Exploring the Inflammatory Pathogenesis of Colorectal Cancer
Abstract
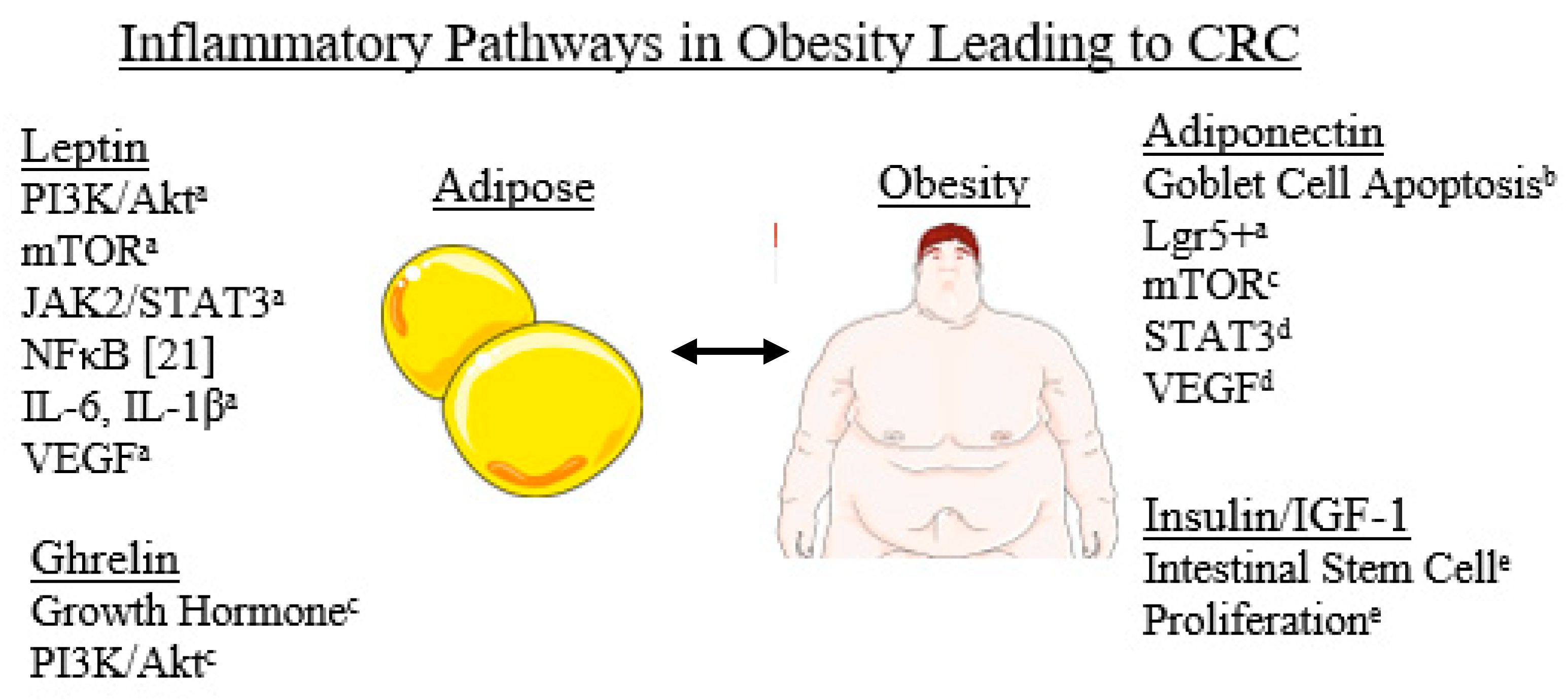
1. Introduction
2. Obesity and Diet, Smoking, Diabetes in Colorectal Carcinoma
2.1. Short Chain Fatty Acids
2.2. Polyphenols/Red Meats
2.3. Other Associations between Obesity and CRC
2.4. Smoking and Diabetes
3. Microbiome
3.1. Direct Impact on Inflammatory Pathways
3.2. Intestinal Barrier Disruption
3.3. Implications of the Microbiota on Genomics, Epigenetics and Metabolism
3.4. Protective Implications of Microbiota and Future Directions
4. Mycobiome
5. The Role of Exosomes
5.1. Exosomes and Intestinal Inflammation
5.2. Exosomes and Colorectal Cancer
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Howlader, N.; Noone, A.M.; Krapcho, M.; Miller, D.; Brest, A.; Yu, M.; Ruhl, J.; Tatalovich, Z.; Mariotto, A.; Lewis, D.R.; et al. SEER Cancer Statistics Review, 1975; National Cancer Institute: Bethesda, MD, USA, April 2021. [Google Scholar]
- Hadjipetrou, A.; Anyfantakis, D.; Galanakis, C.G.; Kastanakis, M.; Kastanakis, S. Colorectal cancer, screening and primary care: A mini literature review. World J. Gastroenterol. 2017, 23, 6049–6058. [Google Scholar] [CrossRef]
- Kedrin, D.; Gala, M.K. Genetics of the Serrated Pathway to Colorectal Cancer. Clin. Transl. Gastroenterol. 2015, 6, e84. [Google Scholar] [CrossRef]
- Teimoorian, F.; Ranaei, M.; Tilaki, K.H.; Shirvani, J.S.; Vosough, Z. Association of Helicobacter pylori Infection with Colon Cancer and Adenomatous Polyps. Iran. J. Pathol. 2018, 13, 325–332. [Google Scholar]
- Wang, R.; Leong, R.W. Primary sclerosing cholangitis as an independent risk factor for colorectal cancer in the context of inflammatory bowel disease: A review of the literature. World J. Gastroenterol. 2014, 20, 8783–8789. [Google Scholar] [CrossRef]
- Hussain, S.P.; Amstad, P.; Raja, K.; Ambs, S.; Nagashima, M.; Bennett, W.P.; Shields, P.G.; Ham, A.J.; Swenberg, J.; Marrogi, A.J.; et al. Increased p53 mutation load in noncancerous colon tissue from ulcerative colitis: A cancer-prone chronic inflammatory disease. Cancer Res. 2000, 60, 60. [Google Scholar]
- Redston, M.S.; Papadopoulos, N.; Caldas, C.; Kinzler, K.W.; Kern, S.E. Common occurrence of APC and K-ras gene mutations in the spectrum of colitis-associated neoplasias. Gastroenterology 1995, 108, 383–392. [Google Scholar] [CrossRef]
- Brentnall, T.A.; Rubin, C.E.; Crispin, D.A.; Stevens, A.; Batchelor, R.H.; Haggitt, R.C.; Bronner, M.P.; Evans, J.P.; McCahill, L.E.; Bilir, N.; et al. A germline substitution in the human MSH2 gene is associated with high-grade dysplasia and cancer in ulcerative colitis. Gastroenterology 1995, 109, 151–155. [Google Scholar] [CrossRef]
- Watanabe, T.; Konishi, T.; Kishimoto, J.; Kotake, K.; Muto, T.; Sugihara, K. Ulcerative colitis-associated colorectal cancer shows a poorer survival than sporadic colorectal cancer: A nationwide Japanese study. Inflamm. Bowel Dis. 2011, 17, 802–808. [Google Scholar] [CrossRef]
- Itzkowitz, S.H.; Yio, X. Inflammation and Cancer—IV. Colorectal Cancer in Inflammatory Bowel Disease: The Role of Inflammation. Am. J. Physiol.–Gastrointest. Liver Physiol. 2004, 287, G7–G17. [Google Scholar] [CrossRef]
- Luo, C.; Zhang, H. The Role of Proinflammatory Pathways in the Pathogenesis of Colitis-Associated Colorectal Cancer. Mediat. Inflamm. 2017, 2017, 1–8. [Google Scholar] [CrossRef]
- Lin, Y.; He, Z.; Ye, J.; Liu, Z.; She, X.; Gao, X.; Liang, R. Progress in Understanding the IL-6/STAT3 Pathway in Colorectal Cancer. OncoTargets Ther. 2020, 13, 13023–13032. [Google Scholar] [CrossRef]
- Jana, A.; Krett, N.L.; Guzman, G.; Khalid, A.; Özden, Ö.; Staudacher, J.; Bauer, J.; Baik, S.H.; Carroll, T.; Yazici, C.; et al. NFkB is essential for activin-induced colorectal cancer migration via upregulation of PI3K-MDM2 pathway. Oncotarget 2017, 8, 37377–37393. [Google Scholar] [CrossRef] [PubMed]
- Schwitalla, S.; Ziegler, P.K.; Horst, D.; Becker, V.; Kerle, I.; Begus-Nahrmann, Y.; Lechel, A.; Rudolph, K.L.; Langer, R.; Slotta-Huspenina, J.; et al. Loss of p53 in Enterocytes Generates an Inflammatory Microenvironment Enabling Invasion and Lymph Node Metastasis of Carcinogen-Induced Colorectal Tumors. Cancer Cell 2013, 23, 93–106. [Google Scholar] [CrossRef]
- Porta, C.; Ippolito, A.; Consonni, F.M.; Carraro, L.; Celesti, G.; Correale, C.; Grizzi, F.; Pasqualini, F.; Tartari, S.; Rinaldi, M.; et al. Protumor Steering of Cancer Inflammation by p50 NF-κB Enhances Colorectal Cancer Progression. Cancer Immunol. Res. 2018, 6, 578–593. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Sica, A. Macrophages, innate immunity and cancer: Balance, tolerance, and diversity. Curr. Opin. Immunol. 2010, 22, 231–237. [Google Scholar] [CrossRef]
- Goday, A.; Barneto, I.; Garcia-Almeida, J.M.; Blasco, A.; Lecube, A.; Gravalos, C.; De Icaya, P.M.; Peñas, R.D.L.; Monereo, S.; Vázquez, L.; et al. Obesity as a risk factor in cancer: A national consensus of the Spanish Society for the Study of Obesity and the Spanish Society of Medical Oncology. Clin. Transl. Oncol. 2015, 17, 763–771. [Google Scholar] [CrossRef] [PubMed]
- Aldaqal, S.M.; Maqbul, A.A.; Alhammad, A.A.; Alghamdi, A.S.; Alharbi, B.A.; Alharbi, M.T.; Alhazmi, O.M.; Zaylaee, Y.O. The Impact of Body Mass Index on the Clinicopathological and Prognostic Factors of Colorectal Cancer in Saudi Arabia. Cureus 2020, 12, 11789. [Google Scholar] [CrossRef]
- Szydło, B.; Kiczmer, P.; Świętochowska, E.; Ostrowska, Z. Role of omentin and chemerin in metabolic syndrome and tumor diseases. Postępy Higieny i Medycyny Doświadczalnej 2016, 70, 844–849. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.R. Obesity-Related Colorectal Cancer: The Role of Leptin. Ann. Coloproctology 2015, 31, 209–210. [Google Scholar] [CrossRef][Green Version]
- Chang, M.-L.; Yang, Z.; Yang, S.-S. Roles of Adipokines in Digestive Diseases: Markers of Inflammation, Metabolic Alteration and Disease Progression. Int. J. Mol. Sci. 2020, 21, 8308. [Google Scholar] [CrossRef]
- Stattin, P.; Lukanova, A.; Biessy, C.; Söderberg, S.; Palmqvist, R.; Kaaks, R.; Olsson, T.; Jellum, E. Obesity and colon cancer: Does leptin provide a link? Int. J. Cancer 2004, 109, 149–152. [Google Scholar] [CrossRef] [PubMed]
- Stattin, P.; Palmqvist, R.; Söderberg, S.; Biessy, C.; Ardnor, B.; Hallmans, G.; Kaaks, R.; Olsson, T. Plasma leptin and colorectal cancer risk: A prospective study in Northern Sweden. Oncol. Rep. 2003, 10, 2015–2021. [Google Scholar] [CrossRef]
- Aleksandrova, K.; Boeing, H.; Jenab, M.; Bueno-De-Mesquita, H.B.; Jansen, E.; van Duijnhoven, F.J.B.; Rinaldi, S.; Fedirko, V.; Romieu, I.; Riboli, E.; et al. Leptin and Soluble Leptin Receptor in Risk of Colorectal Cancer in the European Prospective Investigation into Cancer and Nutrition Cohort. Cancer Res. 2012, 72, 5328–5337. [Google Scholar] [CrossRef]
- Saxena, A.; Baliga, M.S.; Ponemone, V.; Kaur, K.; Larsen, B.; Fletcher, E.; Greene, J.; Fayad, R. Mucus and adiponectin deficiency: Role in chronic inflammation-induced colon cancer. Int. J. Color. Dis. 2013, 28, 1267–1279. [Google Scholar] [CrossRef]
- Moodi, M.; Tavakoli, T.; Tahergorabi, Z. Crossroad between Obesity and Gastrointestinal Cancers: A Review of Molecular Mechanisms and Interventions. Int. J. Prev. Med. 2021, 12, 12. [Google Scholar]
- Moon, H.-S.; Liu, X.; Nagel, J.M.; Chamberland, J.P.; Diakopoulos, K.N.; Brinkoetter, M.T.; Hatziapostolou, M.; Wu, Y.; Robson, S.C.; Iliopoulos, D.; et al. Salutary effects of adiponectin on colon cancer: In vivo and in vitro studies in mice. Gut 2013, 62, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Pourvali, K.; Monji, H. Obesity and intestinal stem cell susceptibility to carcinogenesis. Nutr. Metab. 2021, 18, 1–15. [Google Scholar] [CrossRef]
- Kizil, C.; Kyritsis, N.; Brand, M. Effects of inflammation on stem cells: Together they strive? EMBO Rep. 2015, 16, 416–426. [Google Scholar] [CrossRef]
- Kolb, R.; Sutterwala, F.S.; Zhang, W. Obesity and cancer: Inflammation bridges the two. Curr. Opin. Pharmacol. 2016, 29, 77–89. [Google Scholar] [CrossRef]
- Testa, U.; Pelosi, E.; Castelli, G. Colorectal Cancer: Genetic Abnormalities, Tumor Progression, Tumor Heterogeneity, Clonal Evolution and Tumor-Initiating Cells. Med. Sci. 2018, 6, 31. [Google Scholar] [CrossRef]
- Willett, W.C.; Sacks, F.; Trichopoulou, A.; Drescher, G.; Ferro-Luzzi, A.; Helsing, E.; Trichopoulos, D. Mediterranean diet pyramid: A cultural model for healthy eating. Am. J. Clin. Nutr. 1995, 61, 1402S–1406S. [Google Scholar] [CrossRef]
- Illescas, O.; Rodríguez-Sosa, M.; Gariboldi, M. Mediterranean Diet to Prevent the Development of Colon Diseases: A Meta-Analysis of Gut Microbiota Studies. Nutrients 2021, 13, 2234. [Google Scholar] [CrossRef]
- Gaesser, G.A. Whole Grains, Refined Grains, and Cancer Risk: A Systematic Review of Meta-Analyses of Observational Studies. Nutrients 2020, 12, 3756. [Google Scholar] [CrossRef] [PubMed]
- Carretta, M.D.; Quiroga, J.; López, R.; Hidalgo, M.A.; Burgos, R.A. Participation of Short-Chain Fatty Acids and Their Receptors in Gut Inflammation and Colon Cancer. Front. Physiol. 2021, 12, 662739. [Google Scholar] [CrossRef] [PubMed]
- Venegas, D.P.; de La Fuente, M.K.; Landskron, G.; González, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef]
- Vernia, F.; Longo, S.; Stefanelli, G.; Viscido, A.; Latella, G. Dietary Factors Modulating Colorectal Carcinogenesis. Nutrients 2021, 13, 143. [Google Scholar] [CrossRef]
- Norat, T.; Bingham, S.; Ferrari, P.; Slimani, N.; Jenab, M.; Mazuir, M.; Overvad, K.; Olsen, A.; Tjonneland, A.; Clavel, F.; et al. Meat, Fish, and Colorectal Cancer Risk: The European Prospective Investigation into Cancer and Nutrition. J. Natl. Cancer Inst. 2005, 97, 906–916. [Google Scholar] [CrossRef]
- Hammerling, U.; Laurila, J.B.; Grafström, R.; Ilbäck, N.-G. Consumption of Red/Processed Meat and Colorectal Carcinoma: Possible Mechanisms Underlying the Significant Association. Crit. Rev. Food Sci. Nutr. 2015, 56, 614–634. [Google Scholar] [CrossRef]
- Hunter, D.J. Gene–environment interactions in human diseases. Nat. Rev. Genet. 2005, 6, 287–298. [Google Scholar] [CrossRef] [PubMed]
- Hosoya, T.; Maillard, I.; Engel, J.D. From the cradle to the grave: Activities of GATA-3 throughout T-cell development and differentiation. Immunol. Rev. 2010, 238, 110–125. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Blobel, G.A. GATA Transcription Factors and Cancer. Genes Cancer 2010, 1, 1178–1188. [Google Scholar] [CrossRef] [PubMed]
- Ekine-Afolabi, B.A.; Njan, A.A.; Rotimi, S.O.; Elbehi, A.M.; Cash, E.; Adeyeye, A. The Impact of Diet on the Involvement of Non-Coding RNAs, Extracellular Vesicles, and Gut Microbiome-Virome in Colorectal Cancer Initiation and Progression. Front. Oncol. 2020, 10, 2545. [Google Scholar] [CrossRef] [PubMed]
- Roslan, N.H.; Makpol, S.; Yusof, Y.A.M. A Review on Dietary Intervention in Obesity Associated Colon Cancer. Asian Pac. J. Cancer Prev. 2019, 20, 1309–1319. [Google Scholar] [CrossRef]
- Song, M.; Chan, A.T. Environmental Factors, Gut Microbiota, and Colorectal Cancer Prevention. Clin. Gastroenterol. Hepatol. 2019, 17, 275–289. [Google Scholar] [CrossRef]
- O’Keefe, S.J.D. Diet, Microorganisms and Their Metabolites, and Colon Cancer. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 691–706. [Google Scholar] [CrossRef] [PubMed]
- Gram, I.T.; Park, S.-Y.; Wilkens, L.R.; Haiman, C.; Le Marchand, L. Smoking-Related Risks of Colorectal Cancer by Anatomical Subsite and Sex. Am. J. Epidemiol. 2020, 189, 543–553. [Google Scholar] [CrossRef]
- Huang, C.; Shi, G. Smoking and microbiome in oral, airway, gut and some systemic diseases. J. Transl. Med. 2019, 17, 1–15. [Google Scholar] [CrossRef]
- Kim, M.; Gu, B.; Madison, M.; Song, H.W.; Norwood, K.; Hill, A.A.; Wu, W.-J.; Corry, D.; Kheradmand, F.; Diehl, G.E. Cigarette Smoke Induces Intestinal Inflammation via a Th17 Cell-Neutrophil Axis. Front. Immunol. 2019, 10, 75. [Google Scholar] [CrossRef]
- Lee, G.; Jung, K.-H.; Shin, D.; Lee, C.; Kim, W.; Lee, S.; Kim, J.; Bae, H. Cigarette Smoking Triggers Colitis by IFN-γ+ CD4+ T Cells. Front. Immunol. 2017, 8, 1344. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Fu, X.; Shi, H.; Chen, G.; Dong, P.; Zhang, W. Induction of Murine Macrophage M2 Polarization by Cigarette Smoke Extract via the JAK2/STAT3 Pathway. PLoS ONE 2014, 9, e107063. [Google Scholar] [CrossRef]
- Song, M.; Chan, A.T.; Sun, J. Influence of the Gut Microbiome, Diet, and Environment on Risk of Colorectal Cancer. Gastroenterology 2020, 158, 322–340. [Google Scholar] [CrossRef]
- Piper, M.S.; Saad, R.J. Diabetes Mellitus and the Colon. Curr. Treat. Options Gastroenterol. 2017, 15, 460–474. [Google Scholar] [CrossRef]
- Zhao, M.; Liao, D.; Zhao, J. Diabetes-induced mechanophysiological changes in the small intestine and colon. World J. Diabetes 2017, 8, 249–269. [Google Scholar] [CrossRef] [PubMed]
- Ben-Neriah, Y.; Karin, M. Inflammation Meets Cancer, with NF-ΚB as the Matchmaker. Nat. Immunol. 2011, 12, 715–723. [Google Scholar] [CrossRef]
- Fischbach, S. The Role of TGF-β Signaling in β-Cell Dysfunction and Type 2 Diabetes: A Review. J. Cytol. Histol. 2014, 5, 5. [Google Scholar] [CrossRef]
- Feagins, L.A. Role of transforming growth factor-β in inflammatory bowel disease and colitis-associated colon cancer. Inflamm. Bowel Dis. 2010, 16, 1963–1968. [Google Scholar] [CrossRef] [PubMed]
- Saus, E.; Iraola-Guzmán, S.; Willis, J.R.; Brunet-Vega, A.; Gabaldón, T. Microbiome and colorectal cancer: Roles in carcinogenesis and clinical potential. Mol. Asp. Med. 2019, 69, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, A.; Menon, H.; Chulkina, M.; Yee, N.; Pinchuk, I. Drug–Microbiota Interaction in Colon Cancer Therapy: Impact of Antibiotics. Biomedicines 2021, 9, 259. [Google Scholar] [CrossRef] [PubMed]
- Dalal, N.; Jalandra, R.; Sharma, M.; Prakash, H.; Makharia, G.K.; Solanki, P.R.; Singh, R.; Kumar, A. Omics technologies for improved diagnosis and treatment of colorectal cancer: Technical advancement and major perspectives. Biomed. Pharmacother. 2020, 131, 110648. [Google Scholar] [CrossRef] [PubMed]
- Chen, C. The Inhibitory Effect of Gut Microbiota and Its Metabolites on Colorectal Cancer. J. Microbiol. Biotechnol. 2020, 30, 1607–1613. [Google Scholar] [CrossRef] [PubMed]
- Chew, S.-S.; Tan, L.T.-H.; Law, J.W.-F.; Pusparajah, P.; Goh, B.-H.; Ab Mutalib, N.S.; Lee, L.-H. Targeting Gut Microbial Biofilms—A Key to Hinder Colon Carcinogenesis? Cancers 2020, 12, 2272. [Google Scholar] [CrossRef]
- Marzano, M.; Fosso, B.; Piancone, E.; Defazio, G.; Pesole, G.; De Robertis, M. Stem Cell Impairment at the Host-Microbiota Interface in Colorectal Cancer. Cancers 2021, 13, 996. [Google Scholar] [CrossRef]
- Nosho, K.; Sukawa, Y.; Adachi, Y.; Ito, M.; Mitsuhashi, K.; Kurihara, H.; Kanno, S.; Yamamoto, I.; Ishigami, K.; Igarashi, H.; et al. Association of Fusobacterium Nucleatum with Immunity and Molecular Alterations in Colorectal Cancer. World J. Gastroenterol. 2016, 22, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Li, Q.; Wu, J.; Wu, Y.; Peng, W.; Li, H.; Wang, J.; Tang, X.; Peng, Y.; Fu, X. Fusobacterium nucleatum promotes M2 polarization of macrophages in the microenvironment of colorectal tumours via a TLR4-dependent mechanism. Cancer Immunol. Immunother. 2018, 67, 1635–1646. [Google Scholar] [CrossRef]
- Zhang, S.; Yang, Y.; Weng, W.; Guo, B.; Cai, G.; Ma, Y.; Cai, S. Fusobacterium nucleatum promotes chemoresistance to 5-fluorouracil by upregulation of BIRC3 expression in colorectal cancer. J. Exp. Clin. Cancer Res. 2019, 38, 1–13. [Google Scholar] [CrossRef]
- Yu, T.; Guo, F.; Yu, Y.; Sun, T.; Ma, D.; Han, J.; Qian, Y.; Kryczek, I.; Sun, D.; Nagarsheth, N.; et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell 2017, 170, 548–563.e16. [Google Scholar] [CrossRef] [PubMed]
- Engevik, M.A.; Danhof, H.A.; Ruan, W.; Engevik, A.C.; Chang-Graham, A.L.; Engevik, K.A.; Shi, Z.; Zhao, Y.; Brand, C.K.; Krystofiak, E.S.; et al. Fusobacterium nucleatum Secretes Outer Membrane Vesicles and Promotes Intestinal Inflammation. mBio 2021, 12, e02706-20. [Google Scholar] [CrossRef]
- Chung, L.; Orberg, E.T.; Geis, A.L.; Chan, J.L.; Fu, K.; Shields, C.E.D.; Dejea, C.M.; Fathi, P.; Chen, J.; Finard, B.B.; et al. Bacteroides fragilis Toxin Coordinates a Pro-carcinogenic Inflammatory Cascade via Targeting of Colonic Epithelial Cells. Cell Host Microbe 2018, 23, 203–214.e5. [Google Scholar] [CrossRef] [PubMed]
- Abdulamir, A.S.; Hafidh, R.R.; Mahdi, L.K.; Al-Jeboori, T.; Abubaker, F. Investigation into the controversial association of Streptococcus gallolyticus with colorectal cancer and adenoma. BMC Cancer 2009, 9, 403. [Google Scholar] [CrossRef] [PubMed]
- Lucas, C.; Barnich, N.; Nguyen, H.T.T. Microbiota, Inflammation and Colorectal Cancer. Int. J. Mol. Sci. 2017, 18, 1310. [Google Scholar] [CrossRef]
- Vancamelbeke, M.; Vermeire, S. The intestinal barrier: A fundamental role in health and disease. Expert Rev. Gastroenterol. Hepatol. 2017, 11, 821–834. [Google Scholar] [CrossRef]
- Keita, Å.; Söderholm, J.D. The intestinal barrier and its regulation by neuroimmune factors. Neurogastroenterol. Motil. 2010, 22, 718–733. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H. Mechanisms linking dietary fiber, gut microbiota and colon cancer prevention. World J. Gastrointest. Oncol. 2014, 6, 41–51. [Google Scholar] [CrossRef]
- Rubinstein, M.R.; Wang, X.; Liu, W.; Hao, Y.; Cai, G.; Han, Y.W. Fusobacterium nucleatum Promotes Colorectal Carcinogenesis by Modulating E-Cadherin/β-Catenin Signaling via its FadA Adhesin. Cell Host Microbe 2013, 14, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Pai, S.G.; Carneiro, B.A.; Mota, J.M.; Costa, R.; Leite, C.A.; Barroso-Sousa, R.; Kaplan, J.B.; Chae, Y.K.; Giles, F.J. Wnt/beta-catenin pathway: Modulating anticancer immune response. J. Hematol. Oncol. 2017, 10, 1–12. [Google Scholar] [CrossRef]
- Wu, X.; Su, D. Enterotoxigenic Escherichia coli infection induces tight junction proteins expression in mice. Iran. J. Vet. Res. 2018, 19, 35–40. [Google Scholar]
- Nagpal, R.; Newman, T.M.; Wang, S.; Jain, S.; Lovato, J.F.; Yadav, H. Obesity-Linked Gut Microbiome Dysbiosis Associated with Derangements in Gut Permeability and Intestinal Cellular Homeostasis Independent of Diet. J. Diabetes Res. 2018, 2018, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Bostick, R.M.; Tran, H.Q.; Gewirtz, A.T.; Campbell, P.T.; Fedirko, V. Circulating Biomarkers of Gut Barrier Function: Correlates and Nonresponse to Calcium Supplementation among Colon Adenoma Patients. Cancer Epidemiol. Biomark. Prev. 2016, 25, 318–326. [Google Scholar] [CrossRef]
- Kang, M.; Edmundson, P.; Araujo-Perez, F.; McCoy, A.N.; Galanko, J.; Keku, T. Association of plasma endotoxin, inflammatory cytokines and risk of colorectal adenomas. BMC Cancer 2013, 13, 91. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, M.; Buc, E.; Sauvanet, P.; Darcha, C.; Dubois, D.; Pereira, B.; Déchelotte, P.; Bonnet, R.; Pezet, D.; Darfeuille-Michaud, A. Colonization of the Human Gut by E. coli and Colorectal Cancer Risk. Clin. Cancer Res. 2014, 20, 859–867. [Google Scholar] [CrossRef]
- Arthur, J.C. Microbiota and colorectal cancer: Colibactin makes its mark. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 317–318. [Google Scholar] [CrossRef]
- Dziubańska-Kusibab, P.J.; Berger, H.; Battistini, F.; Bouwman, B.A.M.; Iftekhar, A.; Katainen, R.; Cajuso, T.; Crosetto, N.; Orozco, M.; Aaltonen, L.A.; et al. Colibactin DNA-damage signature indicates mutational impact in colorectal cancer. Nat. Med. 2020, 26, 1063–1069. [Google Scholar] [CrossRef]
- Gargi, A.; Reno, M.; Blanke, S.R. Bacterial Toxin Modulation of the Eukaryotic Cell Cycle: Are All Cytolethal Distending Toxins Created Equally? Front. Cell. Infect. Microbiol. 2012, 2, 124. [Google Scholar] [CrossRef] [PubMed]
- Guerra, L.; Nemec, K.N.; Massey, S.; Tatulian, S.A.; Thelestam, M.; Frisan, T.; Teter, K. A novel mode of translocation for cytolethal distending toxin. Biochim. Biophys. Acta Bioenerg. 2009, 1793, 489–495. [Google Scholar] [CrossRef]
- Fahrer, J.; Huelsenbeck, J.; Jaurich, H.; Dörsam, B.; Frisan, T.; Eich, M.; Roos, W.P.; Kaina, B.; Fritz, G. Cytolethal distending toxin (CDT) is a radiomimetic agent and induces persistent levels of DNA double-strand breaks in human fibroblasts. DNA Repair 2014, 18, 31–43. [Google Scholar] [CrossRef]
- Buc, E.; Dubois, D.; Sauvanet, P.; Raisch, J.; Delmas, J.; Darfeuille-Michaud, A.; Pezet, D.; Bonnet, R. High Prevalence of Mucosa-Associated E. coli Producing Cyclomodulin and Genotoxin in Colon Cancer. PLoS ONE 2013, 8, e56964. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Gharaibeh, R.Z.; Newsome, R.C.; Pope, J.L.; Dougherty, M.; Tomkovich, S.; Pons, B.; Mirey, G.; Vignard, J.; Hendrixson, D.R.; et al. Campylobacter jejuni promotes colorectal tumorigenesis through the action of cytolethal distending toxin. Gut 2018, 68, 289–300. [Google Scholar] [CrossRef]
- El-Aouar Filho, R.A.; Nicolas, A.; de Paula Castro, T.L.; Deplanche, M.; de Carvalho Azevedo, V.A.; Goossens, P.L.; Taieb, F.; Lina, G.; le Loir, Y.; Berkova, N. Heterogeneous Family of Cyclomodulins: Smart Weapons That Allow Bacteria to Hijack the Eukaryotic Cell Cycle and Promote Infections. Front. Cell. Infect. Microbiol. 2017, 7, 208. [Google Scholar] [CrossRef] [PubMed]
- Giamboi-Miraglia, A.; Travaglione, S.; Filippini, P.; Fabbri, A.; Fiorentini, C.; Falzano, L. A multinucleating Escherichia coli cytotoxin perturbs cell cycle in cultured epithelial cells. Toxicol. Vitr. 2007, 21, 235–239. [Google Scholar] [CrossRef]
- Zhang, Z.; Aung, K.M.; Uhlin, B.E.; Wai, S.N. Reversible senescence of human colon cancer cells after blockage of mitosis/cytokinesis caused by the CNF1 cyclomodulin from Escherichia coli. Sci. Rep. 2018, 8, 17780. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, U.S.; Tan, B.W.Q.; Vellayappan, B.A.; Jeyasekharan, A.D. ROS and the DNA damage response in cancer. Redox Biol. 2018, 25, 101084. [Google Scholar] [CrossRef] [PubMed]
- Gunasekera, D.C.; Ma, J.; Vacharathit, V.; Shah, P.; Ramakrishnan, A.; Uprety, P.; Shen, Z.; Sheh, A.; Brayton, C.F.; Whary, M.T.; et al. The development of colitis in Il10−/− mice is dependent on IL-22. Mucosal Immunol. 2020, 13, 493–506. [Google Scholar] [CrossRef]
- Tsoi, H.; Chu, E.S.; Zhang, X.; Sheng, J.; Nakatsu, G.; Ng, S.C.; Chan, A.; Chan, F.K.; Sung, J.J.Y.; Yu, J. Peptostreptococcus anaerobius Induces Intracellular Cholesterol Biosynthesis in Colon Cells to Induce Proliferation and Causes Dysplasia in Mice. Gastroenterology 2017, 152, 1419–1433.e5. [Google Scholar] [CrossRef] [PubMed]
- Barrett, M.; Hand, C.; Shanahan, F.; Murphy, T.; O’Toole, P.W. Mutagenesis by Microbe: The Role of the Microbiota in Shaping the Cancer Genome. Trends Cancer 2020, 6, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Inamura, K. Colorectal Cancers: An Update on Their Molecular Pathology. Cancers 2018, 10, 26. [Google Scholar] [CrossRef] [PubMed]
- Koi, M.; Okita, Y.; Carethers, J.M. Fusobacterium nucleatum Infection in Colorectal Cancer: Linking Inflammation, DNA Mismatch Repair and Genetic and Epigenetic Alterations. J. Anus Rectum Colon 2018, 2, 37–46. [Google Scholar] [CrossRef]
- Allen, J.; Hao, S.; Sears, C.L.; Timp, W. Epigenetic Changes Induced by Bacteroides fragilis Toxin. Infect. Immun. 2019, 87, 87. [Google Scholar] [CrossRef]
- Bishop, K.S.; Xu, H.; Marlow, G. Epigenetic Regulation of Gene Expression Induced by Butyrate in Colorectal Cancer: Involvement of MicroRNA. Genet. Epigenet. 2017, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Bultman, S.J. Interplay between diet, gut microbiota, epigenetic events, and colorectal cancer. Mol. Nutr. Food Res. 2017, 61, 61. [Google Scholar] [CrossRef]
- González-Sánchez, P.; DeNicola, G.M. The microbiome(s) and cancer: Know thy neighbor(s). J. Pathol. 2021, 254, 332–343. [Google Scholar] [CrossRef]
- Donohoe, D.R.; Collins, L.B.; Wali, A.; Bigler, R.; Sun, W.; Bultman, S.J. The Warburg Effect Dictates the Mechanism of Butyrate-Mediated Histone Acetylation and Cell Proliferation. Mol. Cell 2012, 48, 612–626. [Google Scholar] [CrossRef]
- Krautkramer, K.; Kreznar, J.H.; Romano, K.A.; Vivas, E.I.; Barrett-Wilt, G.A.; Rabaglia, M.E.; Keller, M.P.; Attie, A.D.; Rey, F.E.; Denu, J.M. Diet-Microbiota Interactions Mediate Global Epigenetic Programming in Multiple Host Tissues. Mol. Cell 2016, 64, 982–992. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, P.D.; Manicassamy, S.; Munn, D.H.; et al. Activation of Gpr109a, Receptor for Niacin and the Commensal Metabolite Butyrate, Suppresses Colonic Inflammation and Carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Flint, H.J. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 2009, 294, 1–8. [Google Scholar] [CrossRef]
- Mirzaei, R.; Afaghi, A.; Babakhani, S.; Sohrabi, M.R.; Hosseini-Fard, S.R.; Babolhavaeji, K.; Akbari, S.K.A.; Yousefimashouf, R.; Karampoor, S. Role of microbiota-derived short-chain fatty acids in cancer development and prevention. Biomed. Pharmacother. 2021, 139, 111619. [Google Scholar] [CrossRef] [PubMed]
- Hullar, M.A.J.; Fu, B.C. Diet, the Gut Microbiome, and Epigenetics. Cancer J. 2014, 20, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Holst, B.; Williamson, G. A Critical Review of the Bioavailability of Glucosinolates and Related Compounds. Nat. Prod. Rep. 2004, 21, 425–447. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Zhang, J.; Wu, H.; Yu, D.; Fang, X. Akkermansia muciniphila Aspartic Protease Amuc_1434* Inhibits Human Colorectal Cancer LS174T Cell Viability via TRAIL-Mediated Apoptosis Pathway. Int. J. Mol. Sci. 2020, 21, 3385. [Google Scholar] [CrossRef]
- Tavalire, H.F.; Christie, D.M.; Leve, L.D.; Ting, N.; Cresko, W.A.; Bohannan, B.J.M. Shared Environment and Genetics Shape the Gut Microbiome after Infant Adoption. mBio 2021, 12, e00548-21. [Google Scholar] [CrossRef]
- Mima, K.; Kosumi, K.; Baba, Y.; Hamada, T.; Baba, H.; Ogino, S. The microbiome, genetics, and gastrointestinal neoplasms: The evolving field of molecular pathological epidemiology to analyze the tumor–immune–microbiome interaction. Qual. Life Res. 2021, 140, 725–746. [Google Scholar] [CrossRef]
- Chen, W.; Liu, F.; Ling, Z.; Tong, X.; Xiang, C. Human Intestinal Lumen and Mucosa-Associated Microbiota in Patients with Colorectal Cancer. PLoS ONE 2012, 7, e39743. [Google Scholar] [CrossRef] [PubMed]
- Ishida, S.; Kato, K.; Tanaka, M.; Odamaki, T.; Kubo, R.; Mitsuyama, E.; Xiao, J.-Z.; Yamaguchi, R.; Uematsu, S.; Imoto, S.; et al. Genome-wide association studies and heritability analysis reveal the involvement of host genetics in the Japanese gut microbiota. Commun. Biol. 2020, 3, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Uson, P.L.; Riegert-Johnson, D.; Boardman, L.; Kisiel, J.; Mountjoy, L.; Patel, N.; Lizaola-Mayo, B.; Borad, M.J.; Ahn, D.; Sonbol, M.B.; et al. Germline Cancer Susceptibility Gene Testing in Unselected Patients with Colorectal Adenocarcinoma: A Multicenter Prospective Study. Clin. Gastroenterol. Hepatol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Hamada, T.; Keum, N.; Nishihara, R.; Ogino, S. Molecular pathological epidemiology: New developing frontiers of big data science to study etiologies and pathogenesis. J. Gastroenterol. 2017, 52, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Nishihara, R.; Cao, Y.; Chun, E.; Qian, Z.R.; Mima, K.; Inamura, K.; Masugi, Y.; Nowak, J.A.; Nosho, K.; et al. Marine ω-3 Polyunsaturated Fatty Acid Intake and Risk of Colorectal Cancer Characterized by Tumor-Infiltrating T Cells. JAMA Oncol. 2016, 2, 1197–1206. [Google Scholar] [CrossRef] [PubMed]
- Hamada, T.; Zhang, X.; Mima, K.; Bullman, S.; Sukawa, Y.; Nowak, J.A.; Kosumi, K.; Masugi, Y.; Twombly, T.S.; Cao, Y.; et al. Fusobacterium nucleatum in Colorectal Cancer Relates to Immune Response Differentially by Tumor Microsatellite Instability Status. Cancer Immunol. Res. 2018, 6, 1327–1336. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Tabung, F.K.; Zhang, X.; Nowak, J.A.; Qian, Z.R.; Hamada, T.; Nevo, D.; Bullman, S.; Mima, K.; Kosumi, K.; et al. Diets That Promote Colon Inflammation Associate with Risk of Colorectal Carcinomas That Contain Fusobacterium nucleatum. Clin. Gastroenterol. Hepatol. 2018, 16, 1622–1631.e3. [Google Scholar] [CrossRef]
- Mehta, R.S.; Nishihara, R.; Cao, Y.; Song, M.; Mima, K.; Qian, Z.R.; Nowak, J.A.; Kosumi, K.; Hamada, T.; Masugi, Y.; et al. Association of Dietary Patterns with Risk of Colorectal Cancer Subtypes Classified by Fusobacterium nucleatum in Tumor Tissue. JAMA Oncol. 2017, 3, 921–927. [Google Scholar] [CrossRef]
- Hamada, T.; Nowak, J.A.; Milner, D.A.; Song, M.; Ogino, S. Integration of Microbiology, Molecular Pathology, and Epidemiology: A New Paradigm to Explore the Pathogenesis of Microbiome-Driven Neoplasms. J. Pathol. 2019, 247, 615–628. [Google Scholar] [CrossRef]
- Nash, A.K.; Auchtung, T.A.; Wong, M.C.; Smith, D.P.; Gesell, J.R.; Ross, M.C.; Stewart, C.J.; Metcalf, G.A.; Muzny, D.M.; Gibbs, R.A.; et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 2017, 5, 153. [Google Scholar] [CrossRef] [PubMed]
- Hager, C.L.; Ghannoum, M.A. The mycobiome: Role in health and disease, and as a potential probiotic target in gastrointestinal disease. Dig. Liver Dis. 2017, 49, 1171–1176. [Google Scholar] [CrossRef]
- Mims, T.S.; Al Abdallah, Q.; Stewart, J.D.; Watts, S.P.; White, C.T.; Rousselle, T.V.; Gosain, A.; Bajwa, A.; Han, J.; Willis, K.A.; et al. The gut mycobiome of healthy mice is shaped by the environment and correlates with metabolic outcomes in response to diet. Commun. Biol. 2021, 4, 1–11. [Google Scholar] [CrossRef]
- Iliev, I.D.; Funari, V.A.; Taylor, K.D.; Nguyen, Q.; Reyes, C.N.; Strom, S.; Brown, J.; Becker, C.A.; Fleshner, P.R.; Dubinsky, M.; et al. Interactions Between Commensal Fungi and the C-Type Lectin Receptor Dectin-1 Influence Colitis. Science 2012, 336, 1314–1317. [Google Scholar] [CrossRef] [PubMed]
- Castoldi, A.; Andrade-Oliveira, V.; Aguiar, C.F.; Amano, M.; Lee, J.; Miyagi, M.T.; Latância, M.T.; Braga, T.; da Silva, M.B.; Ignácio, A.; et al. Dectin-1 Activation Exacerbates Obesity and Insulin Resistance in the Absence of MyD88. Cell Rep. 2017, 19, 2272–2288. [Google Scholar] [CrossRef] [PubMed]
- Luan, C.; Xie, L.; Yang, X.; Miao, H.; Lv, N.; Zhang, R.; Xiao, X.; Hu, Y.; Liu, Y.; Wu, N.; et al. Dysbiosis of Fungal Microbiota in the Intestinal Mucosa of Patients with Colorectal Adenomas. Sci. Rep. 2015, 5, 7980. [Google Scholar] [CrossRef]
- Gao, R.; Kong, C.; Li, H.; Huang, L.; Qu, X.; Qin, N.; Qin, H. Dysbiosis signature of mycobiota in colon polyp and colorectal cancer. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 2457–2468. [Google Scholar] [CrossRef] [PubMed]
- Coker, O.O.; Nakatsu, G.; Dai, R.Z.; Wu, W.K.; Wong, S.H.; Ng, S.C.; Chan, F.K.; Sung, J.J.Y.; Yu, J. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut 2018, 68, 654–662. [Google Scholar] [CrossRef] [PubMed]
- Hatinguais, R.; Willment, J.A.; Brown, G.D. PAMPs of the Fungal Cell Wall and Mammalian PRRs. In Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2020; Volume 425. [Google Scholar]
- Malik, A.; Sharma, D.; Malireddi, R.; Guy, C.S.; Chang, T.-C.; Olsen, S.R.; Neale, G.; Vogel, P.; Kanneganti, T.-D. SYK-CARD9 Signaling Axis Promotes Gut Fungi-Mediated Inflammasome Activation to Restrict Colitis and Colon Cancer. Immunity 2018, 49, 515–530.e5. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, D.; Hou, Y.; Shen, S.; Wang, T. The adaptor protein CARD9, from fungal immunity to tumorigenesis. Am. J. Cancer Res. 2020, 10, 2203–2225. [Google Scholar]
- Bertin, J.; Guo, Y.; Wang, L.; Srinivasula, S.M.; Jacobson, M.D.; Poyet, J.-L.; Merriam, S.; Du, M.-Q.; Dyer, M.; Robison, K.E.; et al. CARD9 Is a Novel Caspase Recruitment Domain-containing Protein That Interacts with BCL10/CLAP and Activates NF-κB. J. Biol. Chem. 2000, 275, 41082–41086. [Google Scholar] [CrossRef]
- Zhu, Y.; Shi, T.; Lu, X.; Xu, Z.; Qu, J.; Zhang, Z.; Shi, G.; Shen, S.; Hou, Y.; Chen, Y.; et al. Fungal-induced glycolysis in macrophages promotes colon cancer by enhancing innate lymphoid cell secretion of IL-22. EMBO J. 2021, 40, e105320. [Google Scholar] [CrossRef]
- Harrison, C. IL-22: Linking inflammation and cancer. Nat. Rev. Drug Discov. 2013, 12, 505. [Google Scholar] [CrossRef]
- Kryczek, I.; Lin, Y.; Nagarsheth, N.; Peng, D.; Zhao, L.; Zhao, E.; Vatan, L.; Szeliga, W.; Dou, Y.; Owens, S.; et al. IL-22+CD4+ T Cells Promote Colorectal Cancer Stemness via STAT3 Transcription Factor Activation and Induction of the Methyltransferase DOT1L. Immunity 2014, 40, 772–784. [Google Scholar] [CrossRef]
- Omotayo, O.P.; Omotayo, A.O.; Mwanza, M.; Babalola, O.O. Prevalence of Mycotoxins and Their Consequences on Human Health. Toxicol. Res. 2019, 35, 1–7. [Google Scholar] [CrossRef]
- Buszewska-Forajta, M. Mycotoxins, Invisible Danger of Feedstuff with Toxic Effect on Animals. Toxicon 2020, 182, 34–53. [Google Scholar] [CrossRef]
- Lo, E.K.K.; Lee, J.C.-Y.; Turner, P.C.; El-Nezami, H. Low dose of zearalenone elevated colon cancer cell growth through G protein-coupled estrogenic receptor. Sci. Rep. 2021, 11, 7403. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhang, S.; Wang, J.; Shan, A.; Xu, L. Changes in intestinal barrier functions and gut microbiota in rats exposed to zearalenone. Ecotoxicol. Environ. Saf. 2020, 204, 111072. [Google Scholar] [CrossRef] [PubMed]
- Liew, W.-P.-P.; Mohd-Redzwan, S. Mycotoxin: Its Impact on Gut Health and Microbiota. Front. Cell. Infect. Microbiol. 2018, 8, 60. [Google Scholar] [CrossRef]
- Shang, F.-M.; Liu, H.-L. Fusobacterium nucleatum and colorectal cancer: A review. World J. Gastrointest. Oncol. 2018, 10, 71–81. [Google Scholar] [CrossRef]
- Cirauqui, M.L.G.; Nieminen, M.; Frazer, L.N.; Aguirre-Urizar, J.M.; Moragues, M.-D.; Rautemaa, R. Production of carcinogenic acetaldehyde by Candida albicans from patients with potentially malignant oral mucosal disorders. J. Oral Pathol. Med. 2012, 42, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Dunagan, M.; Chaudhry, K.; Samak, G.; Rao, R.K. Acetaldehyde disrupts tight junctions in Caco-2 cell monolayers by a protein phosphatase 2A-dependent mechanism. Am. J. Physiol. Liver Physiol. 2012, 303, G1356–G1364. [Google Scholar] [CrossRef] [PubMed]
- Blagojevic, M.; Camilli, G.; Maxson, M.; Hube, B.; Moyes, D.L.; Richardson, J.P.; Naglik, J.R. Candidalysin triggers epithelial cellular stresses that induce necrotic death. Cell. Microbiol. 2021, 23, e13371. [Google Scholar] [CrossRef] [PubMed]
- Krüger, W.; Vielreicher, S.; Kapitan, M.; Jacobsen, I.D.; Niemiec, M.J. Fungal-Bacterial Interactions in Health and Disease. Pathogens 2019, 8, 70. [Google Scholar] [CrossRef] [PubMed]
- Hold, G.L.; Allen-Vercoe, E. Gut microbial biofilm composition and organisation holds the key to CRC. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 329–330. [Google Scholar] [CrossRef] [PubMed]
- Lakkaraju, A.; Rodriguez-Boulan, E. Itinerant exosomes: Emerging roles in cell and tissue polarity. Trends Cell Biol. 2008, 18, 199–209. [Google Scholar] [CrossRef]
- Keller, S.; Ridinger, J.; Rupp, A.-K.; Janssen, J.W.G.; Altevogt, P. Body fluid derived exosomes as a novel template for clinical diagnostics. J. Transl. Med. 2011, 9, 86. [Google Scholar] [CrossRef]
- Zeringer, E.; Barta, T.; Li, M.; Vlassov, A.V. Strategies for Isolation of Exosomes. Cold Spring Harb. Protoc. 2015, 2015, 319–323. [Google Scholar] [CrossRef]
- Gurunathan, S.; Kang, M.-H.; Kim, J.-H. A Comprehensive Review on Factors Influences Biogenesis, Functions, Therapeutic and Clinical Implications of Exosomes. Int. J. Nanomed. 2021, 16, 1281–1312. [Google Scholar] [CrossRef] [PubMed]
- Ayyar, K.K.; Moss, A.C. Exosomes in Intestinal Inflammation. Front. Pharmacol. 2021, 12, 658505. [Google Scholar] [CrossRef]
- Larabi, A.; Barnich, N.; Nguyen, H.T.T. Emerging Role of Exosomes in Diagnosis and Treatment of Infectious and Inflammatory Bowel Diseases. Cells 2020, 9, 1111. [Google Scholar] [CrossRef]
- Ocansey, D.W.; Zhang, L.; Wang, Y.; Yan, Y.; Qian, H.; Zhang, X.; Xu, W.; Mao, F. Exosome-mediated effects and applications in inflammatory bowel disease. Biol. Rev. 2020, 95, 1287–1307. [Google Scholar] [CrossRef]
- Moein, S.; Vaghari-Tabari, M.; Qujeq, D.; Majidinia, M.; Nabavi, S.M.; Yousefi, B. MiRNAs and inflammatory bowel disease: An interesting new story. J. Cell. Physiol. 2019, 234, 3277–3293. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Shen, J.; Cheng, J.; Fan, X. MicroRNA-21 regulates intestinal epithelial tight junction permeability. Cell Biochem. Funct. 2015, 33, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, F.; He, D.-K.; Fan, X.-M.; Shen, J. MicroRNA-21 is upregulated during intestinal barrier dysfunction induced by ischemia reperfusion. Kaohsiung J. Med. Sci. 2018, 34, 556–563. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Berg, N.; Lee, J.W.; Le, T.-T.; Neudecker, V.; Jing, N.; Eltzschig, H. MicroRNA miR-223 as regulator of innate immunity. J. Leukoc. Biol. 2018, 104, 515–524. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, S.; Yu, Q.; Yang, G.; Guo, J.; Li, M.; Zeng, Z.; He, Y.; Chen, B.; Chen, M. Circulating MicroRNA223 is a New Biomarker for Inflammatory Bowel Disease. Medicine 2016, 95, e2703. [Google Scholar] [CrossRef]
- Wei, M.; Gao, X.; Liu, L.; Li, Z.; Wan, Z.; Dong, Y.; Chen, X.; Niu, Y.; Zhang, J.; Yang, G. Visceral Adipose Tissue Derived Exosomes Exacerbate Colitis Severity via Pro-inflammatory MiRNAs in High Fat Diet Fed Mice. ACS Nano 2020, 14, 5099–5110. [Google Scholar] [CrossRef] [PubMed]
- Carrière, J.; Bretin, A.; Darfeuille-Michaud, A.; Barnich, N.; Nguyen, H.T.T. Exosomes Released from Cells Infected with Crohn’s Disease–associated Adherent-Invasive Escherichia coli Activate Host Innate Immune Responses and Enhance Bacterial Intracellular Replication. Inflamm. Bowel Dis. 2016, 22, 516–528. [Google Scholar] [CrossRef]
- Shimoda, A.; Ueda, K.; Nishiumi, S.; Murata-Kamiya, N.; Mukai, S.-A.; Sawada, S.-I.; Azuma, T.; Hatakeyama, M.; Akiyoshi, K. Exosomes as nanocarriers for systemic delivery of the Helicobacter pylori virulence factor CagA. Sci. Rep. 2016, 6, 18346. [Google Scholar] [CrossRef]
- Deng, Z.; Mu, J.; Tseng, M.; Wattenberg, B.; Zhuang, X.; Egilmez, N.K.; Wang, Q.; Zhang, L.; Norris, J.D.; Guo, H.; et al. Enterobacteria-secreted particles induce production of exosome-like S1P-containing particles by intestinal epithelium to drive Th17-mediated tumorigenesis. Nat. Commun. 2015, 6, 1–15. [Google Scholar] [CrossRef]
- Smyth, E.M.; FitzGerald, G. The Eicosanoids: Prostaglandins, Thromboxanes, Leukotrienes, and Related Compounds; Basic and Clinical Pharmacology 14e; McGraw-Hill Education: New York, NY, USA, 2012. [Google Scholar]
- Guo, S.; Chen, J.; Chen, F.; Zeng, Q.; Liu, W.-L.; Zhang, G. Exosomes derived from Fusobacterium nucleatum-infected colorectal cancer cells facilitate tumour metastasis by selectively carrying miR-1246/92b-3p/27a-3p and CXCL16. Gut 2021, 70, 1507–1519. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Liang, L.; Yang, C.; Zhou, Y.; Chen, Y. Extracellular vesicles of Fusobacterium nucleatum compromise intestinal barrier through targeting RIPK1-mediated cell death pathway. Gut Microbes 2021, 13, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Xu, Y.; Xu, W.; Li, M.; Su, H.; Li, C.; Liu, Z. The exosome secretion inhibitor neticonazole suppresses intestinal dysbacteriosis-induced tumorigenesis of colorectal cancer. Investig. New Drugs 2019, 38, 221–228. [Google Scholar] [CrossRef]
- Huber, V.; Fais, S.; Iero, M.; Lugini, L.; Canese, P.; Squarcina, P.; Zaccheddu, A.; Colone, M.; Arancia, G.; Gentile, M.; et al. Human Colorectal Cancer Cells Induce T-Cell Death Through Release of Proapoptotic Microvesicles: Role in Immune Escape. Gastroenterology 2005, 128, 1796–1804. [Google Scholar] [CrossRef] [PubMed]
- Cafforio, P.; Viggiano, L.; Mannavola, F.; Pellè, E.; Caporusso, C.; Maiorano, E.; Felici, C.; Silvestris, F. pIL6-TRAIL-engineered umbilical cord mesenchymal/stromal stem cells are highly cytotoxic for myeloma cells both in vitro and in vivo. Stem Cell Res. Ther. 2017, 8, 206. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Jia, H.; Bao, X.; Wu, Y.; Zhu, T.; Li, R.; Zhao, H. Tumor exosome promotes Th17 cell differentiation by transmitting the lncRNA CRNDE-h in colorectal cancer. Cell Death Dis. 2021, 12, 1–14. [Google Scholar] [CrossRef]
- Abu, N.; Othman, N.; Ab Razak, N.S.; Bakarurraini, N.A.A.R.; Nasir, S.N.; Soh, J.E.C.; Mazlan, L.; Azman, Z.A.M.; Jamal, R. Extracellular Vesicles Derived from Colorectal Cancer Affects CD8 T Cells: An Analysis Based on Body Mass Index. Front. Cell Dev. Biol. 2020, 8, 564648. [Google Scholar] [CrossRef] [PubMed]
- Takano, Y.; Masuda, T.; Iinuma, H.; Yamaguchi, R.; Sato, K.; Tobo, T.; Hirata, H.; Kuroda, Y.; Nambara, S.; Hayashi, N.; et al. Circulating exosomal microRNA-203 is associated with metastasis possibly via inducing tumor-associated macrophages in colorectal cancer. Oncotarget 2017, 8, 78598–78613. [Google Scholar] [CrossRef]
- Cooks, T.; Pateras, I.S.; Jenkins, L.M.; Patel, K.M.; Robles, A.I.; Morris, J.; Forshew, T.; Appella, E.; Gorgoulis, V.; Harris, C.C. Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat. Commun. 2018, 9, 1–15. [Google Scholar] [CrossRef]
- Menck, K.; Bleckmann, A.; Wachter, A.; Hennies, B.; Ries, L.; Schulz, M.; Balkenhol, M.; Pukrop, T.; Schatlo, B.; Rost, U.; et al. Characterisation of tumour-derived microvesicles in cancer patients’ blood and correlation with clinical outcome. J. Extracell. Vesicles 2017, 6, 1340745. [Google Scholar] [CrossRef]
- Ma, Y.-S.; Wu, T.-M.; Ling, C.-C.; Yu, F.; Zhang, J.; Cao, P.-S.; Gu, L.-P.; Wang, H.-M.; Xu, H.; Li, L.; et al. M2 macrophage-derived exosomal microRNA-155-5p promotes the immune escape of colon cancer by downregulating ZC3H12B. Mol. Ther.–Oncolytics 2021, 20, 484–498. [Google Scholar] [CrossRef] [PubMed]
- Szvicsek, Z.; Oszvald, Á.; Szabó, L.; Sándor, G.O.; Kelemen, A.; Soós, A.Á.; Pálóczi, K.; Harsányi, L.; Tölgyes, T.; Dede, K.; et al. Extracellular vesicle release from intestinal organoids is modulated by Apc mutation and other colorectal cancer progression factors. Cell. Mol. Life Sci. 2019, 76, 2463–2476. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.; Ye, X.; Lu, S.-R.; Li, H.; Liu, H.-Y.; Zhai, Q.; Yu, B. Exosomal miR-30a and miR-222 derived from colon cancer mesenchymal stem cells promote the tumorigenicity of colon cancer through targeting MIA3. J. Gastrointest. Oncol. 2021, 12, 52–68. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Zhou, J.; Wang, H.; Zhang, M.; Yang, X.; Song, W. miR-424-5p promotes the proliferation and metastasis of colorectal cancer by directly targeting SCN4B. Pathol.–Res. Pract. 2020, 216, 152731. [Google Scholar] [CrossRef] [PubMed]
- Mulvey, H.E.; Chang, A.; Adler, J.; Del Tatto, M.; Perez, K.; Quesenberry, P.J.; Chatterjee, D. Extracellular vesicle-mediated phenotype switching in malignant and non-malignant colon cells. BMC Cancer 2015, 15, 571. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Zhong, J.; Zhong, B.; Huang, J.; Jiang, L.; Jiang, Y.; Yuan, J.; Sun, J.; Dai, L.; Yang, C.; et al. Exosomes as potential sources of biomarkers in colorectal cancer. Cancer Lett. 2020, 476, 13–22. [Google Scholar] [CrossRef]
- Chen, H.-L.; Li, J.-J.; Jiang, F.; Shi, W.-J.; Chang, G.-Y. MicroRNA-4461 derived from bone marrow mesenchymal stem cell exosomes inhibits tumorigenesis by downregulating COPB2 expression in colorectal cancer. Biosci. Biotechnol. Biochem. 2020, 84, 338–346. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Zhang, H.; Liu, R.; Deng, T.; Ning, T.; Bai, M.; Yang, Y.; Zhu, K.; Wang, J.; Duan, J.; et al. iRGD-modified exosomes effectively deliver CPT1A siRNA to colon cancer cells, reversing oxaliplatin resistance by regulating fatty acid oxidation. Mol. Oncol. 2021. [Google Scholar] [CrossRef]
- Li, Y.; Gao, Y.; Gong, C.; Wang, Z.; Xia, Q.; Gu, F.; Hu, C.; Zhang, L.; Guo, H.; Gao, S. A33 antibody-functionalized exosomes for targeted delivery of doxorubicin against colorectal cancer. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 1973–1985. [Google Scholar] [CrossRef]
- Liang, G.; Zhu, Y.; Ali, D.J.; Tian, T.; Xu, H.; Si, K.; Sun, B.; Chen, B.; Xiao, Z. Engineered exosomes for targeted co-delivery of miR-21 inhibitor and chemotherapeutics to reverse drug resistance in colon cancer. J. Nanobiotechnol. 2020, 18, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Rezaei, R.; Baghaei, K.; Amani, D.; Piccin, A.; Hashemi, S.M.; Aghdaei, H.A.; Zali, M.R. Exosome-mediated delivery of functionally active miRNA-375-3p mimic regulate epithelial mesenchymal transition (EMT) of colon cancer cells. Life Sci. 2021, 269, 119035. [Google Scholar] [CrossRef] [PubMed]
- Di Rocco, G.; Baldari, S.; Toietta, G. Exosomes and other extracellular vesicles-mediated microRNA delivery for cancer therapy. Transl. Cancer Res. 2017, 6, S1321–S1330. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khalyfa, A.A.; Punatar, S.; Aslam, R.; Yarbrough, A. Exploring the Inflammatory Pathogenesis of Colorectal Cancer. Diseases 2021, 9, 79. https://doi.org/10.3390/diseases9040079
Khalyfa AA, Punatar S, Aslam R, Yarbrough A. Exploring the Inflammatory Pathogenesis of Colorectal Cancer. Diseases. 2021; 9(4):79. https://doi.org/10.3390/diseases9040079
Chicago/Turabian StyleKhalyfa, Ahamed A, Shil Punatar, Rida Aslam, and Alex Yarbrough. 2021. "Exploring the Inflammatory Pathogenesis of Colorectal Cancer" Diseases 9, no. 4: 79. https://doi.org/10.3390/diseases9040079
APA StyleKhalyfa, A. A., Punatar, S., Aslam, R., & Yarbrough, A. (2021). Exploring the Inflammatory Pathogenesis of Colorectal Cancer. Diseases, 9(4), 79. https://doi.org/10.3390/diseases9040079




