Antibiotic Susceptibility, Biofilm Production, and Detection of mecA Gene among Staphylococcus aureus Isolates from Different Clinical Specimens
Abstract
1. Introduction
2. Materials and Methods
2.1. Research Design
2.2. Study Site and Specimens
2.3. Samples for the Study
2.4. Isolation and Identification of S. aureus
2.5. Screening of Biofilm-Producing S. aureus
2.6. Antibiotic Susceptibility Test
2.7. Detection of MRSA by Cefoxitin Disc-Diffusion Method
2.8. Detection of mecA Gene by Polymerase Chain Reaction (PCR)
2.9. Quality Control
2.10. Data Analysis
3. Results
3.1. Growth Pattern and Distribution of Bacteria
3.2. Distribution of MRSA and MSSA in Different Clinical Specimens
3.3. Antibiotic Susceptibility Status and MDR S. aureus
3.4. Distribution of Biofilm Producers among MRSA and MSSA
3.5. Antibiotic Susceptibility Pattern among Different Biofilm Producers
3.6. Detection of mecA Gene in MRSA and MSSA Isolates
4. Discussion
5. Conclusions
6. Strengths and Limitations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tong, S.Y.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G., Jr. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef]
- Singh, A.K.; Prakash, P.; Achra, A.; Singh, G.P.; Das, A.; Singh, R.K. Standardization and classification of in vitro biofilm formation by clinical isolates of Staphylococcus aureus. J. Glob. Infect. Dis. 2017, 9, 93–101. [Google Scholar] [PubMed]
- Lister, J.L.; Horswill, A.R. Staphylococcus aureus biofilms: Recent developments in biofilm dispersal. Front. Cell. Infect. Microbiol. 2014, 4, 178. [Google Scholar] [CrossRef]
- Beenken, K.E.; Dunman, P.M.; McAleese, F.; Macapagal, D.; Murphy, E.; Projan, S.J.; Blevins, J.S.; Smeltzer, M.S. Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 2004, 186, 4665–4684. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S.; Costerton, J.W. Antibiotic resistance of bacteria in biofilms. Lancet 2001, 358, 135–138. [Google Scholar] [CrossRef]
- Dhand, A.; Sakoulas, G. Reduced vancomycin susceptibility among clinical Staphylococcus aureus isolates (‘the MIC Creep’): Implications for therapy. F1000 Med. Rep. 2012, 4, 4. [Google Scholar] [CrossRef] [PubMed]
- Fukunaga, B.T.; Sumida, W.K.; Taira, D.A.; Davis, J.W.; Seto, T.B. Hospital-acquired methicillin-resistant Staphylococcus aureus bacteremia related to medicare antibiotic prescriptions: A state-level analysis. Hawaii J. Med. Public Health 2016, 75, 303–309. [Google Scholar]
- Udo, E.E. Community-acquired methicillin-resistant Staphylococcus aureus: The new face of an old foe? Med. Princ. Pract. 2013, 22 (Suppl. 1), 20–29. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Rosas, G.J.; Merida-Vieyra, J.; Aparicio-Ozores, G.; Lara-Hernandez, A.; De Colsa, A.; Aquino-Andrade, A. Molecular characterization of Staphylococcus aureus obtained from blood cultures of paediatric patients treated in a tertiary care hospital in Mexico. Infect. Drug Resist. 2021, 14, 1545–1556. [Google Scholar] [CrossRef] [PubMed]
- Lakhundi, S.; Zhang, K. Methicillin-resistant Staphylococcus aureus: Molecular characterization, evolution, and epidemiology. Clin. Microbiol. Rev. 2018, 31, 1–103. [Google Scholar] [CrossRef] [PubMed]
- Khanal, L.; Jha, B.K. Prevalence of methicillin resistant Staphylococcus aureus (MRSA) among skin infection cases at a hospital in Chitwan, Nepal. Nepal Med. Coll. J. 2010, 12, 224–228. [Google Scholar] [PubMed]
- Bhomi, U.; Rijal, K.R.; Neupane, B.; Shrestha, S.; Chaudhary, M.; Acharya, D.; Thapa Shrestha, U.; Adhikari, N.; Ghimire, P. Status of inducible clindamycin resistance among macrolide resistant Staphylococcus aureus. Afr. J. Microbiol. Res. 2016, 10, 280–284. [Google Scholar] [CrossRef][Green Version]
- Adhikari, S.; Regmi, R.S.; Pandey, S.; Paudel, P.; Neupane, N.; Chalise, S.; Dubey, A.; Kafle, S.C.; Rijal, K.R. Bacterial etiology of bronchoalveolar lavage fluid in tertiary care patients and antibiogram of the isolates. J. Inst. Sci. Technol. 2021, 26, 99–106. [Google Scholar] [CrossRef]
- Kumari, N.; Mohapatra, T.M.; Singh, Y.I. Prevalence of methicillin-resistant Staphylococcus aureus (MRSA) in a Tertiary-care hospital in Eastern Nepal. J. Nepal Med. Assoc. 2008, 47, 53–56. [Google Scholar] [CrossRef]
- Raut, S.; Bajracharya, K.; Adhikari, J.; Pant, S.S.; Adhikari, B. Prevalence of methicillin resistant Staphylococcus aureus in Lumbini Medical College and Teaching Hospital, Palpa, Western Nepal. BMC Res. Notes 2017, 10, 187. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.; Dhungel, B.; Bastola, A.; Banjara, M.R.; Rijal, K.R.; Ghimire, P. Methicillin resistant Staphylococcus aureus in health careworkers of a tertiary care infectious diseases hospital in Nepal. Tribhuvan Univ. J. Microbiol. 2020, 7, 19–30. [Google Scholar] [CrossRef]
- Rijal, K.R.; Pahari, N.; Shrestha, B.K.; Nepal, A.K.; Paudel, B.; Mahato, P.; Skalko-Basnet, N. Prevalence of methicillin resistant Staphylococcus aureus in school children of Pokhara. Nepal Med. Coll. J. 2008, 10, 192–195. [Google Scholar] [PubMed]
- Shahi, K.; Rijal, K.R.; Adhikari, N.; Thapa-Shrestha, U.; Banjara, M.R.; Sharma, V.K.; Ghimire, P. Methicillin resistant Staphylococcus aureus: Prevalence and antibiogram in various clinical specimens at Alka Hospital. Tribhuvan Univ. J. Microbiol. 2018, 5, 77–82. [Google Scholar] [CrossRef]
- Dhungel, S.; Rijal, K.R.; Yadav, B.; Dhungel, B.; Adhikari, N.; Shrestha, U.T.; Adhikari, B.; Banjara, M.R.; Ghimire, P. Methicillin-resistant Staphylococcus aureus (MRSA): Prevalence, antimicrobial susceptibility pattern, and detection of mecA gene among cardiac patients from a Tertiary Care Heart Center in Kathmandu, Nepal. Infect. Dis. 2021, 14, 11786337211037355. [Google Scholar] [CrossRef]
- American Society for Microbiology. Manual of Clinical Microbiology, 2nd ed.; ASM Press: Washington, DC, USA, 2014. [Google Scholar]
- Cheesbrough, M. District Laboratory Practice in Tropical Countries, 2nd ed.; Cambridge University Press: Cambridge, UK, 2003. [Google Scholar]
- Lamichhane, K.; Adhikari, N.; Bastola, A.; Devkota, L.; Bhandari, P.; Dhungel, B.; Thapa Shrestha, U.; Adhikari, B.; Banjara, M.R.; Rijal, K.R.; et al. Biofilm-producing Candida species causing oropharyngeal candidiasis in HIV patients attending Sukraraj Tropical and Infectious Diseases hospital in Kathmandu, Nepal. HIV AIDS 2020, 12, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Neopane, P.; Nepal, H.P.; Shrestha, R.; Uehara, O.; Abiko, Y. In vitro biofilm formation by Staphylococcus aureus isolated from wounds of hospital-admitted patients and their association with antimicrobial resistance. Int. J. Gen. Med. 2018, 11, 25–32. [Google Scholar] [CrossRef]
- Kuinkel, S.; Acharya, J.; Dhungel, B.; Adhikari, S.; Adhikari, N.; Shrestha, U.T.; Banjara, M.R.; Rijal, K.R.; Ghimire, P. Biofilm Formation and Phenotypic Detection of ESBL, MBL, KPC and AmpC Enzymes and Their Coexistence in Klebsiella spp. Isolated at the National Reference Laboratory, Kathmandu, Nepal. Microbiol. Res. 2021, 12, 683–697. [Google Scholar] [CrossRef]
- Stepanovic, S.V.D.; Hola, V.; Bonaventura, G.D.; Djukic, S.; Cirkovic, I.; Ruzicka, F. Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 2007, 115, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing: Twenty Fifth Informational Supplement Edition; Document M100-S29; CLSI: Wayne, NE, USA, 2019. [Google Scholar]
- Magiorakos, A.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Kandel, S.N.; Adhikari, N.; Dhungel, B.; Shrestha, U.T.; Angbuhang, K.B.; Karki, G.; Adhikari, B.; Banjara, M.R.; Rijal, K.R.; Ghimire, P. Characteristics of Staphylococcus aureus isolated from clinical specimens in a tertiary care hospital, Kathmandu, Nepal. Microbiol. Insights 2020, 13, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Smyth, R.; Kahlmeter, G. Mannitol salt agar-cefoxitin combination as a screening medium for methicillin-resistant. J. Clin. Microbiol. 2005, 43, 3797–3799. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sultan, F.B.; Al Meani, A.S. Prevalence of Staphylococcus aureus toxins genes in clinical and food isolates in Iraq. J. Pharm. Sci. Res. 2019, 11, 636–642. [Google Scholar]
- Vatansever, L.; Sezer, C.; Bilge, N. Carriage rate and methicillin resistance of Staphylococcus aureus in food handlers in Kars City, Turkey. Springerplus 2016, 5, 608. [Google Scholar] [CrossRef][Green Version]
- Oliveira, D.; de Lencastre, H. Methicillin-resistance in Staphylococcus aureus is not affected by the overexpression in trans of the mecA gene repressor: A surprising observation. PLoS ONE 2011, 6, e23287. [Google Scholar] [CrossRef]
- Guragin, N.; Pradhan, A.; Dhungel, B.; Banjara, M.R.; Rijal, K.R.; Ghimire, P. Extended spectrum β-lactamase producing Gram Negative bacterial isolates from urine of patients visiting Everest Hospital, Kathmandu, Nepal. Tribhuvan Univ. J. Microbiol. 2019, 6, 26–31. [Google Scholar] [CrossRef]
- Shrestha, L.; Bhattarai, N.R.; Khanal, B. Comparative evaluation of methods for the detection of biofilm formation in coagulase-negative staphylococci and correlation with antibiogram. Infect. Drug Resist. 2018, 11, 607–613. [Google Scholar] [CrossRef]
- Karki, D.; Dhungel, B.; Bhandari, S.; Kunwar, A.; Joshi, P.R.; Shrestha, B.; Rijal, K.R.; Ghimire, P.; Banjara, M.R. Antibiotic resistance and detection of plasmid mediated colistin resistance mcr-1 gene among Escherichia coli and Klebsiella pneumoniae isolated from clinical samples. Gut Pathog. 2021, 13, 45. [Google Scholar] [CrossRef]
- Sah, R.S.P.; Dhungel, B.; Yadav, B.K.; Adhikari, N.; Thapa Shrestha, U.; Lekhak, B.; Banjara, M.R.; Adhikari, B.; Ghimire, P.; Rijal, K.R. Detection of TEM and CTX-M genes in Escherichia coli isolated from clinical specimens at Tertiary Care Heart Hospital, Kathmandu, Nepal. Diseases 2021, 9, 15. [Google Scholar] [CrossRef]
- Azimi, T.; Maham, S.; Fallah, F.; Azimi, L.; Gholinejad, Z. Evaluating the antimicrobial resistance patterns among major bacterial pathogens isolated from clinical specimens taken from patients in Mofid Children’s Hospital, Tehran, Iran: 2013–2018. Infect. Drug Resist. 2019, 12, 2089–2102. [Google Scholar] [CrossRef]
- Baral, R.; Khanal, B.; Acharya, A. Antimicrobial susceptibility patterns of clinical isolates of S. aureus in Eastern Nepal. Health Renaiss. 2011, 9, 78–82. [Google Scholar] [CrossRef]
- Shrestha, P.D.; Rai, S.; Gaihre, S. Prevalence of hospital acquired infection and its preventive practices among health workers in a tertiary care hospital. J. Nepal Health Res. Counc. 2019, 16, 452–456. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.K.; Acharya, J.; Kattel, H.P.; Koirala, J.; Rijal, B.P.; Pokhrel, B.M. Metallo-β-lactamase producing Gram negative bacterial isolates. J. Nepal Health Res. Counc. 2014, 10, 208–213. [Google Scholar]
- Parajuli, N.P.; Acharya, S.P.; Mishra, S.K.; Parajuli, K.; Rijal, B.P.; Pokharel, B.M. High burden of antimicrobial resistance among Gram negative bacteria causing healthcare associated infections in a critical care unit of Nepal. Antimicrob. Resist. Infect. Control 2017, 6, 28–38. [Google Scholar] [CrossRef]
- Shrestha, D.; Sherchan, S.P.; Gurung, K.; Manandhar, S.; Shrestha, B.; Sherchan, S. Prevalence of multidrug resistant extended-spectrum ß-lactamase-producing bacteria from different clinical specimens in Kathmandu Model Hospital, Kathmandu, Nepal. EC Microbiol. 2016, 4, 676–698. [Google Scholar]
- Nazneen, S.; Mukta, K.; Santosh, C.; Borde, A. Bacteriological trends and antibiotic susceptibility patterns of clinical isolates at Government Cancer Hospital, Marathwada. Indian J. Cancer 2016, 4, 583–586. [Google Scholar] [CrossRef] [PubMed]
- Soltani, B.; Heidari, H.; Ebrahim-Saraie, H.S.; Hadi, N.; Mardaneh, J.; Motamedifar, M. Molecular characteristics of multiple and extensive drug-resistant Acinetobacter baumannii isolates obtained from hospitalized patients in Southwestern Iran. Infez. Med. 2018, 26, 67–76. [Google Scholar] [PubMed]
- Alam, M.; Pillai, P.; Kapur, P.; Pillai, K. Resistant patterns of bacteria isolated from bloodstream infections at a university hospital in Delhi. J. Pharm. Bioallied Sci. 2011, 3, 525. [Google Scholar] [CrossRef] [PubMed]
- Upreti, N.; Rayamajhi, B.; Sherchan, S.P.; Choudhari, M.K.; Banjara, M.R. Prevalence of methicillin resistant Staphylococcus aureus, multidrug resistant and extended spectrum β-lactamase producing gram negative bacilli causing wound infections at a tertiary care hospital of Nepal. Antimicrob. Resist. Infect. Control 2018, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Naimi, H.M.; Raseksh, H.; Noori, A.Z.; Bahaduri, M.A. Determination of antimicrobial susceptibility patterns in S. aureus strains recovered from patients at two main health facilities in Kabul, Afghanistan. BMC Infect. Dis. 2017, 17, 737. [Google Scholar] [CrossRef] [PubMed]
- Sit, P.S.; Teh, C.S.J.; Idris, N.; Sam, I.C.; Syed Omar, S.F.; Sulaiman, H.; Thong, K.L.; Kamarulzaman, A.; Ponnampalavanar, S. Prevalence of methicillin-resistant Staphylococcus aureus (MRSA) infection and the molecular characteristics of MRSA bacteraemia over a two-year period in a tertiary teaching hospital in Malaysia. BMC Infect. Dis. 2017, 17, 1–14. [Google Scholar] [CrossRef]
- Siddiqui, T.M.I.; Khan, M.N.; Fatima, S.; Alam, N.; Masood, R.; Saeed, R.; Naqvi, G.R.; Naqvi, T. Prevalence of mecA: Genotyping screening of community acquired-MRSA isolates in Karachi, Pakistan. Pak. J. Pharm. Sci. 2018, 31, 2091–2094. [Google Scholar]
- Garoy, E.Y.; Gebreab, Y.B.; Achila, O.O.; Tekeste, D.G.; Kesete, R.; Ghirmay, R.; Tesfu, T. Methicillin-resistant S. aureus (MRSA): Prevalence and antimicrobial sensitivity pattern among patients. A multicenter study in Asmara, Eritrea. Can. J. Infect. Dis. Med. Microbiol. 2019, 2019, 8321834. [Google Scholar] [CrossRef]
- Hassoun, A.; Linden, P.K.; Friedman, B. Incidence, prevalence, and management of MRSA bacteremia across patient populations—a review of recent developments in MRSA management and treatment. Crit. Care 2017, 21, 211. [Google Scholar] [CrossRef]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- von Wintersdorff, C.J.; Penders, J.; van Niekerk, J.M.; Mills, N.D.; Majumder, S.; van Alphen, L.B.; Savelkoul, P.H.; Wolffs, P.F. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 2016, 7, 173. [Google Scholar] [CrossRef]
- Rijal, K.R.; Banjara, M.R.; Dhungel, B.; Kafle, S.; Gautam, K.; Ghimire, B.; Ghimire, P.; Dhungel, S.; Adhikari, N.; Shrestha, U.T.; et al. Use of antimicrobials and antimicrobial resistance in Nepal: A nationwide survey. Sci. Rep. 2021, 11, 11554. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, B.; Pokharel, S.; Raut, S.; Adhikari, J.; Thapa, S.; Paudel, K.; Narayan, G.C.; Neupane, S.; Neupane, S.R.; Yadav, R.; et al. Why do people purchase antibiotics over-the-counter? A qualitative study with patients, clinicians and dispensers in central, eastern and western Nepal. BMJ Glob. Health 2021, 5, 6. [Google Scholar]
- Pandey, S.; Raza, M.S.; Bhatta, C.P. Prevalence and antibiotic sensitivity pattern of methicillin- resistant- Staphylococcus aureus in Kathmandu Medical College –Teaching Hospital. J. Inst. Med. 2021, 34, 13–17. [Google Scholar] [CrossRef]
- Ansari, S.; Nepal, H.P.; Gautam, R.; Rayamajhi, N.; Shrestha, S.; Upadhyay, G.; Chapagain, M.L. Threat of drug resistant S. aureus to health in Nepal. BMC Infect. Dis. 2014, 14, 10–30. [Google Scholar] [CrossRef]
- Shrestha, J.; Prajapati, K.G.; Panta, O.P.; Poudel, P.; Khanal, S. Methicillin resistant Staphylococcus aureus isolated from wound infections. Tribhuvan Univ. J. Microbiol. 2018, 5, 19–24. [Google Scholar] [CrossRef]
- Piechota, M.K.B.; Frankowska-Maciejewska, A.; Gruzewska, A.; Woźniak-Kosek, A. Biofilm formation by methicillin-resistant and methicillin-sensitive S. aureus strains from hospitalized patients in Poland. BioMed Res. Int. 2018, 2018, 4657396. [Google Scholar] [CrossRef]
- Omidi, M.F.F.; Saffari, M.; Sedaghat, H.; Zibaei, M.; Khaledi, A. Ability of biofilm production and molecular analysis of spa and ica genes among clinical isolates of methicillin-resistant S. aureus. BMC Res. Notes 2020, 13, 19. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, J.; Ji, Y. Environmental factors modulate biofilm formation by Staphylococcus aureus. Sci. Prog. 2020, 103, 36850419898659. [Google Scholar] [CrossRef]
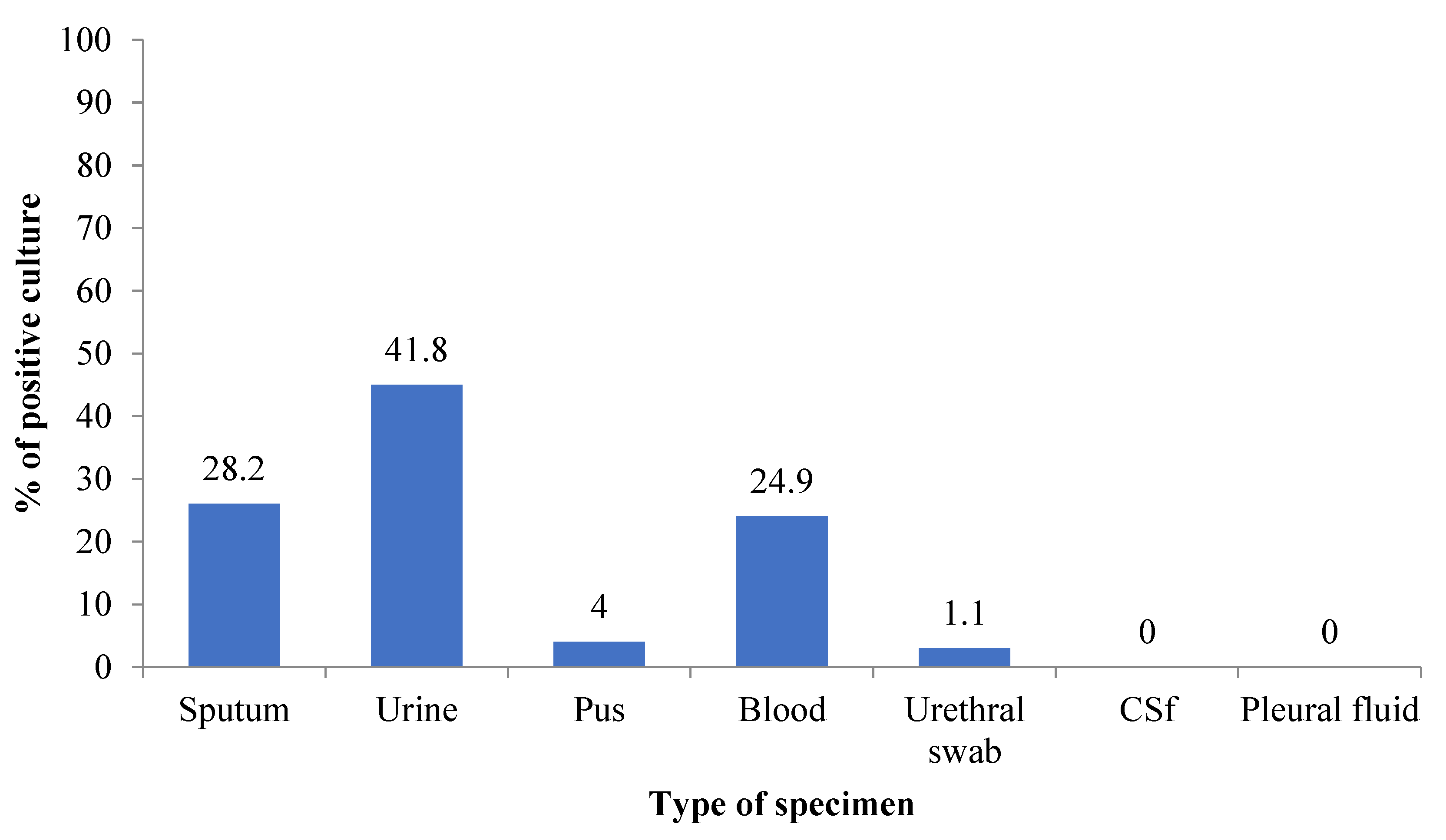
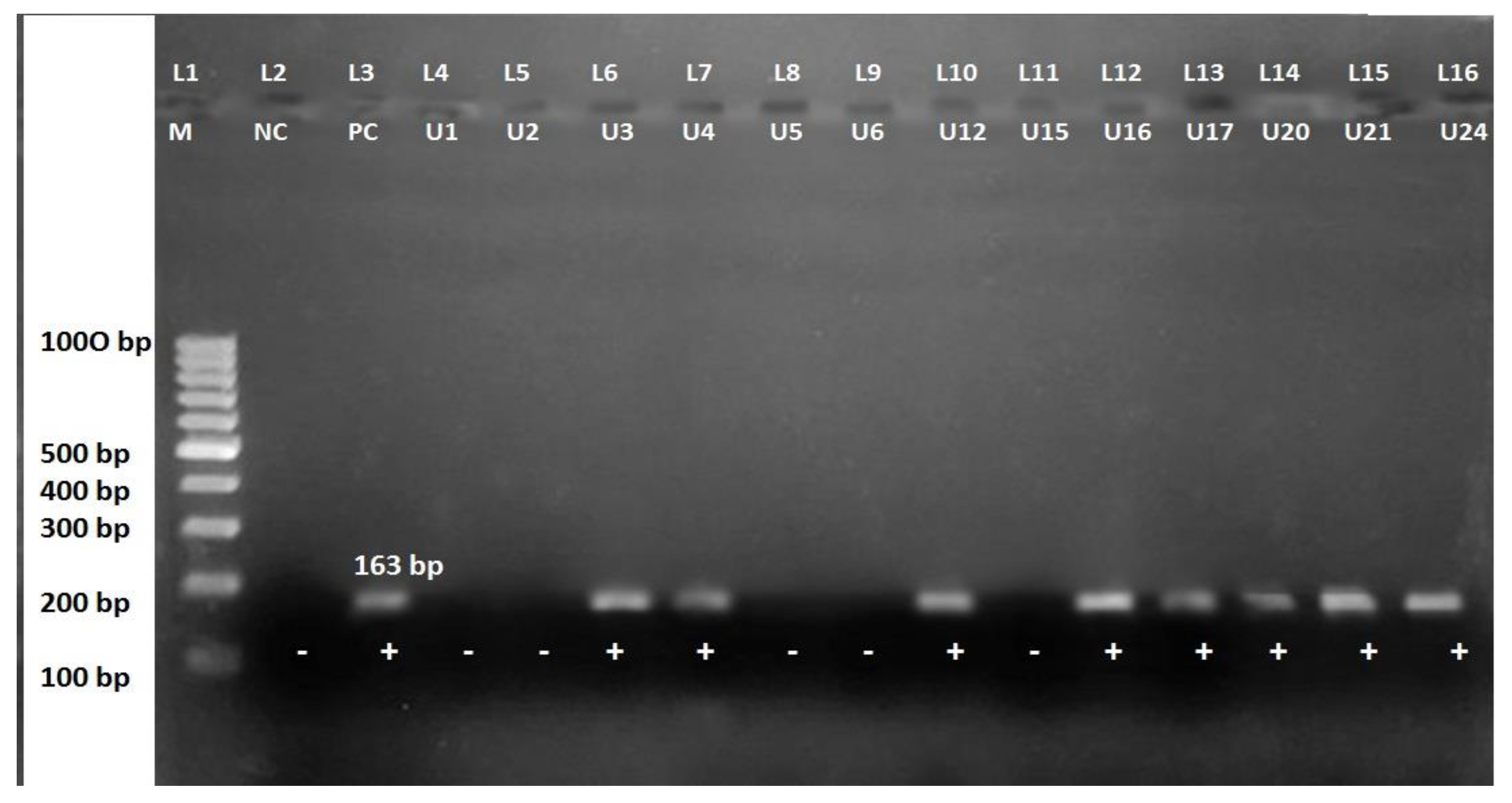
| Bacteria Isolated | Gram-Positive | Gram-Negative | Total (%) | ||
|---|---|---|---|---|---|
| No. | % | No. | % | ||
| S. aureus | 27 | 15.3 | 27 (15.3) | ||
| CoNS | 5 | 2.8 | 5 (2.8) | ||
| S. pneumoniae | 2 | 1.1 | 2 (1.1) | ||
| E. coli | 68 | 38.4 | 68 (38.4) | ||
| P. aeruginosa | 10 | 5.6 | 10 (5.6) | ||
| K. pneumoniae | 15 | 8.5 | 15 (8.5) | ||
| S. Typhi | 13 | 7.3 | 13 (7.3) | ||
| S. Paratyphi | 6 | 3.4 | 6 (3.4) | ||
| Enterobacter spp | 5 | 2.8 | 5 (2.8) | ||
| B. catarrhalis | 9 | 5.1 | 9 (5.1) | ||
| Acinetobacter spp | 3 | 1.7 | 3 (1.7) | ||
| K. oxytoca | 5 | 2.8 | 5 (2.8) | ||
| Providencia spp | 3 | 1.7 | 3 (1.7) | ||
| C. freundii | 1 | 0.6 | 1 (0.6) | ||
| H. influenzae | 1 | 0.6 | 1 (0.6) | ||
| E. faecalis | 1 | 0.6 | 1 (0.6) | ||
| N. gonorrhoeae | 3 | 1.7 | 3 (1.7) | ||
| Total | 34 | 19.2 | 143 | 80.8 | 177 (100) |
| Types of Specimens | MRSA (%) | MSSA (%) | Total (S. aureus) |
|---|---|---|---|
| Sputum (n = 50) | 4 (66.7) | 2 (33.3) | 6 (22.2) |
| Pus (n = 7) | 2 (66.7) | 1 (33.3) | 3 (11.1) |
| Blood (n = 44) | 6 (42.9) | 8 (57.1) | 14 (51.9) |
| Urine (n = 74) | 3 (75) | 1 (33.3) | 4 (14.8) |
| Total | 15 (55.6) | 12 (44.4) | 27 (100) |
| Antibiotics | Type | p-Value | |||
|---|---|---|---|---|---|
| MRSA (n = 15) | MSSA (n = 12) | ||||
| No. of Resistant | % | No. of Resistant | % | ||
| Tetracycline | 1 | 6.6 | 2 | 16.7 | 0.068 |
| Ciprofloxacin | 5 | 33.3 | 5 | 41.6 | |
| Gentamycin | 4 | 26.7 | 2 | 16.7 | |
| Clindamycin | 4 | 26.7 | 1 | 8.3 | |
| Cotrimoxazole | 5 | 33.3 | 2 | 16.7 | |
| Erythromycin | 13 | 86.6 | 8 | 66.6 | |
| Penicillin | 15 | 100.0 | 9 | 75.0 | |
| MDR | 9 | 60.0 | 3 | 25.0 | |
| Type of Biofilm Producer | Type of Strain | Total (%) | |
|---|---|---|---|
| MRSA (%) | MSSA (%) | ||
| Weak biofilm producer | 10 (52.6) | 9 (47.4) | 19 (70.4) |
| Moderate biofilm producer | 5 (71.4) | 2 (28.6) | 7 (25.9) |
| Strong biofilm producer | 0 | 1 (100) | 1 (3.7) |
| Total | 15 | 12 | 27 (100) |
| Antibiotics | Weak Biofilm Producers | Moderate Biofilm Producers | Strong Biofilm Producers | |||
|---|---|---|---|---|---|---|
| Resistant (%) | Sensitive (%) | Resistant (%) | Sensitive (%) | Resistant (%) | Sensitive (%) | |
| Penicillin | 16 (84.2) | 3 (15.8) | 7 (100) | 1 (100) | ||
| Cefoxitin | 9 (47.4) | 10 (52.6) | 6 (85.7) | 1 (14.3) | 1 (100) | |
| Tetracycline | 1 (5.3) | 18 (94.7) | 1 (14.3) | 6 (85.7) | 1 (100) | |
| Ciprofloxacin | 7 (36.8) | 12 (63.2) | 2 (28.6) | 5 (71.4) | 1 (100) | |
| Gentamycin | 4 (21.1) | 15 (78.9) | 1 (14.3) | 6 (85.7) | 1 (100) | |
| Clindamycin | 5 (26.3) | 14 (73.7) | 0 | 7 (100) | 1 (100) | |
| Cotrimoxazole | 3 (15.8) | 16 (84.2) | 3 (42.9) | 4 (57.1) | 1 (100) | |
| Erythromycin | 15 (78.9) | 4 (21.1) | 5 (71.4) | 2 (28.6) | 1 (100) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gaire, U.; Thapa Shrestha, U.; Adhikari, S.; Adhikari, N.; Bastola, A.; Rijal, K.R.; Ghimire, P.; Banjara, M.R. Antibiotic Susceptibility, Biofilm Production, and Detection of mecA Gene among Staphylococcus aureus Isolates from Different Clinical Specimens. Diseases 2021, 9, 80. https://doi.org/10.3390/diseases9040080
Gaire U, Thapa Shrestha U, Adhikari S, Adhikari N, Bastola A, Rijal KR, Ghimire P, Banjara MR. Antibiotic Susceptibility, Biofilm Production, and Detection of mecA Gene among Staphylococcus aureus Isolates from Different Clinical Specimens. Diseases. 2021; 9(4):80. https://doi.org/10.3390/diseases9040080
Chicago/Turabian StyleGaire, Upama, Upendra Thapa Shrestha, Sanjib Adhikari, Nabaraj Adhikari, Anup Bastola, Komal Raj Rijal, Prakash Ghimire, and Megha Raj Banjara. 2021. "Antibiotic Susceptibility, Biofilm Production, and Detection of mecA Gene among Staphylococcus aureus Isolates from Different Clinical Specimens" Diseases 9, no. 4: 80. https://doi.org/10.3390/diseases9040080
APA StyleGaire, U., Thapa Shrestha, U., Adhikari, S., Adhikari, N., Bastola, A., Rijal, K. R., Ghimire, P., & Banjara, M. R. (2021). Antibiotic Susceptibility, Biofilm Production, and Detection of mecA Gene among Staphylococcus aureus Isolates from Different Clinical Specimens. Diseases, 9(4), 80. https://doi.org/10.3390/diseases9040080







