Abstract
Land use change is a major predictor for variations in soil bacteria, which plays a key role in maintaining soil multifunctionality and function of terrestrial ecosystems. The effects of land use change on the soil bacterial community in an alpine region at the Qinghai-Tibetan Plateau (QTP) were still unclear. In this study, we investigated soil bacterial profiles under grazed grassland, enclosed grassland, continuous cropland, rotation cropland and abandoned cropland in the Tianzhu alpine agro-pastoral ecotone. Our results showed that Proteobacteria, Acidobacteria and Actinobacteria were the most three abundant phyla regardless of land use change, accounting for over 60% of the total. Cultivation declined soil bacterial alpha diversity without recovering even after abandonment. Over 73% variations in bacterial diversity can be explained by soil physical and chemical properties. In addition, soil moisture could be the main driver for the difference in bacterial structure between croplands and others. Soil bulk density, pH, organic carbon and total nitrogen contents seem to be the key factors determining the difference in bacterial structure between abandoned croplands and others. Our results have implications for comprehensive understanding about the responses of the soil bacterial community to land use change in alpine regions.
1. Introduction
The Qinghai-Tibetan Plateau (QTP) is a unique geographical area called “the third pole”, widely occupied by alpine meadows with approximately 0.7 million km2 [1,2]. The alpine meadow has powerful ecological service functions; it plays important roles in maintaining soil carbon and water stocks, preserving biodiversity, and providing habitats and foods for livestock [3,4]. However, alpine meadows at the QTP have been experiencing substantial shrinking and seriously degradation under land use change and climate warming [5,6]. This happened particularly at regions located in the intersections of farming area and pasturing area, which are called the alpine agro-pastoral ecotone [7]. In these areas, the increasing needs of cropland caused heavy grassland conversion.
Soil bacteria are well known for their irreplaceable positions in catalyzing biogeochemical reactions and driving global nutrient cycles, playing a key role in shaping the function of terrestrial ecosystems [8,9,10]. Especially in alpine regions, soil bacteria are the dominant soil microbes and more sensitive to environmental change compared with fungi [11]. Previous studies have documented that land use change can strongly affect soil bacterial community through affecting soil properties in various ecosystems across the world, such as semi-arid area, wetlands, tropical uplands, tropical forests, agro-ecosystems, etc. [12,13,14,15,16,17]. However, how bacterial community and diversity responds to land use change in the alpine meadow zone at the QTP remains poorly understood. The studies on soil bacteria in alpine meadows at the QTP were mainly focused on the effects of nitrogen addition, grazing practices, grazing prohibition and grassland degradation [18,19,20].
Conversion from grassland to cropland increases the diversity of bacterial communities in north China and semi-arid grassland [21,22]; in contrast, cultivation decreases the soil bacterial abundance in the agro-pastoral ecotone in northeast China [23]. These controversial results across different ecotones lead to large uncertainty in evaluating the effects of cultivation on soil bacterial diversity in the alpine region. In the present study, we examined the soil bacterial composition and structure across grazed grassland, enclosed grassland, continuous cropland, rotation cropland and abandoned cropland in a typical alpine agro-pastoral ecotone at the northeast boarder of the QTP. Our hypotheses are (1) grassland converted to cropland would decrease soil bacterial abundance but increase soil bacterial diversity; (2) after abandoned, the soil bacterial community should return towards to the original state before cultivation; and (3) variations in soil properties driven by land use change determine soil bacterial shifting.
2. Materials and Methods
2.1. Study Area
Our study site (37°40′ N, 102°32′ E, ~3000 m a.s.l.) was in the Tianzhu alpine agro-pastoral ecotone. The average annual air temperature is −0.1 °C, with the average temperature ranging from −18.3 °C in January to 12.7 °C in July. The average annual precipitation is 416 mm. There is no frost-free period in this region; the annual sunshine time is 2600 h, and the plant growing season lasts about 130 days. The soil type is alpine meadow (Chernozem) with basic properties such as soil thickness ranging from 40~80 cm, pH 7.0~8.2 and soil organic matter content 10~16%.
2.2. Details of Selected Land Use Patterns
Grazed grassland, enclosed grassland, continuous cropland, rotation cropland and abandoned cropland were selected as five land use patterns in our study area. The descriptions of the five land use patterns were described in detail in our previous study [7]. Briefly, the altitudes of the five land use patterns were all around 2900 m and ranged from 2909 to 2942 m, all within an area with a ~1.2 km diameter. These five land use patterns were for yak and sheep grazing (i.e., grazed grassland in our study) before the middle of last century until converted to the current land use practices decades ago. The grazed grassland was an all-year pasture with medium to high grazing intensity, and dominant species were Kobresia humilis and Stipa capillata L. in this site. To set up an enclosed grassland, part of the grazed grassland had been fenced over 20 years, the dominant species had been in succession to Elymus nutans, Griseb and Poa annua L., which belongs to gramineae. Two croplands were converted to the current practice over 20 and 7 years, respectively; one cropland was for continuous oat cultivation, the other was for highland barley-oat rotation, both with N, P, K fertilizer application. Abandoned cropland was ploughed up for crop production since 1950s and without any disturbance for eight years prior to our study.
2.3. Soil Collection
Four replicated plots (2 m × 10 m) were randomly established within each land use pattern (1000 m2 for each except 240 m2 for enclosed grassland) in September 2016. We collected the top 20 cm of soil profiles with litter, stone and roots free by an auger with an internal diameter of 5 cm. Six soil samples from each plot were taken and homogenized mixed to form one replicate, dividing two parts for analyzing soil properties and bacterial communities, respectively. Subsamples for testing soil physical and chemical characters were air-dried and subsamples for bacterial analysis were immediately taken back to the lab and stored at −80 °C.
2.4. Soil Physicochemical Analysis
Soil organic carbon (SOC) contents were detected by a TOC analyzer (multi N/C UV, Analytik Jena AG, Jena, Germany), and TN was determined by the Kjeldahl method [24]. Soil pH was measured in a soil/water ratio 1:1 [25]. Bulk density (BD) was taken separately by a press coring device (50.46 cm diameter, 50 cm height, 100 cm3) with a sleeve of steel cylinders. Soil temperature (ST) and moisture (SM) were detected directly in the field by automatic hygrothermographs (L99-TWS-1, Hangzhou Loggertech Co., Ltd., Hangzhou, China).
2.5. Soil Bacterial Analysis
DNA was extracted from each soil sample by Soil DNA Extraction Kit (Omega Bio-Tek, Atlanta, GA, USA), then V4 regions of the 16S rRNA gene were amplified from the extracted DNA by primers (515F and 806R) designed to amplify bacteria. After detected and purified, the PCR products were barcoded, sequenced and prepared for a DNA library (TruSeq®® DNA PCR-Free Sample Preparation Kit), then performed via paired-end sequencing based on Illumina HiSeq platform by Novogene Co., Ltd. Sequence data from Illumina HiSeq platform were cleaned and filtered, and then clustered into different operational taxonomic units (OTUs) according to 97% sequence similarity. These data were normalized based on the sample with the least sequences and used for the subsequent analysis of bacterial diversity.
2.6. Statistical Analysis
A venn diagram was used to visualize shared and unique OTUs under the five land use patterns in R package “VennDiagram”. Alpha diversity (Observed species, Chao 1, ACE, Simpson index and Shannon index) were calculated with QIIME (version 1.7.0). Correlations between alpha diversity and soil properties using the Pearson correlation coefficient were performed with the “Hmisc” package in R. Redundancy analysis (RDA), nonmetric multidimensional scaling (NMDS), analysis of similarities (ANOSIM) values and Variation partition analysis (VPA) were all performed in R package “vegan”. We used one-way ANOVA with Fishers Least Significant Difference (LSD) to test the effects of land use change on soil properties and bacterial indices within package of “agricolae” in RStudio.
3. Results
3.1. Soil Bacterial Taxonomic Classification and Composition
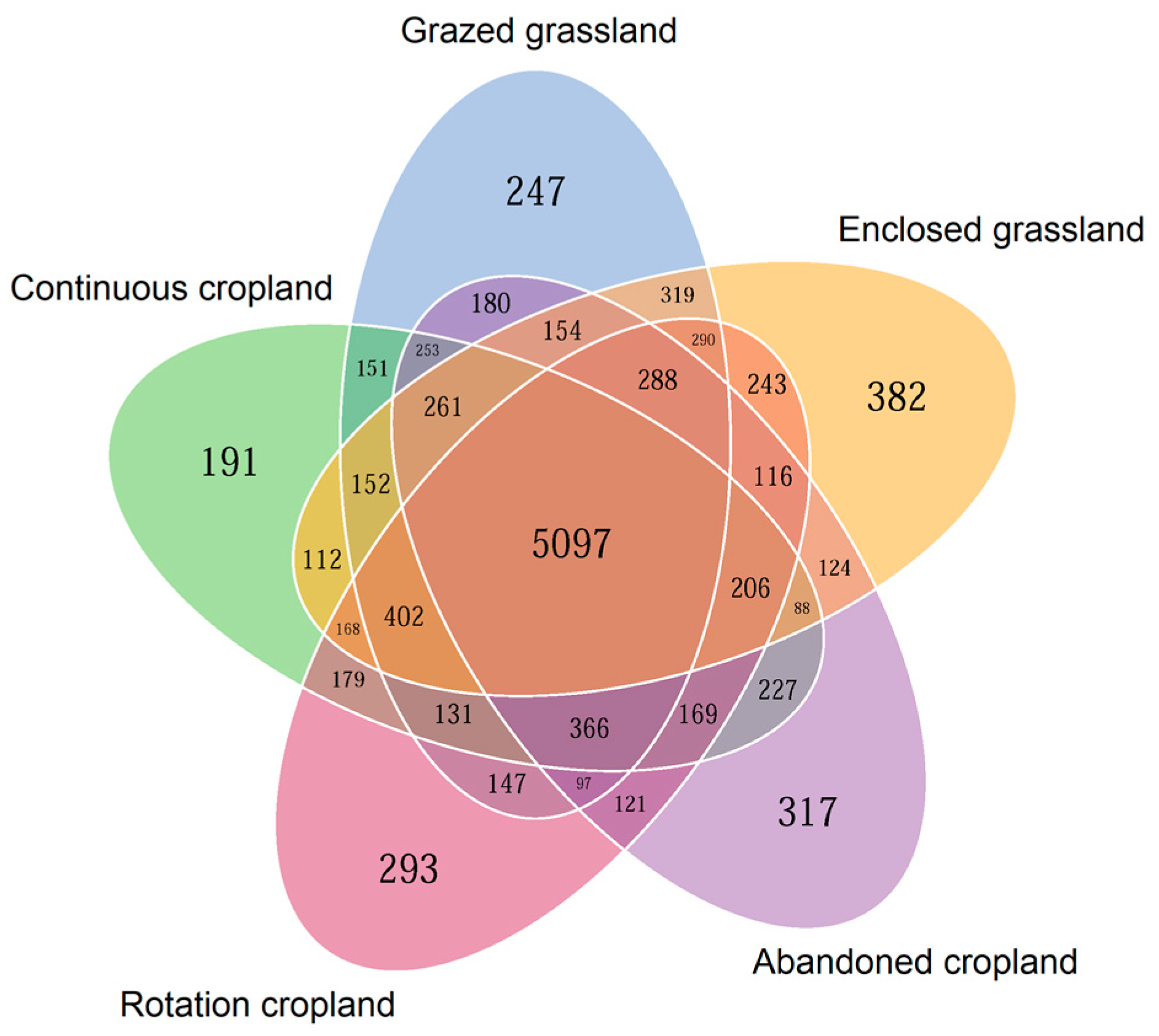
A total of 5097 shared OTUs were found in all five land use patterns, accounting for more than 90% of total OTUs under different land use patterns (Figure 1). The quantity of unique bacteria was shown a decreasing order of enclosed grassland > abandoned cropland > rotation cropland > grazed grassland > continuous cropland, with OTUs of 382, 317, 293, 247 and 191, respectively.
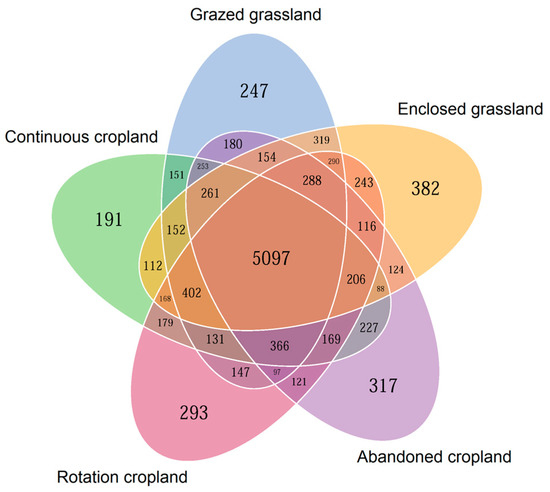
Figure 1.
Venn diagrams demonstrating the averaged shared and unique bacterial sequences associated with different land use patterns (n = 4).
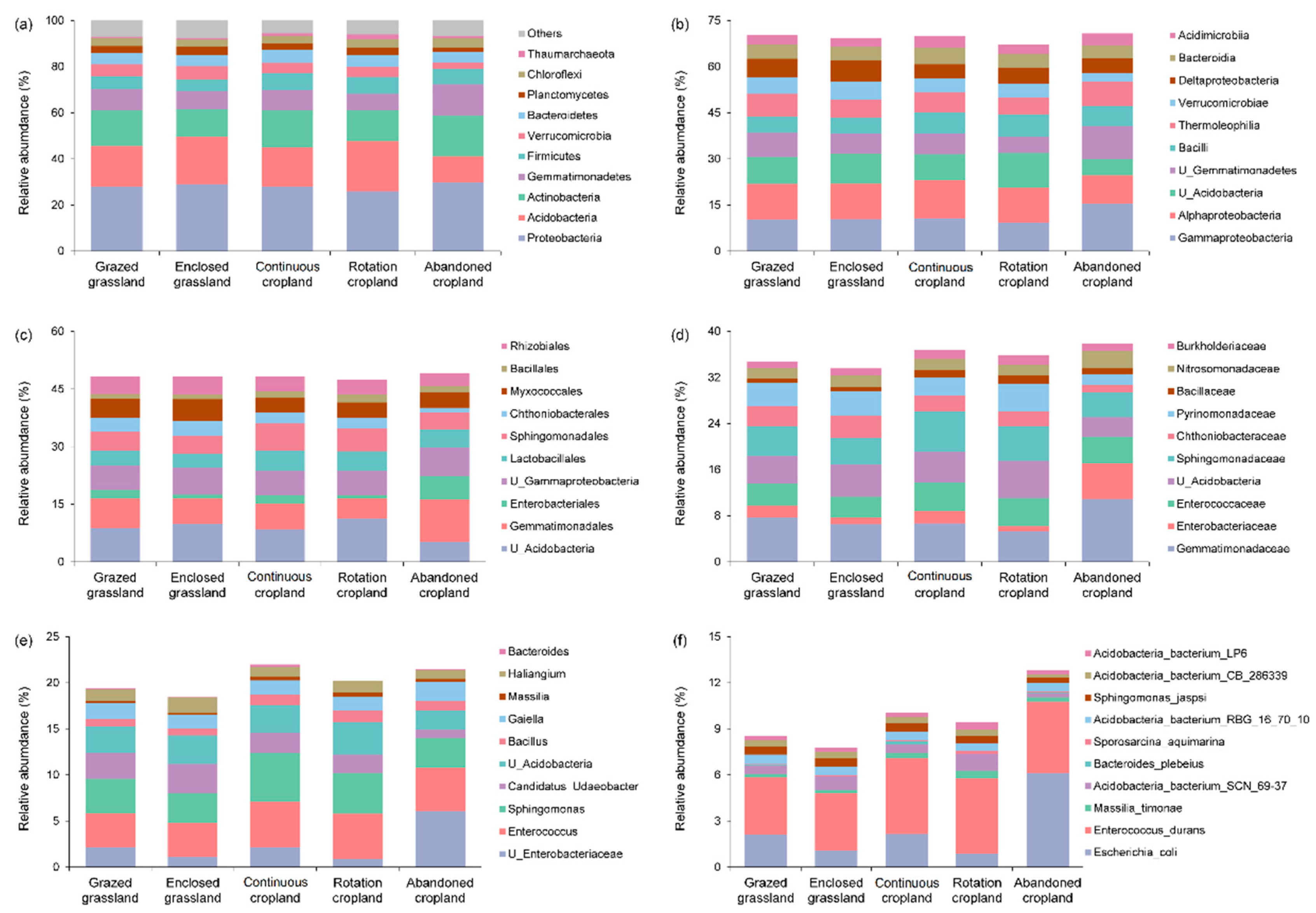
There were more than 60 identified phyla in five land use patterns. The top 10 abundant bacterial phyla at 0~20 cm soil depths were same across the five land use patterns in Tianzhu alpine agro-pastoral ecotone (Figure 2a), and the top 10 abundant bacterial phyla were Proteobacteria, Acidobacteria, Actinobacteria, Gemmatimonadetes, Firmicutes, Verrucomicrobia, Bacteroidetes, Planctomycetes, Chloroflexi and Thaumarchaeota. We found that Proteobacteria was the dominant bacterial phylum in Tianzhu alpine agro-pastoral ecotone regardless of land use change, with relative abundance ranging from 26~30%. The three most abundant bacterial phylum accounted for around 60% of the total. The relative abundance of Firmicutes was significantly lower in grasslands (grazed and enclosed) relative to croplands (continuous and rotation), with averaged relative abundance of 5% and 7%, respectively (p < 0.05). The relative abundances of Acidobacteria (11.4%, p < 0.05) and Verrucomicrobia (2.8%, p < 0.05) were significantly lower in abandoned cropland, while the relative abundance of Gemmatimonadetes (13.4%, p < 0.05) was significantly higher in abandoned cropland.
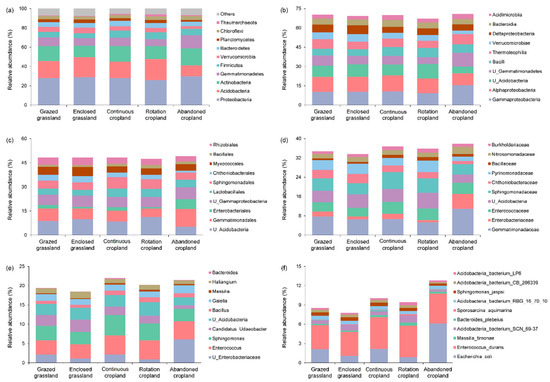
Figure 2.
The averaged relative abundance of top 10 bacterial taxonomic groups at phylum (a), class (b), order (c), family (d), genus (e) and species (f) levels across the five land use patterns (n = 4).
Across all taxonomic levels, the dominant soil bacterial groups were identical regardless of land use change (Figure 2). However, the total relative abundance of top 10 dominant bacterial groups was decreasing as the orders of phylum (92~94%), class (67~71%), order (48~49%), family (34~38%), genus (18~22%) and species (8~13%).
3.2. Soil Bacterial Alpha Diversity
The five indices of bacterial alpha diversity for each land use pattern were shown in Table 1. We found that although there were no significant differences among land use patterns based on ACE and Chao1 indices, Shannon and Simpson indices indicated that conversion from grasslands to croplands decreased soil bacterial diversity. Enclosed grassland had the highest soil bacterial alpha diversity compared with other land use patterns, while soil bacterial alpha diversity was significantly lower under abandoned cropland relative to the other four land use patterns.

Table 1.
The averaged bacterial diversity for each sample site (n = 4). Different letters indicate the significance between land-use patterns.
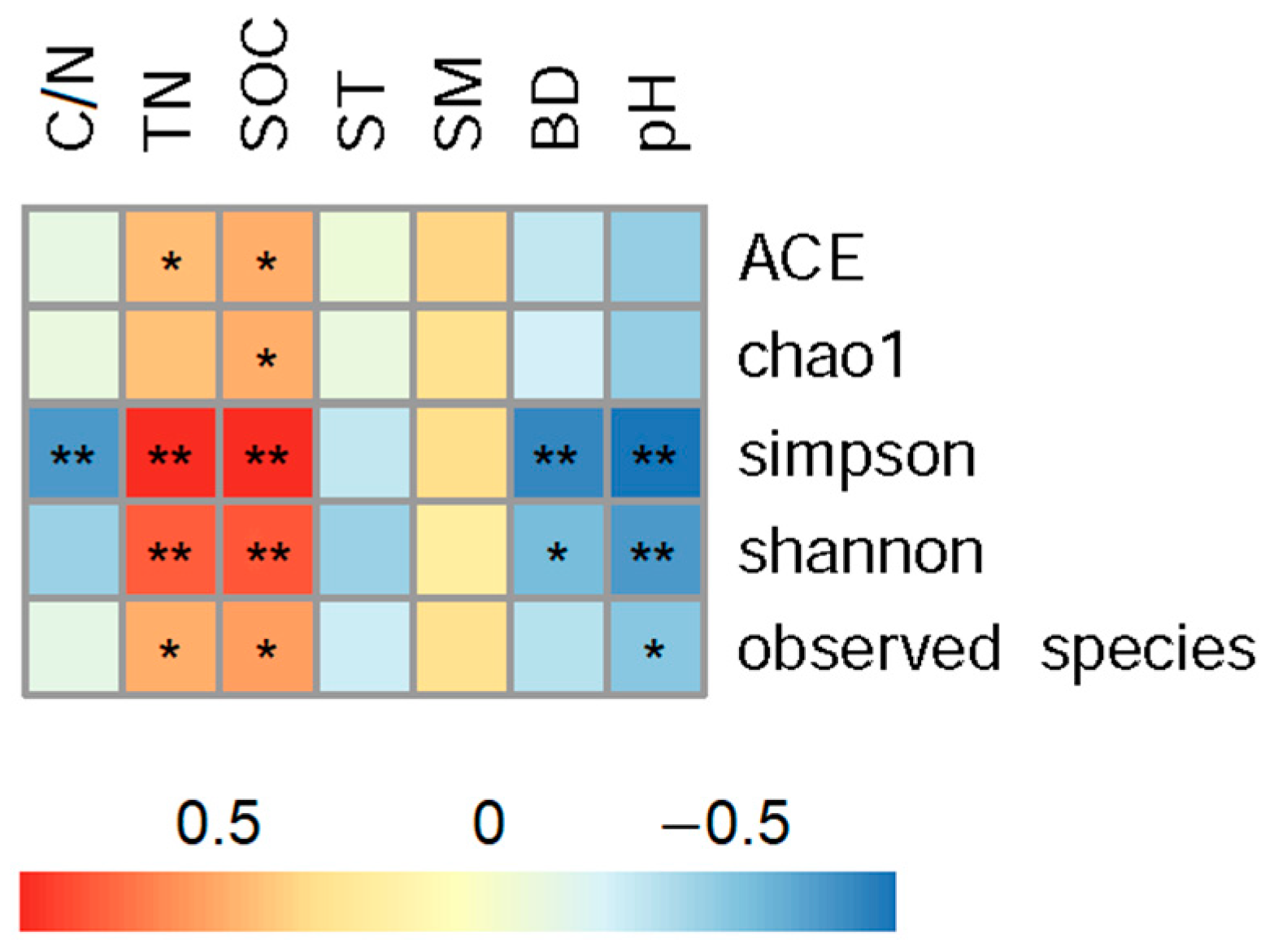
The correlation analysis showed that indices of soil bacterial alpha diversity were significantly correlated to soil properties (Figure 3). Concretely, both Shannon and Simpson indices had extremely significant positive correlations with soil organic carbon and total nitrogen contents, had significant negative correlations with soil bulk density and pH.
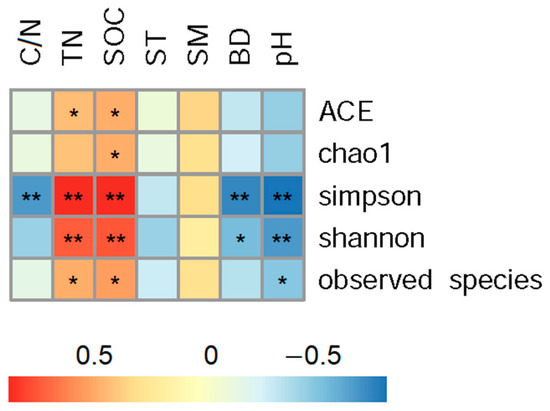
Figure 3.
Correlation analysis between alpha diversity and soil characters across all land use patterns in Tianzhu alpine agro-pastoral ecotone. “*” indicates p < 0.05 and “**” indicates p < 0.01. SOC and TN represents soil organic carbon and total nitrogen contents; C/N means SOC and TN ratio; ST and SM represent soil temperature and moisture, and BD means bulk density.
3.3. Soil Bacterial Beta Diversity
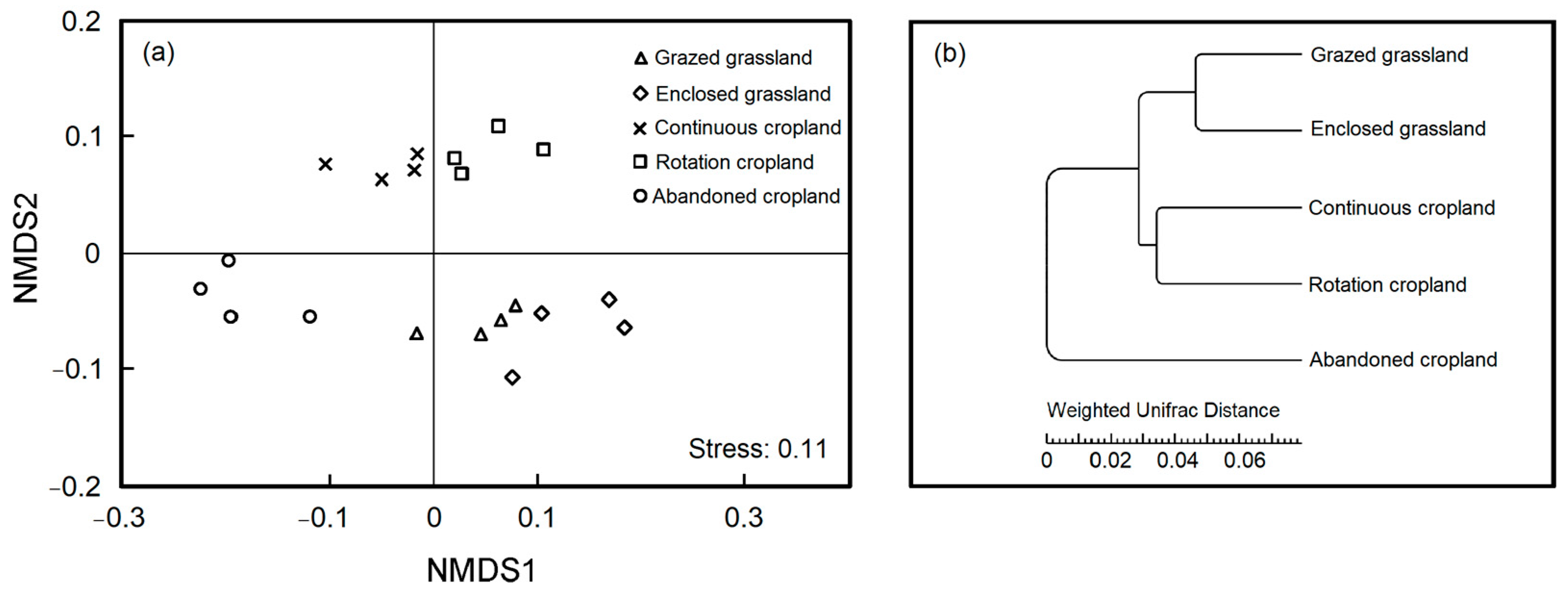
The NMDS and hierarchical clustering analysis revealed the resemblance and difference of bacterial composition between land use patterns (Figure 4). Cultivation from grasslands substantially affected soil bacterial beta diversity that land use patterns were distinct from one another in two axes (Figure 4a, Stress = 0.11). By algorithm of weighted unifrac distance, we found that soil bacteria in grazed grassland and enclosed grassland were clustered together, and soil bacteria in continuous cropland and rotation cropland were clustered together, separated from those in abandoned cropland (Figure 4b).
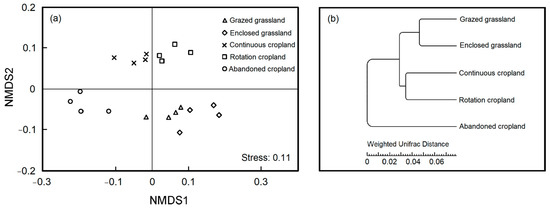
Figure 4.
Non-metric multi-dimensional scaling (NMDS) ordination of relative abundance (a) and clustering by weighted unifrac distance (b) for soil bacterial OTUs under the five land use patterns.
We applied Anosim analysis to detect the significance of land use effects on soil bacterial beta diversity (Table 2). R values were all significantly positive, and ranged from 0.28~0.98, indicating the difference between land use patterns was significantly stronger relative to the difference within land use patterns.

Table 2.
Anosim analysis between the five land use patterns for soil bacterial communities under the five land use patterns in Tianzhu alpine agro-pastoral ecotone. Data represent R values and significance were indicated by “**” (p < 0.01).
3.4. Relationships between Soil Properties and Bacterial Variations
Our results in Table 3 showed that cultivation substantially decreased soil organic carbon contents and total nitrogen contents, and even after the land was abandoned, soil organic carbon and total nitrogen declined further. Soil organic carbon and total nitrogen contents in rotation cropland were 3.1% and 0.26%, significantly lower than that in continuous cropland. Cultivation substantially increased soil pH and bulk density, while it significantly decreased soil moisture.

Table 3.
The averaged soil characters for each sample site (n = 4). Note that SOC and TN represent soil organic carbon and total nitrogen contents; BD means bulk density, and SM represent soil moisture. Different letters indicate the significance between land-use patterns.
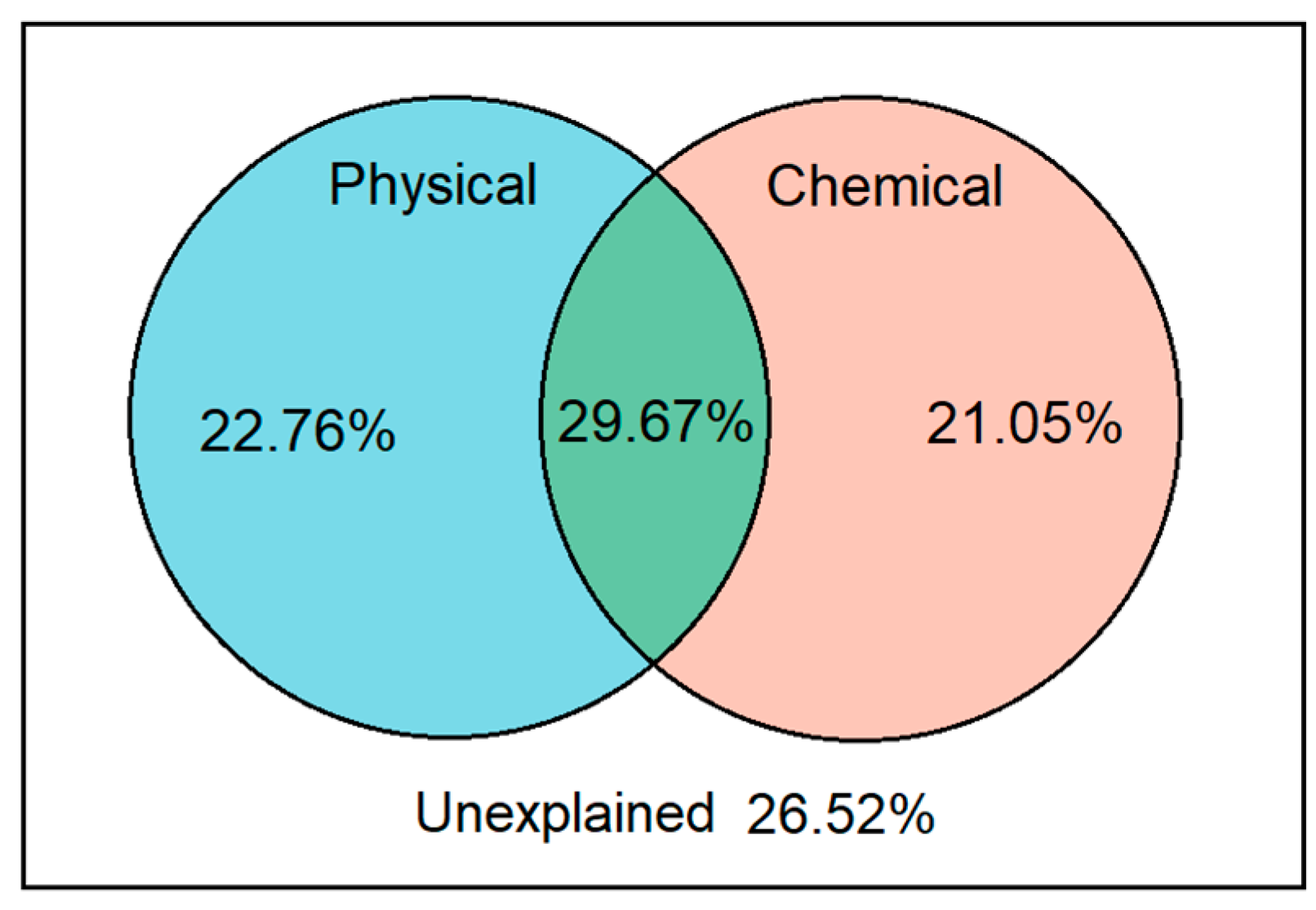
From VPA analysis, our results illustrated that the soil physical and chemical properties explained over 70% variances of soil bacteria under different land use patterns in Tianzhu alpine agro-pastoral ecotone (Figure 5). The interpretation for variation in soil bacteria by soil physical and chemical factors were similar, with degrees of 22.8% and 21.1%, respectively. In total, 26.5% variance in soil bacteria remaining were unexplained based on our results.
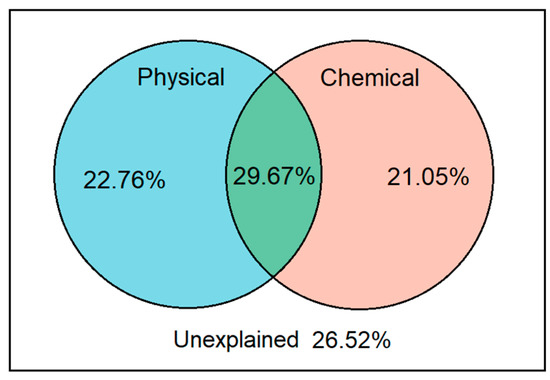
Figure 5.
Variation partition analysis (VPA) for soil bacterial communities across the five land use patterns in the Tianzhu alpine agro-pastoral ecotone.
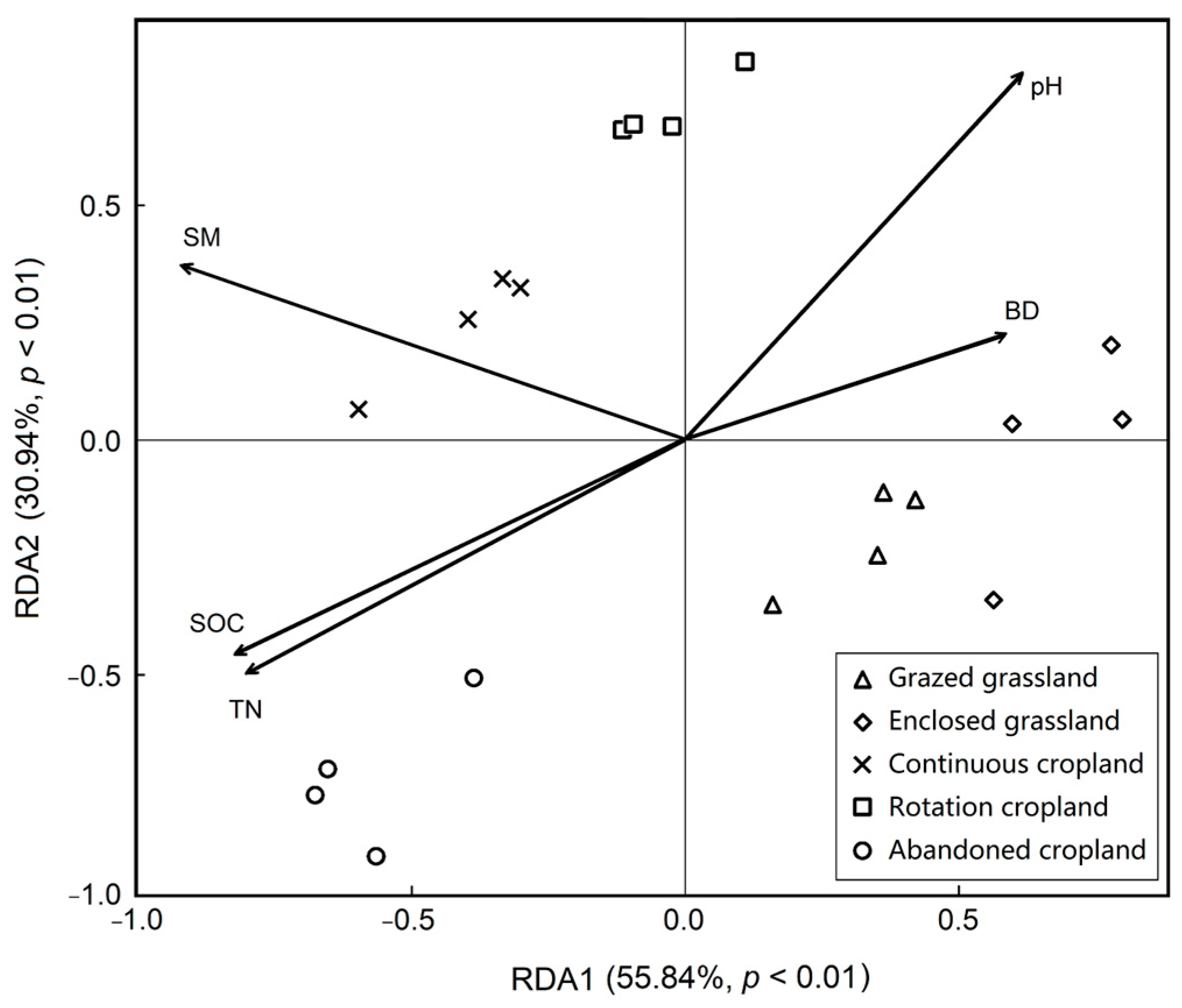
To specify which soil properties are more important to soil bacteria in our study, the RDA was performed to explore the relationships between soil properties and bacteria further under the land use patterns (Figure 6). Our results showed that soil organic content, total nitrogen content, soil moisture, soil bulk density and pH explained 86.78% variance of soil bacteria with land use change, with 55.84% for the first constraint axis and 30.94% for the second constraint axis, respectively. Soil bacteria under abandoned cropland was positively correlated to soil organic carbon and total nitrogen contents, whilst it had negative relationships with soil pH and bulk density.
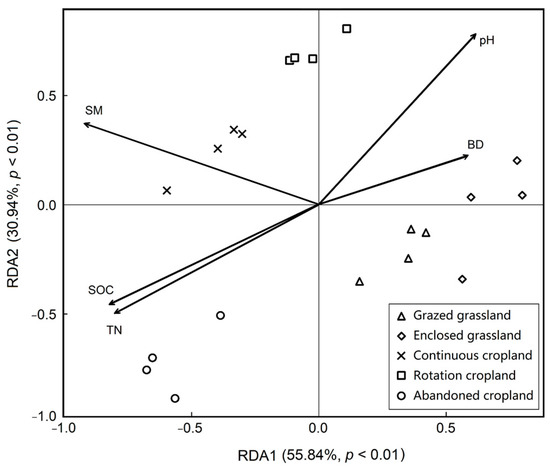
Figure 6.
Redundancy analysis (RDA) of the soil bacterial and edaphic factors under the five land use patterns. SM means soil moisture, BD means bulk density, SOC means soil organic carbon and TN means total nitrogen.
4. Discussion
Our results support that the most three abundant soil bacterial phyla are Proteobacteria, Acidobacteria and Actinobacteria across the whole world [10], even though sites are located far apart with various climates and ecosystems [26,27]. However, despite finding the same most dominant phyla across the five land use patterns in our study, their proportional abundances were variant. That is, the most dominant bacterial species are not closely linked to land use change, while the proportional abundances of dominant bacterial species could be influenced by land use change.
The abandoned cropland had the lowest relative abundance of Acidobacteria, corresponding with the highest soil pH in this land use practice. This finding is in line with that Acidobacteria prefer living in soil with lower pH condition [28]. Moreover, our results show that croplands and abandoned croplands had significantly higher relative abundance of Firmicutes phylum, which has better resistance to poor nutrition and worse environment [29]. This reflects that grassland conversion could possibly induce soil degradation in the Tianzhu alpine agro-pastoral contone. Notably, like the Firmicutes phylum, Gemmatimonadetes also have better activity under the harsh environment, especially within dry conditions and even drought stress [30]. The Gemmatimonadetes and Firmicutes phyla show much higher proportional abundances in our study area compared to the averaged level globally [10], suggesting that not only croplands and abandoned croplands, but also the whole alpine region may be experiencing environmental degradation.
Our findings that soil bacterial diversity is substantially different under the five land use patterns are in concordance with soil bacterial diversity regulated by land use change across many ecoregions [16,17,31]. Although our study did not find significant difference in bacterial diversity between grazed and enclosed grassland, the enclosed grassland had an increasing trend compared to grazed grassland, consistent with previous studies that grazing could cause a decline in the diversity of soil bacteria; however, this effect could be influenced by grazing intensity and lasting time of grazing prohibition [32,33,34,35]. Therefore, we suggest a long-term experiment with different grazing intensities should be conducted for further studies. In addition, although abandonment could recover soil bacterial diversity [36], we found Shannon and Simpson indices under abandoned cropland were substantially lower than that under croplands in the Tianzhu alpine agro-pastoral ecotone. This could be explained by the fact that the vegetation was completely different between grasslands and abandoned croplands, while restoration of bacterial diversity is dependant on the recovering of above-ground vegetation [37]. Notably, the higher soil bacterial diversity does not mean the better functioning of terrestrial ecosystem and vice versa. The actual effects of bacterial diversity depended on the complementary or antagonistic relationships between different groups [38,39].
Soil property, climate and plants are supposed to be the most critical factors for shaping soil microbial structure [11]. In our study, soil physical and chemical properties explained over 70% variations of soil bacterial diversity. Since the same climatic conditions are in the Tianzhu alpine agro-pstoral ecotone, we could infer that the unexplained variations of soil bacterial diversity depend on changes in above-ground vegetation [37]. Due to redundancy analysis, we suggest that soil moisture could be the main driver for the difference in bacterial structure between croplands and others. Soil bulk density, pH, organic carbon and total nitrogen contents seems to be the key factors determining difference in bacterial structure between abandoned croplands and others in Tianzhu alpine agro-pastoral ecotone.
5. Conclusions
In this study, we examined our hypotheses and found that cultivation did not change the most abundant bacterial groups at phylum, class, order, genus and species levels. In addition, cultivation declined soil bacterial diversity without recovery even after abandonment. Moreover, the key soil predictor for the soil bacteria community was different between land uses. Overall, our findings illustrated that the land use change alters soil bacterial diversity, but not dominant species through affecting key edaphic characters in an alpine agro-pastoral ecotone. However, due to the limitations of this study, we suggest more sampling locations should be included for reaching common conclusions.
Author Contributions
Conceptualization, C.S. and X.S.; Methodology, X.H. and J.S.; Formal analysis, X.H.; Investigation, X.H. and W.S.; Resources, J.S. and W.S.; Writing—original draft, X.H.; Writing—review and editing, C.S.; Supervision, C.S. and X.S.; Funding acquisition, C.S. and X.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Natural Science Foundation of Jiangsu Province, grant number BK20210831 and 21KJB210002, the National Natural Science Foundation of China, grant number 32201919 and A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Data Availability Statement
The data that support the findings of this study are available from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ding, M.; Zhang, Y.; Sun, X.; Liu, L.; Wang, Z.; Bai, W. Spatiotemporal variation in alpine grassland phenology in the Qinghai-Tibetan Plateau from 1999 to 2009. Chin. Sci. Bull. 2013, 58, 396–405. [Google Scholar] [CrossRef]
- Wang, J.; Wang, X.; Liu, G.; Wang, G.; Zhang, C. Grazing-to-fencing conversion affects soil microbial composition, functional profiles by altering plant functional groups in a Tibetan alpine meadow. Appl. Soil Ecol. 2021, 166, 104008. [Google Scholar] [CrossRef]
- Yang, Y.; Li, H.; Zhang, L.; Zhu, J.; He, H.; Wei, Y.; Li, Y. Characteristics of soil water percolation and dissolved organic carbon leaching and their response to long-term fencing in an alpine meadow on the Tibetan plateau. Environ. Earth Sci. 2016, 75, 1471. [Google Scholar] [CrossRef]
- Yang, T.; Adams, J.; Shi, Y.; He, J.; Jing, X.; Chen, L.; Tedersoo, L.; Chu, H. Soil fungal diversity in natural grasslands of the Tibetan Plateau: Associations with plant diversity and productivity. New Phytol. 2017, 215, 756–765. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Song, Z.; Groenigen, K.; Xu, Z.; Huang, B.; Zhang, Y.; Hang, X.; Tan, S.; Zhang, D.; Zhang, W. Grassland conversion along a climate gradient in northwest China: Implications for soil carbon and nutrients. Soil Use Manag. 2020, 36, 410–419. [Google Scholar] [CrossRef]
- Li, C.; Jong, R.; Schmid, B.; Wulf, H.; Schaepman, M. Changes in grassland cover and in its spatial heterogeneity indicate degradation on the Qinghai-Tibetan Plateau. Ecol. Indic. 2020, 119, 106641. [Google Scholar] [CrossRef]
- Huang, X.; Lu, X.; Zhou, G.; Shi, Y.; Zhang, D.; Zhang, W.; Bai, S. How land-use change affects soil respiration in an alpine agro-pastoral ecotone. Catena 2022, 214, 106291. [Google Scholar] [CrossRef]
- Bardgett, R.; Putten, W. Belowground biodiversity and ecosystem functioning. Nature 2014, 515, 505–511. [Google Scholar] [CrossRef]
- Bodelier, P. Interactions between nitrogenous fertilizers and methane cycling in wetland and upland soils. Curr. Opin. Environ. Sustain. 2011, 3, 379–388. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Maestre, F.; Reich, P.; Jeffries, T.; Gaitan, J.; Encinar, D.; Berdugo, M.; Campbell, C.; Singh, B. Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat. Commun. 2016, 7, 10541. [Google Scholar] [CrossRef]
- Chen, D.; Cheng, J.; Chu, P.; Bai, Y. Regional-scale patterns of soil microbes and nematodes across grasslands on the Mongolian plateau: Relationships with climate, soil, and plants. Ecography 2015, 38, 622–631. [Google Scholar] [CrossRef]
- Dai, H.; Zhang, H.; Li, Z.; Liu, K.; Zamanian, K. Tillage Practice Impacts on the Carbon Sequestration Potential of Topsoil Microbial Communities in an Agricultural Field. Agronomy 2021, 11, 60. [Google Scholar] [CrossRef]
- Le, H.; Rochelle-Newall, E.; Ribolzi, O.; Janeau, J.; Huon, S.; Latsachack, K.; Pommier, T. Land use strongly influences soil organic carbon and bacterial community export in runoff in tropical uplands. Land Degrad. Dev. 2020, 31, 118–132. [Google Scholar] [CrossRef]
- Lu, M.; Ren, Y.; Wang, S.; Tian, K.; Sun, X.; Peng, S. Contribution of soil variables to bacterial community composition following land use change in Napahai plateau wetlands. J. Environ. Manag. 2019, 246, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Rodrigures, J.; Pellizari, V.; Mueller, R.; Baek, K.; Jesus, E.; Paula, F.; Mirza, B.; Hamaoui, G.; Tsai, S.; Feigl, B.; et al. Conversion of the Amazon rainforest to agriculture results in biotic homogenization of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2013, 110, 988–993. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Luo, C.; Jiang, L.; Song, M.; Zhang, D.; Li, J.; Li, Y.; Ostle, N.; Zhang, G. Land-use changes alter soil bacterial composition and diversity in tropical forest soil in China. Sci. Total Environ. 2020, 712, 136526. [Google Scholar] [CrossRef]
- Viruel, E.; Fontana, C.; Puglisi, E.; Nasca, J.; Banegas, N.; Cocconcelli, P. Land-use change affects the diversity and functionality of soil bacterial communities in semi-arid Chaco region, Argentina. Appl. Soil Ecol. 2022, 172, 104362. [Google Scholar] [CrossRef]
- Li, Y.; Wang, S.; Jiang, L.; Zhang, L.; Cui, S.; Meng, F.; Wang, X.; Li, X.; Zhou, Y. Changes of soil microbial community under different degraded gradients of alpine meadow. Agric. Ecosyst. Environ. 2016, 222, 213–222. [Google Scholar] [CrossRef]
- Qi, X.; Wang, C.; He, T.; Ding, F.; Zhang, X.; An, L.; Xu, S. Bacterial community changes and their responses to nitrogen addition among diferent alpine grassland types at the eastern edge of Qinghai–Tibetan Plateau. Arch. Microbiol. 2021, 203, 5963–5974. [Google Scholar] [CrossRef]
- Yin, Y.; Wang, Y.; Li, S.; Liu, Y.; Zhao, W.; Ma, Y.; Bao, G. Soil microbial character response to plant community variation after grazing prohibition for 10 years in a Qinghai-Tibetan alpine meadow. Plant Soil. 2021, 458, 175–189. [Google Scholar] [CrossRef]
- He, Y.; Xu, M.; Qi, Y.; Dong, Y.; He, X.; Li, J.; Liu, X.; Sun, L. Differential Responses of Soil Microbial Community to Four-Decade Long Grazing and Cultivation in a Semi-Arid Grassland. Sustainability 2017, 9, 128. [Google Scholar] [CrossRef]
- Bai, Z.; Zheng, L.; Bai, Z.; Jia, A.; Wang, M. Long-term cultivation alter soil bacterial community in a forest-grassland transition zone. Front. Microbiol. 2022, 13, 1001781. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Li, S.; Wang, Z.; Zhang, Y.; Wang, K. Effects of grassland converted to cropland on soil microbial biomass and community from agro-pastoral ecotone in Northern China. Grassl. Sci. 2022, 68, 36–43. [Google Scholar] [CrossRef]
- Bremner, J. Nitrogen-total. In Methods of Soil Analysis; Sparks, D.L., Ed.; SSSA: Madison, WI, USA, 1996; pp. 532–535. [Google Scholar]
- Mclean, E.O. Soil pH and Lime Requirement. In Methods of Soil Analysis; Page, A.L., Ed.; SSSA: Madison, WI, USA, 1982; pp. 199–224. [Google Scholar]
- Yang, F.; Wu, J.; Zhang, D.; Chen, Q.; Zhang, Q.; Cheng, X. Soil bacterial community composition and diversity in relation to edaphic properties and plant traits in grasslands of southern China. Appl. Soil Ecol. 2018, 128, 43–53. [Google Scholar] [CrossRef]
- Viruel, E.; Fontana, C.; Bassi, D.; Puglisi, E.; Radrizzani, A.; Martinez Calsina, L.; Banegas, N.; Cocconcelli, P. Silvopastoral systems in dry Chaco, Argentina: Impact on soil chemical parameters and bacterial communities. Soil Use Manag. 2020, 37, 866–878. [Google Scholar] [CrossRef]
- Sait, M.; Davis, K.; Janssen, P. Effect of pH on isolation and distribution of members of subdivision 1 of the phylum Acidobacteria occurring in soil. Appl. Environ. Microbiol. 2006, 72, 1852–1857. [Google Scholar] [CrossRef]
- Bukar, M.; Sodipo, O.; Dawkins, K.; Ramirez, R.; Kaldapa, J.; Tarfa, M.; Esiobu, N. Microbiomes of top and sub-layers of semi-arid soils in north-eastern Nigeria are rich in firmicutes and proteobacteria with surprisingly high diversity of rare species. Adv. Microbiol. 2019, 9, 1024–1118. [Google Scholar] [CrossRef]
- Cao, J.; Wang, H.; Holden, N.; Adamowski, J.; Biswas, A.; Zhang, X.; Feng, Q. Soil properties and microbiome of annual and perennial cultivated grasslands on the Qinghai–Tibetan Plateau. Land Degrad. Dev. 2021, 32, 5306–5321. [Google Scholar] [CrossRef]
- He, B.; Duan, X.; Rong, L.; Zhang, R.; Li, Y.; Lu, H. Land use controls soil bacterial diversity in the dry-hot valley region, Southern China. Arch. Agron. Soil Sci. 2020, 66, 694–705. [Google Scholar] [CrossRef]
- Cui, Y.; Dong, Y.; Liu, H.; Sun, Z. Short-term grazing exclusions reduced soil organic carbon but not bacterial diversity in the sagebrush desert, Northwest China. Glob. Ecol. Conserv. 2021, 31, e01872. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, G.; Song, Z.; Wang, J.; Guo, L. Interactions of soil bacteria and fungi with plants during long-term grazing exclusion in semiarid grasslands. Soil Biol. Biochem. 2018, 124, 47–58. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, X.; Hao, X.; Alexander, T.; Shi, X.; Long, J.; Thomas, B. Heavy grazing over 64 years reduced soil bacterial diversity in the foothills of the Rocky Mountains, Canada. Appl. Soil Ecol. 2020, 147, 103361. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, J.; Hao, Y.; Wang, Y. Intermediate grazing intensities by sheep increase soil bacterial diversities in an Inner Mongolian steppe. Biol. Fertil. Soils 2010, 46, 817–824. [Google Scholar] [CrossRef]
- Zhang, K.; Cheng, X.; Shu, X.; Liu, Y.; Zhang, Q. Linking soil bacterial and fungal communities to vegetation succession following agricultural abandonment. Plant Soil 2018, 431, 19–36. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, S.; Niu, K. Relationships between soil fungal diversity, plant community functional traits, and soil attributes in Tibetan alpine meadows. Chin. J. Appl. Environ. Biol. 2020, 26, 1–9. (In Chinese) [Google Scholar]
- Eisenhauer, N.; Schulz, W.; Scheu, S.; Jousset, A.; Pfrender, M. Niche dimensionality links biodiversity and invisibility of microbial communities. Funct. Ecol. 2013, 27, 282–288. [Google Scholar] [CrossRef]
- Foster, K.; Bell, T. Competition, not cooperation, dominates interactions among culturable microbial species. Curr. Biol. 2012, 22, 1845–1850. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).