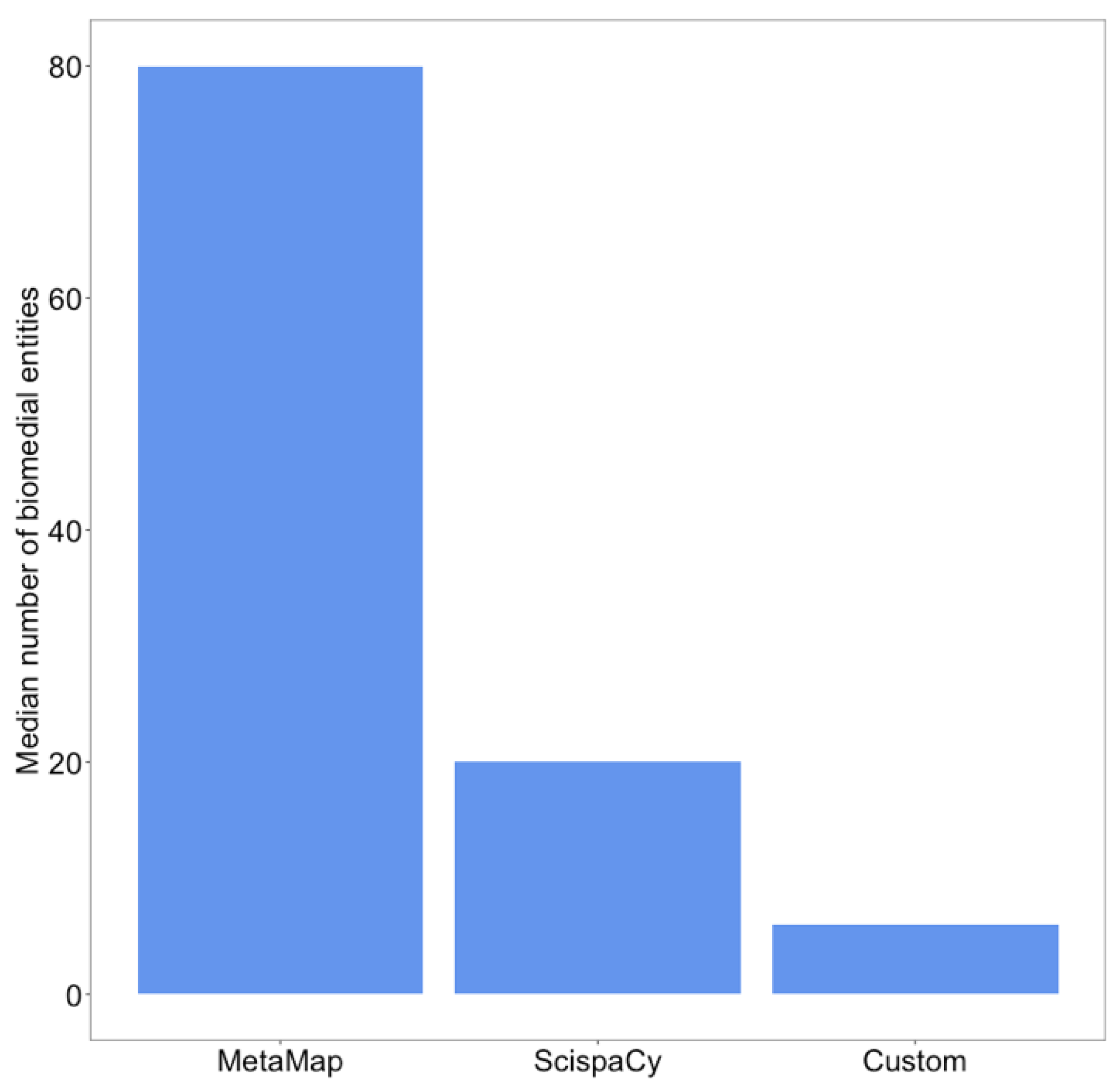
3.1. Biomedical Information Extraction Is Useful for COVID-19 Fake News Detection
3.2. Biomedical Information Extraction Improved the Information Features-Based COVID-19 Fake News Detection
3.3. Information-Based Models Incorporating Biomedical Information Have a Higher Power in Identifying COVID-19 Fake News Than Linguistics-Based and Sentiment-Based Models
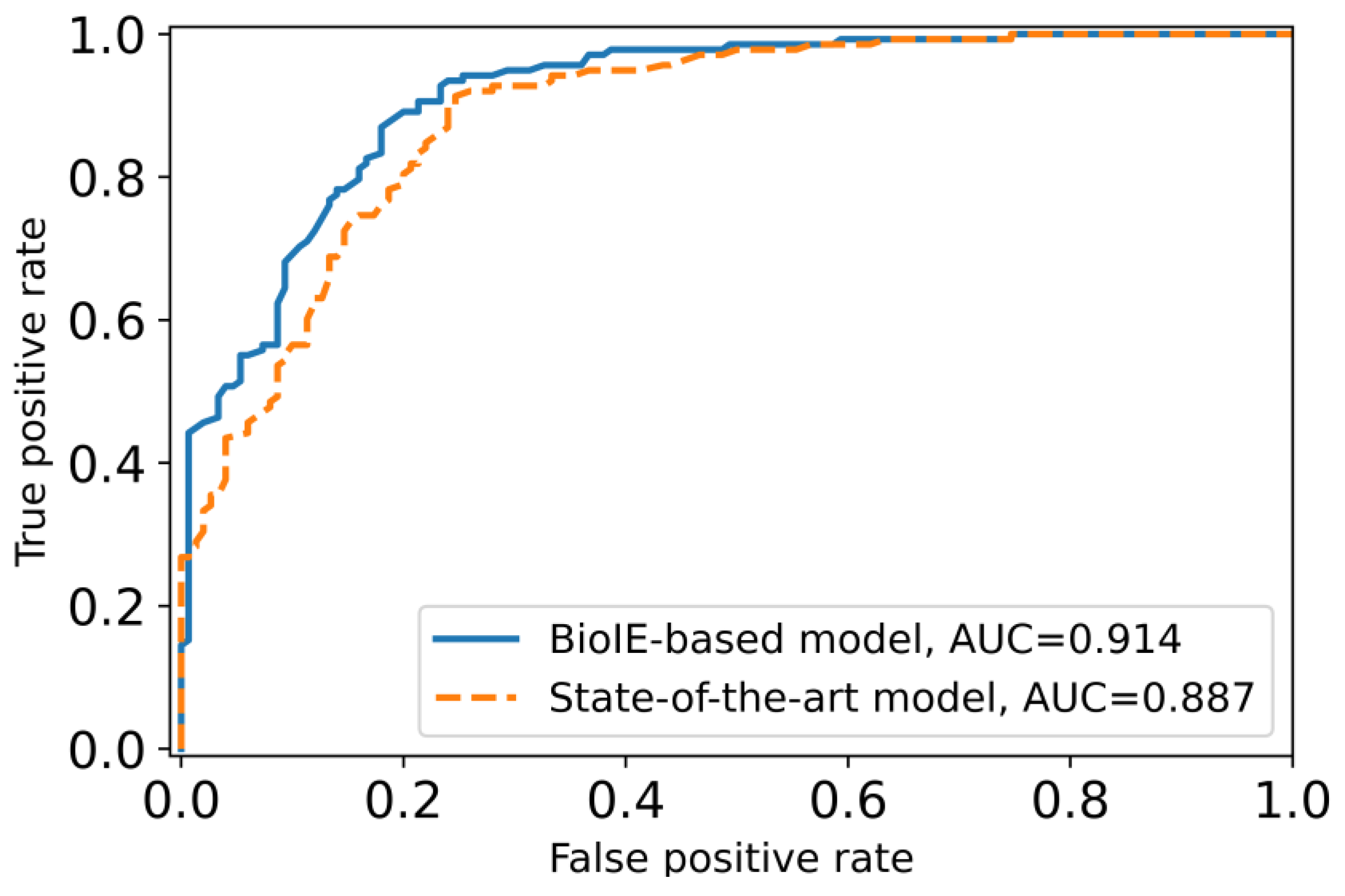
3.4. A Novel Biomedical Information-Driven Multi-Modality Model Outperforms a State-of-the-Art Multi-Modality Model
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bang, Y.; Ishii, E.; Cahyawijaya, S.; Ji, Z.; Fung, P. Model generalization on COVID-19 fake news detection. In Proceedings of the International Workshop on Combating Online Hostile Posts in Regional Languages during Emergency Situation, Online, 8 February 2021; pp. 128–140. [Google Scholar]
- Pennycook, G.; McPhetres, J.; Zhang, Y.; Lu, J.G.; Rand, D.G. Fighting COVID-19 misinformation on social media: Experimental evidence for a scalable accuracy-nudge intervention. Psychol. Sci. 2020, 31, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Radwan, E.; Radwan, A.; Radwan, W. The role of social media in spreading panic among primary and secondary school students during the COVID-19 pandemic: An online questionnaire study from the Gaza Strip, Palestine. Heliyon 2020, 6, e05807. [Google Scholar] [CrossRef] [PubMed]
- Freeman, D.; Waite, F.; Rosebrock, L.; Petit, A.; Causier, C.; East, A.; Jenner, L.; Teale, A.-L.; Carr, L.; Mulhall, S. Coronavirus conspiracy beliefs, mistrust, and compliance with government guidelines in England. Psychol. Med. 2022, 52, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Pierri, F.; Perry, B.; DeVerna, M.R.; Yang, K.-C.; Flammini, A.; Menczer, F.; Bryden, J. The impact of online misinformation on US COVID-19 vaccinations. arXiv 2021, arXiv:2104.10635. [Google Scholar]
- Orellana, C.I. Health workers as hate crimes targets during COVID-19 outbreak in the Americas. Rev. Salud Pública 2020, 22, 253–257. [Google Scholar]
- Kim, J.Y.; Kesari, A. Misinformation and Hate Speech: The Case of Anti-Asian Hate Speech During the COVID-19 Pandemic. J. Online Trust Saf. 2021, 1, 1–14. [Google Scholar] [CrossRef]
- Rocha, Y.M.; de Moura, G.A.; Desidério, G.A.; de Oliveira, C.H.; Lourenço, F.D.; de Figueiredo Nicolete, L.D. The impact of fake news on social media and its influence on health during the COVID-19 pandemic: A systematic review. J. Public Health 2021, 1–10. [Google Scholar] [CrossRef]
- Ahmad, A.R.; Murad, H.R. The impact of social media on panic during the COVID-19 pandemic in Iraqi Kurdistan: Online questionnaire study. J. Med. Internet Res. 2020, 22, e19556. [Google Scholar] [CrossRef]
- Secosan, I.; Virga, D.; Crainiceanu, Z.P.; Bratu, L.M.; Bratu, T. Infodemia: Another enemy for romanian frontline healthcare workers to fight during the COVID-19 outbreak. Medicina 2020, 56, 679. [Google Scholar] [CrossRef]
- World Health Organization. Novel Coronavirus (2019-nCoV) Situation Report-13. Available online: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200202-sitrep-13-ncov-v3.pdf (accessed on 5 December 2022).
- Zarocostas, J. How to fight an infodemic. Lancet 2020, 395, 676. [Google Scholar] [CrossRef]
- Bavel, J.J.V.; Baicker, K.; Boggio, P.S.; Capraro, V.; Cichocka, A.; Cikara, M.; Crockett, M.J.; Crum, A.J.; Douglas, K.M.; Druckman, J.N. Using social and behavioural science to support COVID-19 pandemic response. Nat. Hum. Behav. 2020, 4, 460–471. [Google Scholar] [CrossRef] [PubMed]
- Habersaat, K.B.; Betsch, C.; Danchin, M.; Sunstein, C.R.; Böhm, R.; Falk, A.; Brewer, N.T.; Omer, S.B.; Scherzer, M.; Sah, S. Ten considerations for effectively managing the COVID-19 transition. Nat. Hum. Behav. 2020, 4, 677–687. [Google Scholar] [CrossRef] [PubMed]
- van Der Linden, S.; Roozenbeek, J.; Compton, J. Inoculating against fake news about COVID-19. Front. Psychol. 2020, 11, 566790. [Google Scholar] [CrossRef] [PubMed]
- Tashtoush, Y.; Alrababah, B.; Darwish, O.; Maabreh, M.; Alsaedi, N. A Deep Learning Framework for Detection of COVID-19 Fake News on Social Media Platforms. Data 2022, 7, 65. [Google Scholar] [CrossRef]
- Zhang, X.; Ghorbani, A.A. An overview of online fake news: Characterization, detection, and discussion. Inf. Process. Manag. 2020, 57, 102025. [Google Scholar] [CrossRef]
- Varma, R.; Verma, Y.; Vijayvargiya, P.; Churi, P.P. A systematic survey on deep learning and machine learning approaches of fake news detection in the pre-and post-COVID-19 pandemic. Int. J. Intell. Comput. Cybern. 2021, 14, 617–646. [Google Scholar] [CrossRef]
- Choraś, M.; Demestichas, K.; Giełczyk, A.; Herrero, Á.; Ksieniewicz, P.; Remoundou, K.; Urda, D.; Woźniak, M. Advanced Machine Learning techniques for fake news (online disinformation) detection: A systematic mapping study. Appl. Soft Comput. 2021, 101, 107050. [Google Scholar] [CrossRef]
- Abdelminaam, D.S.; Ismail, F.H.; Taha, M.; Taha, A.; Houssein, E.H.; Nabil, A. Coaid-deep: An optimized intelligent framework for automated detecting covid-19 misleading information on twitter. IEEE Access 2021, 9, 27840–27867. [Google Scholar] [CrossRef] [PubMed]
- Al-Rakhami, M.S.; Al-Amri, A.M. Lies kill, facts save: Detecting COVID-19 misinformation in twitter. IEEE Access 2020, 8, 155961–155970. [Google Scholar] [CrossRef]
- Bangyal, W.H.; Qasim, R.; Ahmad, Z.; Dar, H.; Rukhsar, L.; Aman, Z.; Ahmad, J. Detection of fake news text classification on COVID-19 using deep learning approaches. Comput. Math. Methods Med. 2021, 2021, 5514220. [Google Scholar] [CrossRef]
- Endo, P.T.; Santos, G.L.; de Lima Xavier, M.E.; Nascimento Campos, G.R.; de Lima, L.C.; Silva, I.; Egli, A.; Lynn, T. Illusion of Truth: Analysing and Classifying COVID-19 Fake News in Brazilian Portuguese Language. Big Data Cogn. Comput. 2022, 6, 36. [Google Scholar] [CrossRef]
- Khan, S.; Hakak, S.; Deepa, N.; Prabadevi, B.; Dev, K.; Trelova, S. Detecting COVID-19-Related Fake News Using Feature Extraction. Front. Public Health 2021, 9, 788074. [Google Scholar] [CrossRef] [PubMed]
- Iwendi, C.; Mohan, S.; Khan, S.; Ibeke, E.; Ahmadian, A.; Ciano, T. Covid-19 fake news sentiment analysis. Comput. Electr. Eng. 2022, 101, 107967. [Google Scholar] [CrossRef] [PubMed]
- Alenezi, M.N.; Alqenaei, Z.M. Machine learning in detecting COVID-19 misinformation on twitter. Future Internet 2021, 13, 244. [Google Scholar] [CrossRef]
- Fauzi, A.; Setiawan, E.; Baizal, Z. Hoax news detection on Twitter using term frequency inverse document frequency and support vector machine method. J. Phys. Conf. Ser. 2019, 1192, 012025. [Google Scholar] [CrossRef]
- Kong, S.H.; Tan, L.M.; Gan, K.H.; Samsudin, N.H. Fake news detection using deep learning. In Proceedings of the 2020 IEEE 10th Symposium on Computer Applications & Industrial Electronics (ISCAIE), Penang, Malaysia, 18–19 April 2020; pp. 102–107. [Google Scholar]
- Baarir, N.F.; Djeffal, A. Fake news detection using machine learning. In Proceedings of the 2020 2nd International Workshop on Human-Centric Smart Environments for Health and Well-Being (IHSH), Boumerdes, Algeria, 9–10 February 2021; pp. 125–130. [Google Scholar]
- Goldani, M.H.; Momtazi, S.; Safabakhsh, R. Detecting fake news with capsule neural networks. Appl. Soft Comput. 2021, 101, 106991. [Google Scholar] [CrossRef]
- Bogale Gereme, F.; Zhu, W. Fighting fake news using deep learning: Pre-trained word embeddings and the embedding layer investigated. In Proceedings of the 2020 The 3rd International Conference on Computational Intelligence and Intelligent Systems, Tokyo, Japan, 13–15 November 2020; pp. 24–29. [Google Scholar]
- Qaiser, S.; Ali, R. Text mining: Use of TF-IDF to examine the relevance of words to documents. Int. J. Comput. Appl. 2018, 181, 25–29. [Google Scholar] [CrossRef]
- Khattak, F.K.; Jeblee, S.; Pou-Prom, C.; Abdalla, M.; Meaney, C.; Rudzicz, F. A survey of word embeddings for clinical text. J. Biomed. Inform. 2019, 100, 100057. [Google Scholar] [CrossRef] [PubMed]
- Alonso, M.A.; Vilares, D.; Gómez-Rodríguez, C.; Vilares, J. Sentiment analysis for fake news detection. Electronics 2021, 10, 1348. [Google Scholar] [CrossRef]
- Daley, B.P. Leveraging Machine Learning for Automatically Classifying Fake News in the COVID-19 Outbreak. 2020. Available online: https://scholarworks.boisestate.edu/icur/2020/Poster_Session/118/ (accessed on 22 August 2022).
- Zhou, Z.; Guan, H.; Bhat, M.M.; Hsu, J. Fake news detection via NLP is vulnerable to adversarial attacks. arXiv 2019, arXiv:1901.09657. [Google Scholar]
- Lazer, D.M.; Baum, M.A.; Benkler, Y.; Berinsky, A.J.; Greenhill, K.M.; Menczer, F.; Metzger, M.J.; Nyhan, B.; Pennycook, G.; Rothschild, D. The science of fake news. Science 2018, 359, 1094–1096. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Sukumaran, R.; John, K.; Teki, S. Hostility detection and covid-19 fake news detection in social media. arXiv 2021, arXiv:2101.05953. [Google Scholar]
- Brennen, J.S.; Simon, F.M.; Howard, P.N.; Nielsen, R.K. Types, Sources, and Claims of COVID-19 Misinformation; University of Oxford: Oxford, UK, 2020. [Google Scholar]
- Posetti, J.; Bontcheva, K. Disinfodemic: Deciphering COVID-19 Disinformation. Policy Brief. 2020, Volume 1. Available online: https://en.unesco.org/covid19/disinfodemic/brief1 (accessed on 25 August 2022).
- Charquero-Ballester, M.; Walter, J.G.; Nissen, I.A.; Bechmann, A. Different types of COVID-19 misinformation have different emotional valence on Twitter. Big Data Soc. 2021, 8, 20539517211041279. [Google Scholar] [CrossRef]
- Liu, F.; Chen, J.; Jagannatha, A.; Yu, H. Learning for biomedical information extraction: Methodological review of recent advances. arXiv 2016, arXiv:1606.07993. [Google Scholar]
- Zhou, M.; Wang, Q.; Zheng, C.; John Rush, A.; Volkow, N.D.; Xu, R. Drug repurposing for opioid use disorders: Integration of computational prediction, clinical corroboration, and mechanism of action analyses. Mol. Psychiatry 2021, 26, 5286–5296. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Chen, Y.; Xu, R. A drug-side effect context-sensitive network approach for drug target prediction. Bioinformatics 2019, 35, 2100–2107. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Zheng, C.; Xu, R. Combining phenome-driven drug-target interaction prediction with patients’ electronic health records-based clinical corroboration toward drug discovery. Bioinformatics 2020, 36, i436–i444. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Xu, R. Mining comorbidities of opioid use disorder from FDA adverse event reporting system and patient electronic health records. BMC Med. Inform. Decis. Mak. 2022, 22, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Xu, R. The Alzheimer’s comorbidity phenome: Mining from a large patient database and phenome-driven genetics prediction. JAMIA Open 2019, 2, 131–138. [Google Scholar] [CrossRef]
- Zheng, C.; Xu, R. Large-scale mining disease comorbidity relationships from post-market drug adverse events surveillance data. BMC Bioinform. 2018, 19, 85–93. [Google Scholar] [CrossRef]
- Friedman, C.; Hripcsak, G.; Shagina, L.; Liu, H. Representing information in patient reports using natural language processing and the extensible markup language. J. Am. Med. Inform. Assoc. 1999, 6, 76–87. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Liu, F.; Simpson, P.; Antieau, L.; Bennett, A.; Cimino, J.J.; Ely, J.; Yu, H. AskHERMES: An online question answering system for complex clinical questions. J. Biomed. Inform. 2011, 44, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Aronson, A.R. Effective mapping of biomedical text to the UMLS Metathesaurus: The MetaMap program. In Proceedings of the AMIA Symposium, Washington, DC, USA, 3–7 November 2001; p. 17. [Google Scholar]
- Aronson, A.R.; Lang, F.M. An overview of MetaMap: Historical perspective and recent advances. J. Am. Med. Inf. Assoc. 2010, 17, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Feng, Y.; Wang, X.; Wu, Y.; Zhang, Y.; Jiang, M.; Wang, J.; Xu, H. A comparison of conditional random fields and structured support vector machines for chemical entity recognition in biomedical literature. J. Cheminform. 2015, 7, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Leaman, R.; Wei, C.-H.; Lu, Z. tmChem: A high performance approach for chemical named entity recognition and normalization. J. Cheminform. 2015, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Neumann, M.; King, D.; Beltagy, I.; Ammar, W. ScispaCy: Fast and robust models for biomedical natural language processing. arXiv 2019, arXiv:1902.07669. [Google Scholar]
- Github. Fake News Dataset. Available online: https://raw.githubusercontent.com/susanli2016/NLP-with-Python/master/data/corona_fake.csv (accessed on 14 May 2022).
- Hussain, S.-A.; Sezgin, E.; Krivchenia, K.; Luna, J.; Rust, S.; Huang, Y. A natural language processing pipeline to synthesize patient-generated notes toward improving remote care and chronic disease management: A cystic fibrosis case study. JAMA Open 2021, 4, ooab084. [Google Scholar] [CrossRef]
- Bodenreider, O. The unified medical language system (UMLS): Integrating biomedical terminology. Nucleic Acids Res. 2004, 32, D267–D270. [Google Scholar] [CrossRef]
- Bada, M.; Eckert, M.; Evans, D.; Garcia, K.; Shipley, K.; Sitnikov, D.; Baumgartner, W.A.; Cohen, K.B.; Verspoor, K.; Blake, J.A. Concept annotation in the CRAFT corpus. BMC Bioinform. 2012, 13, 1–20. [Google Scholar] [CrossRef]
- Huang, M.-S.; Lai, P.-T.; Tsai, R.T.-H.; Hsu, W.-L. Revised JNLPBA corpus: A revised version of biomedical NER corpus for relation extraction task. arXiv 2019, arXiv:1901.10219. [Google Scholar]
- Li, J.; Sun, Y.; Johnson, R.J.; Sciaky, D.; Wei, C.-H.; Leaman, R.; Davis, A.P.; Mattingly, C.J.; Wiegers, T.C.; Lu, Z. BioCreative V CDR task corpus: A resource for chemical disease relation extraction. Database 2016, 2016, baw068. [Google Scholar] [CrossRef]
- Pyysalo, S.; Ohta, T.; Rak, R.; Rowley, A.; Chun, H.-W.; Jung, S.-J.; Choi, S.-P.; Tsujii, J.i.; Ananiadou, S. Overview of the cancer genetics and pathway curation tasks of bionlp shared task 2013. BMC Bioinform. 2015, 16, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Kaggle. Available online: https://www.kaggle.com/datasets/finalepoch/medical-ner (accessed on 1 June 2022).
- Honnibal, M.; Montani, I. spaCy 2: Natural language understanding with Bloom embeddings, convolutional neural networks and incremental parsing. Appear 2017, 7, 411–420. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Liu, H.; Motoda, H.; Setiono, R.; Zhao, Z. Feature selection: An ever evolving frontier in data mining. In Proceedings of the Feature Selection in Data Mining, Hyderabad, India, 21 June 2010; pp. 4–13. [Google Scholar]
- Saeys, Y.; Inza, I.; Larranaga, P. A review of feature selection techniques in bioinformatics. Bioinformatics 2007, 23, 2507–2517. [Google Scholar] [CrossRef]
- Louppe, G.; Wehenkel, L.; Sutera, A.; Geurts, P. Understanding variable importances in forests of randomized trees. Adv. Neural Inf. Process. Syst. 2013, 26. Available online: https://proceedings.neurips.cc/paper/2013/hash/e3796ae838835da0b6f6ea37bcf8bcb7-Abstract.html (accessed on 6 October 2022).
- Molina, M.D.; Sundar, S.S.; Le, T.; Lee, D. “Fake news” is not simply false information: A concept explication and taxonomy of online content. Am. Behav. Sci. 2021, 65, 180–212. [Google Scholar] [CrossRef]
- Xu, R.; Wang, Q. Large-scale extraction of accurate drug-disease treatment pairs from biomedical literature for drug repurposing. BMC Bioinform. 2013, 14, 1–11. [Google Scholar] [CrossRef]
- Xu, R.; Li, L.; Wang, Q. Towards building a disease-phenotype knowledge base: Extracting disease-manifestation relationship from literature. Bioinformatics 2013, 29, 2186–2194. [Google Scholar] [CrossRef]
- Xu, R.; Li, L.; Wang, Q. dRiskKB: A large-scale disease-disease risk relationship knowledge base constructed from biomedical text. BMC Bioinform. 2014, 15, 1–13. [Google Scholar] [CrossRef]
- Xu, R.; Wang, Q. A Knowledge-Driven Approach in Constructing a Large-Scale Drug-Side Effect Relationship Knowledge Base for Computational Drug Discovery. In Proceedings of the Bioinformatics Research and Applications: 10th International Symposium, ISBRA 2014, Zhangjiajie, China, 28–30 June 2014; p. 391. [Google Scholar]
- Westerlund, M. The emergence of deepfake technology: A review. Technol. Innov. Manag. Rev. 2019, 9, 39–52. [Google Scholar] [CrossRef]
- Abonizio, H.Q.; de Morais, J.I.; Tavares, G.M.; Barbon Junior, S. Language-independent fake news detection: English, Portuguese, and Spanish mutual features. Future Internet 2020, 12, 87. [Google Scholar] [CrossRef]
- Guibon, G.; Ermakova, L.; Seffih, H.; Firsov, A.; Le Noé-Bienvenu, G. Multilingual fake news detection with satire. In Proceedings of the CICLing: International Conference on Computational Linguistics and Intelligent Text Processing, La Rochelle, France, 7–13 April 2019. [Google Scholar]
- Lee, J.-W.; Kim, J.-H. Fake Sentence Detection Based on Transfer Learning: Applying to Korean COVID-19 Fake News. Appl. Sci. 2022, 12, 6402. [Google Scholar] [CrossRef]
- Digan, W.; Névéol, A.; Neuraz, A.; Wack, M.; Baudoin, D.; Burgun, A.; Rance, B. Can reproducibility be improved in clinical natural language processing? A study of 7 clinical NLP suites. J. Am. Med. Inform. Assoc. 2021, 28, 504–515. [Google Scholar] [CrossRef] [PubMed]

| Studies | Data Sources | Linguistics | Sentiment | NER | Biomedical |
|---|---|---|---|---|---|
| Alenezi et al. [26] | Twitter, WHO, CDC, etc. | No | No | No | No |
| Tashtoush et al. [16] | WHO, UN, Google Fact Check, etc. | No | No | No | No |
| Bangyal et al. [22] | Facebook, Instagram, etc. | No | No | No | No |
| Endo et al. [23] | Brazilian Ministry of Health, Boatos.org, etc. | Yes | No | No | No |
| Al-Rakhami et al. [21] | Yes | No | No | No | |
| Daley et al. [35] | Politifact.com | Yes | Yes | No | No |
| Gupta et al. [38] | Yes | No | Yes | No | |
| Iwendi et al. [25] | Facebook, Twitter, The New York Times, etc. | Yes | Yes | Yes | No |
| Khan et al. [24] | Facebook, Twitter, The New York Times, etc. | Yes | Yes | Yes | No |
| Our study | Facebook, Twitter, The New York Times, etc. | Yes | Yes | Yes | Yes |
| Models | Hyperparameters | Levels |
|---|---|---|
| Logistic regression | Inverse of regularization strength and norm of the penalty | 0.1, 1.0, and 10.0 L1 and L2 |
| AdaBoost | Maximum number of estimators and learning rate | 50, 100, and 200 0.1, 0.5, and 1.0 |
| Bagging | Maximum number of estimators | 10, 50, and 100 |
| Decision tree | Minimum samples to split and minimum samples to be at a leaf | 2.0, 5.0, and 10.0 1.0, 2.0, and 4.0 |
| SVC_rbf | Regularization parameter | 0.1, 1.0, 5.0, and 10 |
| SVC_poly | Regularization parameter | 0.1, 1.0, 5.0, and 10 |
| SVC_sigmoid | Regularization parameter | 0.1, 1.0, 5.0, and 10 |
| kNN | Number of neighbors and weight function | 3.0, 5.0, 7.0, and 9.0 Uniform and distance |
| Naïve bayes | Additive smoothing parameter | 1.0, 5.0, and 10.0 |
| Random forest | Number of trees and number of features | 50, 100, and 200, Sqrt and Log2 |
| SGDClassifier_L1 | Learning rate | Constant, optimal, and invscaling |
| SGDClassifier_L2 | Learning rate | Constant, optimal, and invscaling |
| SGDClassifier_EN | Learning rate | Constant, optimal, and invscaling |
| LinearSVC_L1 | Regularization parameter | 0.1, 1.0, 10.0, and 100.0 |
| LinearSVC_L2 | Regularization parameter | 0.1, 1.0, 10.0, and 100.0 |
| Models | Hyperparameters | ScispaCy | MetaMap | Custom | Combine 1 | Combine 2 | Combine 3 | All |
|---|---|---|---|---|---|---|---|---|
| Logistic regression | Inverse of regularization strength and norm of the penalty | 10 L2 | 0.1 L2 | 0.1 L2 | 0.1 L2 | 1 L2 | 0.1 L2 | 0.1 L2 |
| AdaBoost | Maximum number of estimators and learning rate | 200 1.0 | 100 1.0 | 50 0.1 | 200 0.5 | 100 1.0 | 50 1.0 | 50 1.0 |
| Bagging | Maximum number of estimators | 100 | 50 | 10 | 100 | 100 | 100 | 100 |
| Decision tree | Minimum samples to split and minimum samples to be at a leaf | 10 4 | 10 4 | 2 2 | 10 2 | 10 4 | 5 4 | 10 4 |
| SVC_rbf | Regularization parameter | 5 | 5 | 5 | 1 | 1 | 5 | 1 |
| SVC_poly | Regularization parameter | 5 | 1 | 10 | 1 | 10 | 1 | 1 |
| SVC_sigmoid | Regularization parameter | 0.1 | 0.1 | 1 | 0.1 | 0.1 | 0.1 | 0.1 |
| kNN | Number of neighbors and weight function | 9 distance | 9 distance | 5 uniform | 9 distance | 9 distance | 9 distance | 7 distance |
| Naïve bayes | Additive smoothing parameter | 10.0 | 1.0 | 10.0 | 1.0 | 10.0 | 1.0 | 1.0 |
| Random forest | Number of trees and number of features | 100 Log2 | 200 Log2 | 100 Sqrt | 200 Log2 | 200 Log2 | 200 Sqrt | 200 Sqrt |
| SGDClassifier_L1 | Learning rate | constant | optimal | invscaling | optimal | optimal | optimal | optimal |
| SGDClassifier_L2 | Learning rate | constant | optimal | invscaling | constant | constant | optimal | optimal |
| SGDClassifier_EN | Learning rate | optimal | invscaling | invscaling | optimal | invscaling | constant | optimal |
| LinearSVC_L1 | Regularization parameter | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| LinearSVC_L2 | Regularization parameter | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| ScispaCy | MetaMap | Custom | Combine 1 | Combine 2 | Combine 3 | All | |
|---|---|---|---|---|---|---|---|
| Logistic regression | 0.712 | 0.824 | 0.629 | 0.821 | 0.722 | 0.825 | 0.826 |
| AdaBoost | 0.713 | 0.823 | 0.621 | 0.839 | 0.752 | 0.813 | 0.847 |
| Bagging | 0.736 | 0.863 | 0.645 | 0.853 | 0.764 | 0.852 | 0.849 |
| Decision tree | 0.658 | 0.722 | 0.646 | 0.703 | 0.637 | 0.730 | 0.747 |
| SVC_rbf | 0.741 | 0.844 | 0.669 | 0.836 | 0.740 | 0.844 | 0.844 |
| SVC_poly | 0.716 | 0.776 | 0.568 | 0.781 | 0.748 | 0.777 | 0.789 |
| SVC_sigmoid | 0.698 | 0.774 | 0.560 | 0.783 | 0.709 | 0.777 | 0.786 |
| kNN | 0.709 | 0.824 | 0.659 | 0.815 | 0.744 | 0.829 | 0.830 |
| Naïve bayes | 0.667 | 0.673 | 0.618 | 0.677 | 0.672 | 0.673 | 0.676 |
| Random forest | 0.738 | 0.879 | 0.668 | 0.877 | 0.775 | 0.882 | 0.882 |
| SGDClassifier_L1 | 0.598 | 0.723 | 0.526 | 0.705 | 0.679 | 0.710 | 0.699 |
| SGDClassifier_L2 | 0.598 | 0.723 | 0.526 | 0.745 | 0.643 | 0.710 | 0.699 |
| SGDClassifier_EN | 0.663 | 0.760 | 0.526 | 0.705 | 0.723 | 0.724 | 0.699 |
| LinearSVC_L1 | 0.667 | 0.743 | 0.602 | 0.743 | 0.658 | 0.746 | 0.746 |
| LinearSVC_L2 | 0.663 | 0.743 | 0.595 | 0.736 | 0.658 | 0.746 | 0.739 |
| ScispaCy | MetaMap | Custom | Combine 1 | Combine 2 | Combine 3 | All | |
|---|---|---|---|---|---|---|---|
| Acc | 0.667 | 0.795 | 0.628 | 0.778 | 0.698 | 0.781 | 0.799 |
| AUC (ROC) | 0.738 | 0.879 | 0.668 | 0.877 | 0.775 | 0.882 | 0.882 |
| F1 | 0.681 | 0.794 | 0.649 | 0.778 | 0.695 | 0.784 | 0.801 |
| Recall | 0.647 | 0.76 | 0.647 | 0.747 | 0.660 | 0.760 | 0.780 |
| Specificity | 0.642 | 0.762 | 0.617 | 0.747 | 0.667 | 0.755 | 0.774 |
| Precision | 0.693 | 0.832 | 0.639 | 0.812 | 0.733 | 0.809 | 0.829 |
| Number of Trees | Number of Features | |
|---|---|---|
| NER (baseline) | Log2 | 200 |
| All | Sqrt | 200 |
| NER + ScispaCy | Sqrt | 200 |
| NER + MetaMap | Sqrt | 200 |
| NER + Custom | log2 | 200 |
| NER + Combine 1 | log2 | 200 |
| NER + Combine 2 | Sqrt | 200 |
| NER + Combine 3 | log2 | 200 |
| NER + All | Sqrt | 200 |
| Acc. | AUC | F1 | Recall | Spec. | Precision | |
|---|---|---|---|---|---|---|
| NER (baseline) | 0.701 | 0.754 | 0.709 | 0.700 | 0.683 | 0.719 |
| All | 0.799 | 0.882 | 0.801 | 0.780 | 0.774 | 0.829 |
| NER + ScispaCy | 0.781 | 0.830 | 0.765 | 0.727 | 0.732 | 0.807 |
| NER + MetaMap | 0.833 | 0.890 | 0.815 | 0.780 | 0.781 | 0.854 |
| NER + Custom | 0.743 | 0.807 | 0.753 | 0.753 | 0.732 | 0.753 |
| NER + Combine 1 | 0.823 | 0.906 | 0.826 | 0.807 | 0.800 | 0.846 |
| NER + Combine 2 | 0.760 | 0.838 | 0.767 | 0.747 | 0.740 | 0.789 |
| NER + Combine 3 | 0.792 | 0.890 | 0.789 | 0.747 | 0.753 | 0.836 |
| NER + All | 0.819 | 0.899 | 0.814 | 0.787 | 0.784 | 0.843 |
| Number of Trees | Number of Features | |
|---|---|---|
| Linguistics | Log2 | 200 |
| Sentiment | Sqrt | 200 |
| Linguistics + Sentiment | Sqrt | 200 |
| NER | Log2 | 200 |
| BioIE | Sqrt | 200 |
| BioIE + NER | Sqrt | 200 |
| Acc. | AUC | F1 | Recall | Spec. | Precision | |
|---|---|---|---|---|---|---|
| Linguistics | 0.674 | 0.785 | 0.667 | 0.627 | 0.641 | 0.712 |
| Sentiment | 0.628 | 0.673 | 0.649 | 0.660 | 0.617 | 0.639 |
| Linguistics + Sentiment | 0.753 | 0.834 | 0.751 | 0.713 | 0.719 | 0.793 |
| NER | 0.701 | 0.754 | 0.709 | 0.700 | 0.683 | 0.719 |
| BioIE | 0.799 | 0.882 | 0.801 | 0.780 | 0.774 | 0.829 |
| BioIE + NER | 0.819 | 0.899 | 0.814 | 0.787 | 0.784 | 0.843 |
| Number of Trees | Number of Features | |
|---|---|---|
| BioIE-driven model | Sqrt | 200 |
| State-of-the-art model | Sqrt | 200 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fifita, F.; Smith, J.; Hanzsek-Brill, M.B.; Li, X.; Zhou, M. Machine Learning-Based Identifications of COVID-19 Fake News Using Biomedical Information Extraction. Big Data Cogn. Comput. 2023, 7, 46. https://doi.org/10.3390/bdcc7010046
Fifita F, Smith J, Hanzsek-Brill MB, Li X, Zhou M. Machine Learning-Based Identifications of COVID-19 Fake News Using Biomedical Information Extraction. Big Data and Cognitive Computing. 2023; 7(1):46. https://doi.org/10.3390/bdcc7010046
Chicago/Turabian StyleFifita, Faizi, Jordan Smith, Melissa B. Hanzsek-Brill, Xiaoyin Li, and Mengshi Zhou. 2023. "Machine Learning-Based Identifications of COVID-19 Fake News Using Biomedical Information Extraction" Big Data and Cognitive Computing 7, no. 1: 46. https://doi.org/10.3390/bdcc7010046
APA StyleFifita, F., Smith, J., Hanzsek-Brill, M. B., Li, X., & Zhou, M. (2023). Machine Learning-Based Identifications of COVID-19 Fake News Using Biomedical Information Extraction. Big Data and Cognitive Computing, 7(1), 46. https://doi.org/10.3390/bdcc7010046





