1. Introduction
Rice is the principal food grain consumed by more than half of the world’s population [
1]. However, rice (
Oryza sativa L.) production faces biotic and abiotic constraints worldwide [
2]. The most common biotic constraints include blast, sheath rot, and brown spot in Burundi [
3,
4]. Among abiotic constraints, drought stress is the major one in rain-fed ecologies [
2]. For numerous soils, at least two weeks without rainfall induces noticeable negative differences in drought sensitivity during the vegetative stage, and at least seven days without rainfall causes severe drought damage during the reproductive stage [
5]. Drought can cause yield losses of up to 21% under mild drought, up to 51% under moderate drought, and up to 90.6% in severe cases [
6], depending on the grown variety, growth stage, degree, and duration of the stress.
Reduced grain yield is a result of morphological responses such as increases in leaf rolling, stomata closure, and leaf tip drying; molecular responses that include changes in gene expression (up/down regulation of transcripts) and the activation of relevant transcription factors and signaling pathways; and physiological and biochemical responses such as reductions in transpiration, photosynthesis, chlorophyll content, membrane stability, stomatal conductance, and increases in osmoprotectants [
7]. Drought stress reduces the performance of rice varieties that are grown worldwide [
8,
9,
10].
In East Africa, rice-growing areas are exposed to severe drought [
10]. In Burundi, the major constraints on rice production include inputs, flooding, and drought, which accounted for 41%, 30%, and 29%, respectively [
11]. In Burundi, irrigated lowland rice is grown in Moso and mostly in the Imbo region. Most farmers in Imbo do not regularly obtain water for irrigation due to insufficient or destroyed infrastructures. Imbo is generally semiarid with low and irregular rainfall that can reach up to 500 mm per year [
12]. Neither of the current rice varieties grown in Burundi can withstand such a complicated rainfall pattern. Thus, screening rice varieties suitable for Burundi becomes a priority using an appropriate approach.
A modified conventional breeding approach was suggested to integrate phenotyping, genotyping, and a strategy to screen many lines among which selection can be made [
13]. The approach improved the assessment of plant responses to drought stress.
Three levels of drought stress corresponding to 5% (m
3/m
3), 10.6% (m
3/m
3), and 16% (m
3/m
3) soil moisture content, representing severe, moderate, and low drought, respectively, were used by Singh et al. [
14] to assess the response of rice cultivars to early-season drought stress. Different methods of screening rice genotypes for drought tolerance have been used by researchers through classical markers or DNA/molecular markers [
15,
16]. Depending on the specific objective, each type of marker may present advantages and/or disadvantages. Currently, single nucleotides polymorphic markers (SNPs) are mostly used due to their high frequency, low mutation rates, and high-throughput nature [
17]. Molecular markers have been a very useful tool mostly in Asian countries (Philippines, India, Nepal, Malaysia, etc.) where different quantitative trait loci (QTLs) for drought tolerance have been identified [
16,
18].
Polygenic architectures were reported for many traits under both drought and irrigated environments. Previous studies have demonstrated that conditional neutrality is more common than antagonistic pleiotropy [
19]. This provides an explanation as to why rice breeders successfully developed drought tolerant rice lines and varieties without a yield penalty in irrigated environments [
20]. Shamsudin et al. [
21] found a positive interaction between qDTY2.2 and qDTY12.1 in the developed rice lines through marker assisted breeding.
According to IRRI, QTLs with large effects that are qDTY1.1, qDTY2.2, qDTY3.1, qDTY3.2, and qDTY12.1 may be used to improve rice varieties for grain yield under reproductive-stage drought in lowland areas [
18]. Introgression or pyramiding of some of these QTLs was successfully achieved, especially in Asia, where drought-tolerant varieties have been released [
18,
22].
The International Rice Research Institute (IRRI) released genotypes IR 86781-3-3-1-1 and IR 81412-B-B-82-1 in the Philippines, and IR 82077-B-B-71-1, IR 82589-B-B-84-3, and IR 83388-B-B-108-3 in Malawi, Bangladesh, and Nepal, respectively, for drought tolerance in lowland ecosystems. IR 79913-B-176-B-4 and IR 55423-01 are upland varieties that were released in the Philippines and India, respectively [
13]. However, there is no information about the use of these genotypes in rice improvement for drought tolerance. Furthermore, there is no report on the release of drought-tolerant rice varieties in Burundi [
8].
Therefore, there is a need to effectively utilize the identified QTLs for drought tolerance in developing drought-tolerant rice lines to meet the preferences of producers in Burundi. To enhance breeding efforts, potentially drought-tolerant genotypes were collected from Asia and Eastern and Southern African countries. This study aimed to identify genotypes that have putative QTLs associated with drought tolerance.
3. Discussion
This study detected significant differences in leaf rolling and leaf drying among genotypes under drought stress. Furthermore, differences in plant height, number of panicles per plant, one thousand grain weight, and grain yield were significant, suggesting genetic diversity among the tested genotypes. This implies the possibility of selecting most drought-tolerant lines for their further use by farmers or by the breeding program. Comparable results were reported by Mohd Ikmal et al. [
23] in BC1 F4 lines. Spikelet fertility and yield components were significantly reduced by drought stress. Similarly, all cultivars subjected to drought stress exhibited a significant grain yield reduction in a study conducted by Adhikari et al. [
9]. The effects of drought on morphological and agronomic traits, including leaf area, panicle length, plant height, tillering ability, and efficiency, results in decreased yield [
24].
The performance of the evaluated genotypes was better under irrigated conditions than under drought stress conditions. The current results agree with the findings of previous studies on drought where the best cultivars under nonstress conditions exhibited poor performance under stress conditions [
9,
18]. The reduction in performance of a given genotype increases with drought intensity [
14]. This validates the significant efforts that breeders and geneticists have put into coping with water scarcity by finding QTLs and genes for drought tolerance and deploying them in genotypes with different backgrounds [
13,
18,
22]. Therefore, growing drought-tolerant rice varieties is an alternative genetic adaptive strategy to increase rice yield and production in areas where farmers have limited access to water for irrigation [
8].
A positive correlation between yield and the number of filled grains per panicle and the number of panicles per hill indicates that a higher number of panicles per plant and a higher number of filled grains per panicle lead to higher yield. Abd Allah et al. [
25] reported that the number of panicles per plant, the number of filled grains per panicle, and 100 grain weight are key traits in improving yield under both irrigated and drought stress conditions. The negative correlation between yield and plant phenotypic acceptability is due to the nature of the scale for scoring phenotypic acceptability where higher scores correspond to poor performance [
5]. The results imply that the phenotypically desirable genotypes also had higher yields.
Strong, positive, and highly significant correlations between the yield, the yield BLUP, and the STI for yield suggest that high yielding genotypes can be selected based on BLUP or STI. However, the results of this study showed that the correlation coefficient between the yield and the BLUP was greater than the one between yield and STI for yield under drought stress. Furthermore, some genotypes with high STI had low yield under both drought and irrigated conditions (
Table 3 and
Tables S2 and S3).
Best linear unbiased prediction (BLUP) was reported to be the most efficient prediction method among the commonly used methods for selection [
26]. Breeding values imply the ability to perform well in crosses, and they have been recommended to select genotypes with high performance in most of the desirable traits [
13].
A significantly negative correlation between the number of days to flowering and the number of filled grains per panicle shows that the longer the cycle of the genotypes, the fewer the number of filled grains per panicle due to the increase in drought intensity at the reproductive stage. Guimarães et al. [
27] stated that late-flowering genotypes had high spikelet sterility. The negative and nonsignificant correlation between yield and brown spot shows that yield was slightly affected by this disease. Severe cases of this disease were reported with a yield loss of 50–90% in Bengal [
17].
Promising lines were found among screened genotypes during the current study. The genotype IR 108044-B-B-B-3-B-B was classified as the best in the field based on yield BLUP. This genotype has three of the targeted QTLs for drought tolerance, including qDTY2.2, which was significantly associated with all the phenotypic data, and qDTY1.1, which was found in a few genotypes and was significantly associated with the yield BLUPs. In the same way, the genotype IR 92522-45-3-1-4 was ranked fourth based on the yield BLUP from the field where it was under severe drought stress and had three of the targeted QTLs for drought tolerance, including qDTY2.2. Similarly, the genotype BRRI DHAN 55 was under severe drought stress but was ranked seventh in yield BLUP and had three of the major targeted QTLs for drought tolerance, including qDTY1.1. The BLUPs and high yield of these genotypes under drought can enable them to be considered parents for drought tolerance, which can be used to improve existing rice varieties. Dhawan et al. [
28] used Nagina 22 as a drought-tolerant parent, for which the yield was 1.77 t/ha under drought stress.
The yield of genotypes IR 97013-8-1-3-2-B, IR 13240-108-2-2-3, WAHIWAHI, and IR 97013-19-1-3-1-B was reduced by rodents that strongly attacked them a few days before pesticide application. The high scores of leaf rolling and leaf drying from the field experiment indicate the presence of a high intensity of drought during this study. High leaf drying induced a reduced yield, confirmed by the negative correlation between these traits. Similar results were obtained by Bocco et al. [
24] where more plants with high leaf drying provided lower yields.
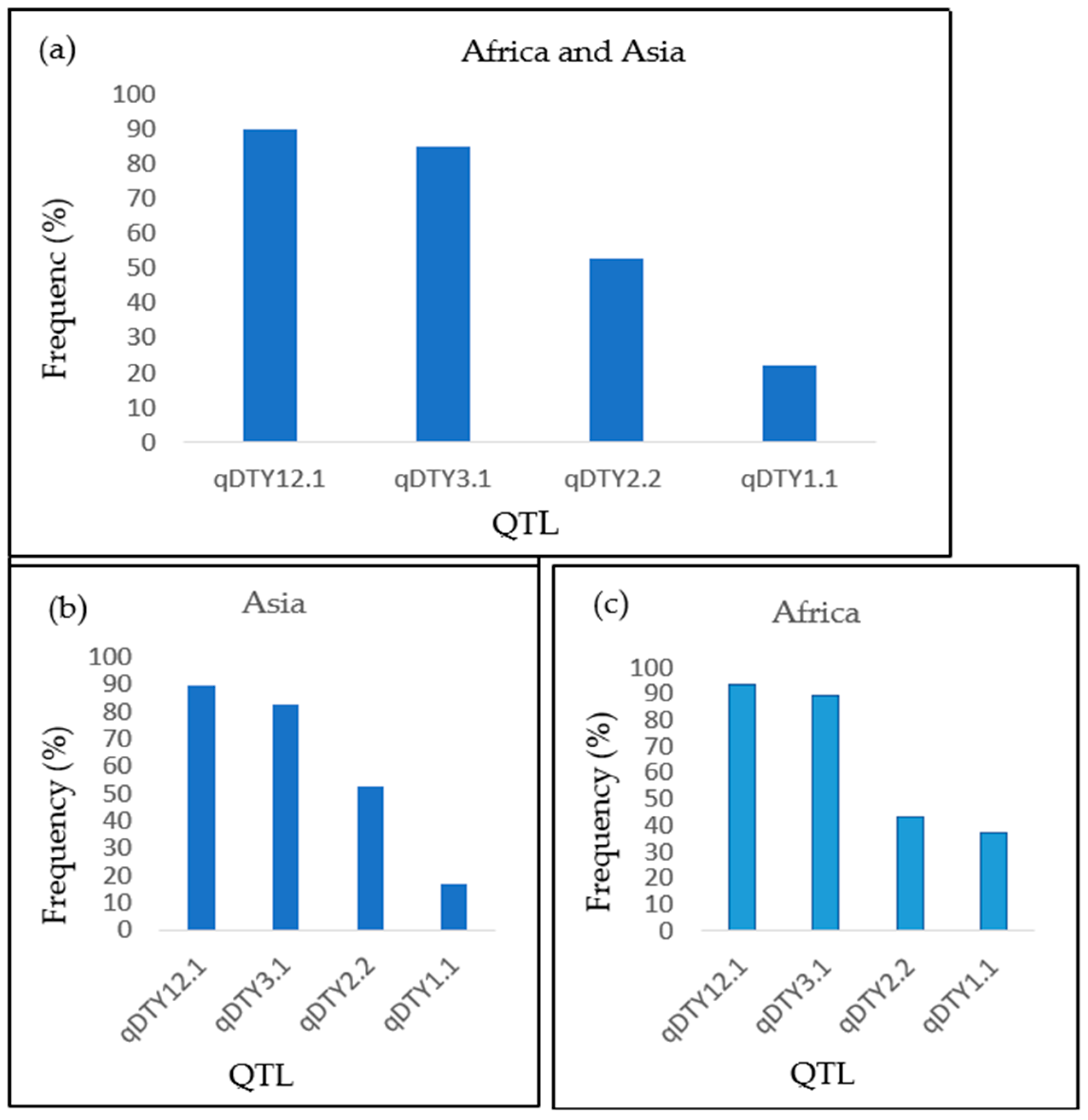
The majority of evaluated genotypes had at least two QTLs for drought tolerance providing some yield advantage under drought stress. Appropriate QTL combinations is a good approach for improving drought tolerance [
16,
21,
29]. In the current study, some genotypes with all the four major QTLs were among the worst genotypes under drought stress. Indeed, most of these genotypes with low yield under drought stress were low-yielding even under no stress conditions. Researchers have reported that a high yield potential under no stress is a good indicator of a high yield advantage under drought stress [
21]. Another reason could be the interaction between these QTLs, even if conditional neutrality was reported to be more common than antagonistic pleiotropy [
19]. The QTLs qDTY2.2 and qDTY3.1 pyramided with qDTY12.1 significantly increased the yield of lines having qDTY12.1 in the study of Shamsudin et al. [
21]. Therefore, pyramiding the best combination of alleles with favorable interactions is the best strategy to improve the performance of rice varieties under drought stress [
13]. In this study, the QTLs qDTY12.1, qDTY3.1, and qDTY2.2 were present in more than 50% of the evaluated genotypes. Shamsudin et al. [
21] found qDTY12.1, qDTY3.1, and qDTY2.2 in 82%, 36%, and 18% of selected pyramided lines, respectively. The analysis of genetic diversity of 60 rice genotypes detected qDTY12.1 and qDTY2.2 in 43.3% and 6.67% of evaluated genotypes, respectively [
30].
During the current study, only qDTY2.2 was significantly associated with all the phenotypic data. This indicates that qDTY2.2 is a major QTL for drought tolerance that can be used for the improvement of preferred varieties in Burundi. However, qDTY1.1 was also significantly associated with the yield BLUPs only; further study using other genotypes in Burundi shall help to confirm our findings. Kadam et al. [
31] demonstrated the complexity of yield traits under drought stress by detecting very many different QTLs between years and treatments, even by comparing with previous studies using the same genotypes. Overlapping transcriptions between the water-use efficiency and the days to flowering revealed a genetic basis for a trade-off between drought avoidance and drought escape in rice [
19]. Therefore, a statistical power analysis accompanying a proper genotypic and phenotypic sampling is very important for QTL studies [
32,
33]. Drought tolerance is a complex trait that is characterized by low heritability, genotype-by-environment interactions, genetic interactions, and polygenic effects [
16]. Furthermore, drought and heat are reported to often occur together. The genes regulating tolerance to these stresses are different but share some signaling pathways [
34]. Several genetic management approaches have been suggested by researchers to increase rice production in a changing climate [
34,
35]. Rice improvement for drought tolerance may continue through pyramiding major QTLs for drought tolerance by crossing elite x elite cultivars or by marker-assisted backcrossing [
21,
29] involving landraces or wild rice as a source of drought tolerance [
36] followed by multi-environmental trials to select for a specific environment or location [
16].