The Microbiome as a Therapeutic Target for Multiple Sclerosis: Can Genetically Engineered Probiotics Treat the Disease?
Abstract
1. Introduction
2. The Crosstalk between the Intestinal Microbiome and the Host
2.1. The Intestinal Microbiome
2.2. Impact of the Intestinal Microbiome on the Immune System
2.3. Impact of the Intestinal Microbiome beyond the Immune System
3. The Intestinal Microbiome and MS
3.1. Experimental Evidence That Associates the Microbiome with MS
3.2. Clinical and Epidemiological Evidence That Associate the Microbiome with MS
4. Interventions of the Intestinal Microbiota as a Treatment of MS
4.1. Experimental Interventions of the Microbiota
4.1.1. Antibiotics
4.1.2. Bacteriophage Therapy
4.1.3. Fecal Microbiota Transplantation (FMT)
4.1.4. Microbial-Derived Products
4.1.5. Probiotics
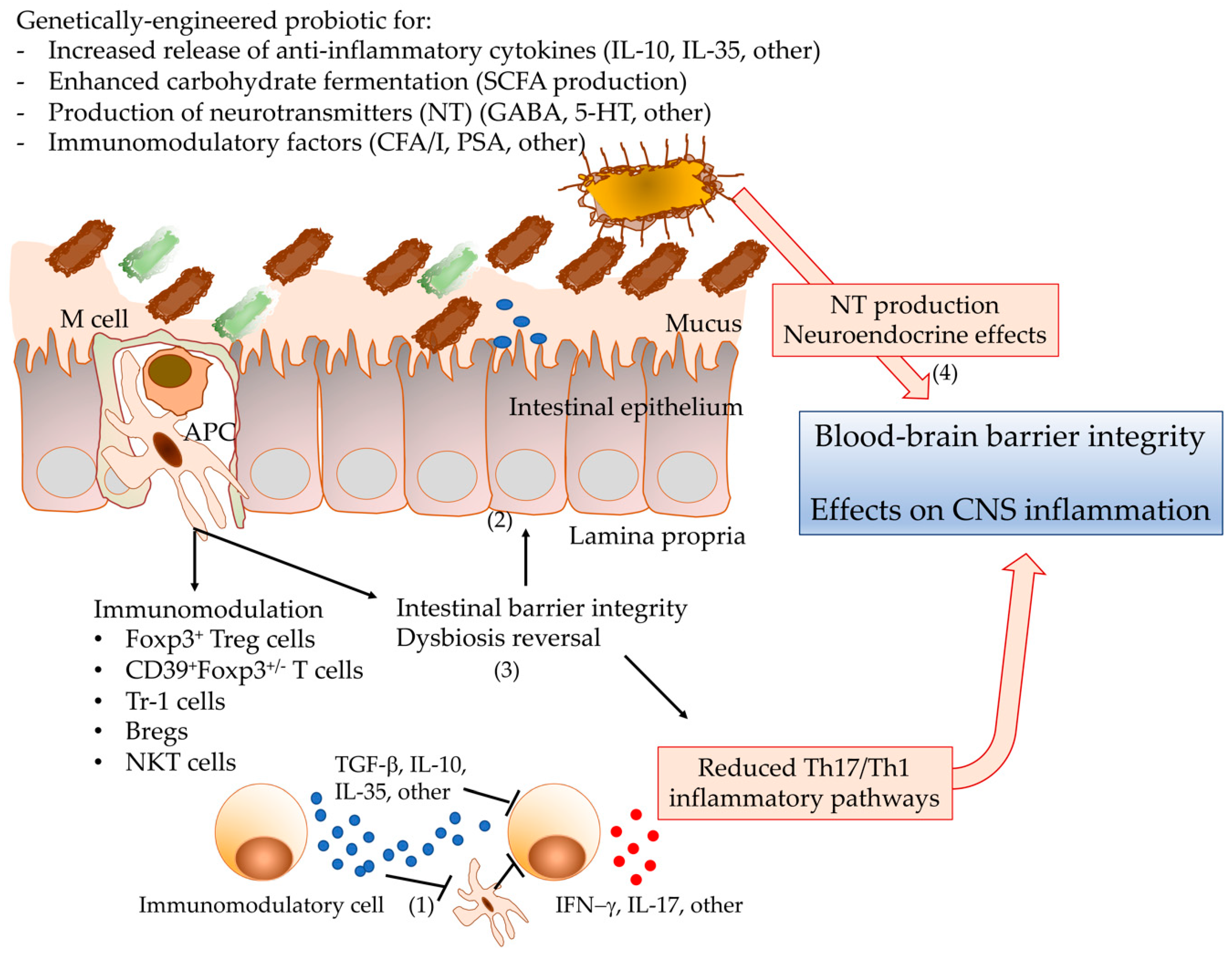
4.2. Genetic Design of Probiotics with Enhanced Protective Phenotype?
4.2.1. Probiotic Strain Choice for Engineering?
4.2.2. Tools for Probiotic Engineering
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Financial Disclosures
References
- Dobson, R.; Giovannoni, G. Multiple sclerosis—A review. Eur. J. Neurol. 2019, 26, 27–40. [Google Scholar] [CrossRef]
- Cross, A.H.; Naismith, R.T. Refining the use of MRI to predict multiple sclerosis. Lancet Neurol. 2018, 17, 105–106. [Google Scholar] [CrossRef]
- Franklin, R.J.M.; Goldman, S.A. Glia Disease and Repair—Remyelination. Cold Spring Harb. Perspect. Biol. 2015, 7, a020594. [Google Scholar] [CrossRef]
- Cawley, N.; Solanky, B.S.; Muhlert, N.; Tur, C.; Edden, R.A.E.; Wheeler-Kingshott, C.A.M.; Miller, D.H.; Thompson, A.J.; Ciccarelli, O. Reduced gamma-aminobutyric acid concentration is associated with physical disability in progressive multiple sclerosis. Brain 2015, 138, 2584–2595. [Google Scholar] [CrossRef]
- De Angelis, F.; Plantone, D.; Chataway, J. Pharmacotherapy in Secondary Progressive Multiple Sclerosis: An Overview. CNS Drugs 2018, 32, 499–526. [Google Scholar] [CrossRef]
- Lucchinetti, C.; Brück, W.; Parisi, J.; Scheithauer, B.; Rodriguez, M.; Lassmann, H. Heterogeneity of multiple sclerosis lesions: Implications for the pathogenesis of demyelination. Ann. Neurol. 2000, 47, 707–717. [Google Scholar] [CrossRef]
- Pierson, E.R.; Goverman, J.M. GM-CSF is not essential for experimental autoimmune encephalomyelitis but promotes brain-targeted disease. JCI Insight 2017, 2, e92362. [Google Scholar] [CrossRef]
- Aram, J.; Francis, A.; Tanasescu, R.; Constantinescu, C.S. Granulocyte-Macrophage Colony-Stimulating Factor as a Therapeutic Target in Multiple Sclerosis. Neurol. Ther. 2019, 8, 45–57. [Google Scholar] [CrossRef]
- Scalfari, A.; Knappertz, V.; Cutter, G.; Goodin, D.S.; Ashton, R.; Ebers, G.C. Mortality in patients with multiple sclerosis. Neurology 2013, 81, 184–192. [Google Scholar] [CrossRef]
- Wilkins, L.W. The prevalence of MS in the United States: A population-based estimate using health claims data. Neurology 2019, 93, 688. [Google Scholar] [CrossRef]
- Ahlgren, C.; Odén, A.; Lycke, J. High nationwide prevalence of multiple sclerosis in Sweden. Mult. Scler. 2011, 17, 901–908. [Google Scholar] [CrossRef] [PubMed]
- Compston, A.; Coles, A. Multiple sclerosis. Lancet 2008, 372, 1502–1517. [Google Scholar] [CrossRef]
- Ventura, R.E.; Iizumi, T.; Battaglia, T.; Liu, M.; Perez-Perez, G.I.; Herbert, J.; Blaser, M.J. Gut microbiome of treatment-naïve MS patients of different ethnicities early in disease course. Sci. Rep. 2019, 9, 16396. [Google Scholar] [CrossRef] [PubMed]
- Rivas-Rodríguez, E.; Amezcua, L. Ethnic Considerations and Multiple Sclerosis Disease Variability in the United States. Neurol. Clin. 2018, 36, 151–162. [Google Scholar] [CrossRef]
- Deschasaux, M.; Bouter, K.E.; Prodan, A.; Levin, E.; Groen, A.K.; Herrema, H.; Tremaroli, V.; Bakker, G.J.; Attaye, I.; Pinto-Sietsma, S.-J.; et al. Depicting the composition of gut microbiota in a population with varied ethnic origins but shared geography. Nat. Med. 2018, 24, 1526–1531. [Google Scholar] [CrossRef]
- Harbo, H.F.; Lie, B.A.; Sawcer, S.; Celius, E.G.; Dai, K.-Z.; Oturai, A.; Hillert, J.; Lorentzen, A.R.; Laaksonen, M.; Myhr, K.-M.; et al. Genes in the HLA class I region may contribute to the HLA class II-associated genetic susceptibility to multiple sclerosis. Tissue Antigens 2004, 63, 237–247. [Google Scholar] [CrossRef]
- International Multiple Sclerosis Genetics Consortium A systems biology approach uncovers cell-specific gene regulatory effects of genetic associations in multiple sclerosis. Nat. Commun. 2019, 10, 2236. [CrossRef]
- Gregory, S.G.; Schmidt, S.; Seth, P.; Oksenberg, J.R.; Hart, J.; Prokop, A.; Caillier, S.J.; Ban, M.; Goris, A.; Barcellos, L.F.; et al. Interleukin 7 receptor alpha chain (IL7R) shows allelic and functional association with multiple sclerosis. Nat. Genet. 2007, 39, 1083–1091. [Google Scholar] [CrossRef]
- Zhang, D.; Wang, L.; Zhang, R.; Li, S. Association of Vitamin D Receptor Gene Polymorphisms and the Risk of Multiple Sclerosis: A Meta Analysis. Arch. Med. Res. 2019, 50, 350–361. [Google Scholar] [CrossRef]
- Joscelyn, J.; Kasper, L.H. Digesting the emerging role for the gut microbiome in central nervous system demyelination. Mult. Scler. 2014, 20, 1553–1559. [Google Scholar] [CrossRef]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.-C.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103. [Google Scholar] [CrossRef] [PubMed]
- Lederberg, J.; Mccray, A. The Scientist: Óme Sweet Ómics—A Genealogical Treasury of Words. Scientist 2001, 15, 8. [Google Scholar]
- Whipps, J.; Lewis, K.; Cooke, R. Mycoparasitism and plant disease control. In Burge, NM (Editor) Fungi in Biological Control Systems; Manchester University Press: Manchester, UK, 1988; pp. 161–187. [Google Scholar]
- Marchesi, J.R.; Ravel, J. The vocabulary of microbiome research: A proposal. Microbiome 2015, 3, 31. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.R.; Pop, M.; DeBoy, R.T.; Eckburg, P.B.; Turnbaugh, P.J.; Samuel, B.S.; Gordon, J.I.; Relman, D.A.; Fraser-Liggett, C.M.; Nelson, K.E. Metagenomic Analysis of the Human Distal Gut Microbiome. Science 2006, 312, 1355–1359. [Google Scholar] [CrossRef] [PubMed]
- Frank, D.N.; St. Amand, A.L.; Feldman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 2007, 104, 13780–13785. [Google Scholar] [CrossRef] [PubMed]
- Faith, J.J.; Guruge, J.L.; Charbonneau, M.; Subramanian, S.; Seedorf, H.; Goodman, A.L.; Clemente, J.C.; Knight, R.; Heath, A.C.; Leibel, R.L.; et al. The long-term stability of the human gut microbiota. Science 2013, 341, 1237439. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Knight, R.; Gordon, J.I. The effect of diet on the human gut microbiome: A metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 2009, 1, 6ra14. [Google Scholar] [CrossRef]
- Aleman, F.D.D.; Valenzano, D.R. Microbiome evolution during host aging. PLoS Pathog. 2019, 15, e1007727. [Google Scholar] [CrossRef]
- de la Cuesta-Zuluaga, J.; Kelley, S.T.; Chen, Y.; Escobar, J.S.; Mueller, N.T.; Ley, R.E.; McDonald, D.; Huang, S.; Swafford, A.D.; Knight, R.; et al. Age- and Sex-Dependent Patterns of Gut Microbial Diversity in Human Adults. mSystems 2019, 4, e00261-19. [Google Scholar] [CrossRef]
- Rausch, P.; Rehman, A.; Kunzel, S.; Hasler, R.; Ott, S.J.; Schreiber, S.; Rosenstiel, P.; Franke, A.; Baines, J.F. Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proc. Natl. Acad. Sci. USA 2011, 108, 19030–19035. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Tang, M.S.; Easton, A.V.; Devlin, J.C.; Chua, L.L.; Cho, I.; Moy, F.M.; Khang, T.F.; Lim, Y.A.L.; Loke, P. Linking the effects of helminth infection, diet and the gut microbiota with human whole-blood signatures. PLoS Pathog. 2019, 15, e1008066. [Google Scholar] [CrossRef] [PubMed]
- Langdon, A.; Crook, N.; Dantas, G. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med. 2016, 8, 39. [Google Scholar] [CrossRef] [PubMed]
- Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [CrossRef] [PubMed]
- Gensollen, T.; Iyer, S.S.; Kasper, D.L.; Blumberg, R.S. How colonization by microbiota in early life shapes the immune system. Science 2016, 352, 539–544. [Google Scholar] [CrossRef]
- Kiyono, H.; McGhee, J.R.; Wannemuehler, M.J.; Michalek, S.M. Lack of oral tolerance in C3H/HeJ mice. J. Exp. Med. 1982, 155, 605–610. [Google Scholar] [CrossRef] [PubMed]
- Wannemuehler, M.J.; Kiyono, H.; Babb, J.L.; Michalek, S.M.; McGhee, J.R. Lipopolysaccharide (LPS) regulation of the immune response: LPS converts germfree mice to sensitivity to oral tolerance induction. J. Immunol. 1982, 129, 959–965. [Google Scholar]
- Sudo, N.; Sawamura, S.; Tanaka, K.; Aiba, Y.; Kubo, C.; Koga, Y. The requirement of intestinal bacterial flora for the development of an IgE production system fully susceptible to oral tolerance induction. J. Immunol. 1997, 159, 1739–1745. [Google Scholar]
- Smythies, L.E.; Sellers, M.; Clements, R.H.; Mosteller-Barnum, M.; Meng, G.; Benjamin, W.H.; Orenstein, J.M.; Smith, P.D. Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bacteriocidal activity. J. Clin. Investig. 2005, 115, 66–75. [Google Scholar] [CrossRef]
- Dasgupta, S.; Erturk-Hasdemir, D.; Ochoa-Reparaz, J.; Reinecker, H.-C.; Kasper, D.L. Plasmacytoid dendritic cells mediate anti-inflammatory responses to a gut commensal molecule via both innate and adaptive mechanisms. Cell Host Microbe 2014, 15, 413–423. [Google Scholar] [CrossRef]
- Mazmanian, S.K.; Liu, C.H.; Tzianabos, A.O.; Kasper, D.L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 2005, 122, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Ni, X.; Sun, R.; Zeng, B.; Wei, H.; Tian, Z.; Wei, H. Commensal Bacteria-Dependent CD8αβ+ T Cells in the Intestinal Epithelium Produce Antimicrobial Peptides. Front. Immunol. 2018, 9, 1065. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, K.; Nishimura, J.; Shima, T.; Umesaki, Y.; Yamamoto, M.; Onoue, M.; Yagita, H.; Ishii, N.; Evans, R.; Honda, K.; et al. ATP drives lamina propria T(H)17 cell differentiation. Nature 2008, 455, 808–812. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, I.I.; Atarashi, K.; Manel, N.; Brodie, E.L.; Shima, T.; Karaoz, U.; Wei, D.; Goldfarb, K.C.; Santee, C.A.; Lynch, S.V.; et al. Induction of Intestinal Th17 Cells by Segmented Filamentous Bacteria. Cell 2009, 139, 485–498. [Google Scholar] [CrossRef]
- Gaboriau-Routhiau, V.; Rakotobe, S.; Lécuyer, E.; Mulder, I.; Lan, A.; Bridonneau, C.; Rochet, V.; Pisi, A.; De Paepe, M.; Brandi, G.; et al. The Key Role of Segmented Filamentous Bacteria in the Coordinated Maturation of Gut Helper T Cell Responses. Immunity 2009, 31, 677–689. [Google Scholar] [CrossRef]
- Cerf-Bensussan, N.; Gaboriau-Routhiau, V. The immune system and the gut microbiota: Friends or foes? Nat. Publ. Group 2010, 10, 724–733. [Google Scholar] [CrossRef]
- Choi, V.M.; Herrou, J.; Hecht, A.L.; Teoh, W.P.; Turner, J.R.; Crosson, S.; Wardenburg, J.B. Activation of Bacteroides fragilis toxin by a novel bacterial protease contributes to anaerobic sepsis in mice. Nat. Med. 2016, 22, 563–567. [Google Scholar] [CrossRef]
- Wu, S.; Lim, K.-C.; Huang, J.; Saidi, R.F.; Sears, C.L. Bacteroides fragilis enterotoxin cleaves the zonula adherens protein, E-cadherin. Proc. Natl. Acad. Sci. USA 1998, 95, 14979–14984. [Google Scholar] [CrossRef]
- Zhao, Y.; Lukiw, W.J. Microbiome-generated amyloid and potential impact on amyloidogenesis in Alzheimer’s disease (AD). J. Nat. Sci. 2015, 1, e138. [Google Scholar]
- Zhao, Y.; Dua, P.; Lukiw, W. Microbial Sources of Amyloid and Relevance to Amyloidogenesis and Alzheimer’s Disease (AD). J. Alzheimers Dis. Parkinsonism 2015, 5, 177. [Google Scholar] [CrossRef]
- Zhan, X.; Stamova, B.; Jin, L.-W.; DeCarli, C.; Phinney, B.; Sharp, F.R. Gram-negative bacterial molecules associate with Alzheimer disease pathology. Neurology 2016, 87, 2324–2332. [Google Scholar] [CrossRef] [PubMed]
- Laman, J.D.; ’t Hart, B.A.; Power, C.; Dziarski, R. Bacterial Peptidoglycan as a Driver of Chronic Brain Inflammation. Trends Mol. Med. 2020, 26, 670–682. [Google Scholar] [CrossRef] [PubMed]
- Moreno, B.; Jukes, J.-P.; Vergara-Irigaray, N.; Errea, O.; Villoslada, P.; Perry, V.H.; Newman, T.A. Systemic inflammation induces axon injury during brain inflammation. Ann. Neurol. 2011, 70, 932–942. [Google Scholar] [CrossRef] [PubMed]
- Schrijver, I.A. Bacterial peptidoglycan and immune reactivity in the central nervous system in multiple sclerosis. Brain 2001, 124, 1544–1554. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Oshima, K.; Suda, W.; Nagano, Y.; Nishikawa, H.; Fukuda, S.; Saito, T.; Narushima, S.; Hase, K.; et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013, 500, 232–236. [Google Scholar] [CrossRef]
- Round, J.L.; Mazmanian, S.K. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. USA 2009, 107, 12204–12209. [Google Scholar] [CrossRef]
- Wang, K.; Dong, H.; Qi, Y.; Pei, Z.; Yi, S.; Yang, X.; Zhao, Y.; Meng, F.; Yu, S.; Zhou, T.; et al. Lactobacillus casei regulates differentiation of Th17/Treg cells to reduce intestinal inflammation in mice. Can. J. Vet. Res. 2017, 81, 122–128. [Google Scholar]
- Mangalam, A.; Shahi, S.K.; Luckey, D.; Karau, M.; Marietta, E.; Luo, N.; Choung, R.S.; Ju, J.; Sompallae, R.; Gibson-Corley, K.; et al. Human Gut-Derived Commensal Bacteria Suppress CNS Inflammatory and Demyelinating Disease. Cell Rep. 2017, 20, 1269–1277. [Google Scholar] [CrossRef]
- Ochoa-Repáraz, J.; Mielcarz, D.W.; Wang, Y.; Begum-Haque, S.; Dasgupta, S.; Kasper, D.L.; Kasper, L.H. A polysaccharide from the human commensal Bacteroides fragilis protects against CNS demyelinating disease. Mucosal. Immunol. 2010, 3, 487–495. [Google Scholar] [CrossRef]
- Wang, Y.; Telesford, K.M.; Ochoa-Reparaz, J.; Haque-Begum, S.; Christy, M.; Kasper, E.J.; Wang, L.; Wu, Y.; Robson, S.C.; Kasper, D.L.; et al. An intestinal commensal symbiosis factor controls neuroinflammation via TLR2-mediated CD39 signalling. Nat. Commun. 2014, 5, 4432. [Google Scholar] [CrossRef]
- Lavasani, S.; Dzhambazov, B.; Nouri, M.; Fåk, F.; Buske, S.; Molin, G.; Thorlacius, H.; Alenfall, J.; Jeppsson, B.; Weström, B. A Novel Probiotic Mixture Exerts a Therapeutic Effect on Experimental Autoimmune Encephalomyelitis Mediated by IL-10 Producing Regulatory T Cells. PLoS ONE 2010, 5, e9009. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Song, L.; Wang, Y.; Liu, C.; Zhang, L.; Zhu, S.; Liu, S.; Duan, L. Beneficial effect of butyrate-producing Lachnospiraceae on stress-induced visceral hypersensitivity in rats. J. Gastroenterol. Hepatol. 2019, 34, 1368–1376. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly-Y, M.; Glickman, J.N.; Garrett, W.S. The Microbial Metabolites, Short-Chain Fatty Acids, Regulate Colonic Treg Cell Homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, S.; Toh, H.; Hase, K.; Oshima, K.; Nakanishi, Y.; Yoshimura, K.; Tobe, T.; Clarke, J.M.; Topping, D.L.; Suzuki, T.; et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 2011, 469, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, S.; Macfarlane, G.T. Regulation of short-chain fatty acid production. Proc. Nutr. Soc. 2003, 62, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Comstock, L.E. Importance of Glycans to the Host-Bacteroides Mutualism in the Mammalian Intestine. Cell Host Microbe 2009, 5, 522–526. [Google Scholar] [CrossRef]
- Warner, B.B. The contribution of the gut microbiome to neurodevelopment and neuropsychiatric disorders. Pediatr. Res. 2019, 85, 216–224. [Google Scholar] [CrossRef]
- Vuong, H.E.; Yano, J.M.; Fung, T.C.; Hsiao, E.Y. The Microbiome and Host Behavior. Annu. Rev. Neurosci. 2017, 40, 21–49. [Google Scholar] [CrossRef]
- Kelly, J.R.; Borre, Y.; O’ Brien, C.; Patterson, E.; El Aidy, S.; Deane, J.; Kennedy, P.J.; Beers, S.; Scott, K.; Moloney, G.; et al. Transferring the blues: Depression-associated gut microbiota induces neurobehavioural changes in the rat. J. Psychiatr. Res. 2016, 82, 109–118. [Google Scholar] [CrossRef]
- Cryan, J.F.; O’Riordan, K.J.; Cowan, C.S.M.; Sandhu, K.V.; Bastiaanssen, T.F.S.; Boehme, M.; Codagnone, M.G.; Cussotto, S.; Fulling, C.; Golubeva, A.V.; et al. The Microbiota-Gut-Brain Axis. Physiol. Rev. 2019, 99, 1877–2013. [Google Scholar] [CrossRef]
- Sundman, E.; Olofsson, P.S. Neural control of the immune system. Adv. Physiol. Educ. 2014, 38, 135–139. [Google Scholar] [CrossRef]
- Breit, S.; Kupferberg, A.; Rogler, G.; Hasler, G. Vagus Nerve as Modulator of the Brain–Gut Axis in Psychiatric and Inflammatory Disorders. Front. Psychiatry 2018, 9, 44. [Google Scholar] [CrossRef] [PubMed]
- Corcoran, C.; Connor, T.J.; O’Keane, V.; Garland, M.R. The Effects of Vagus Nerve Stimulation on Pro- and Anti-Inflammatory Cytokines in Humans: A Preliminary Report. NIM 2005, 12, 307–309. [Google Scholar] [CrossRef] [PubMed]
- De Herdt, V.; Bogaert, S.; Bracke, K.R.; Raedt, R.; De Vos, M.; Vonck, K.; Boon, P. Effects of vagus nerve stimulation on pro- and anti-inflammatory cytokine induction in patients with refractory epilepsy. J. Neuroimmunol. 2009, 214, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Bonaz, B.; Picq, C.; Sinniger, V.; Mayol, J.F.; Clarençon, D. Vagus nerve stimulation: From epilepsy to the cholinergic anti-inflammatory pathway. Neurogastroenterol. Motil. 2013, 25, 208–221. [Google Scholar] [CrossRef] [PubMed]
- Goehler, L.E.; Gaykema, R.P.A.; Hansen, M.K.; Anderson, K.; Maier, S.F.; Watkins, L.R. Vagal immune-to-brain communication: A visceral chemosensory pathway. Auton. Neurosci. 2000, 85, 49–59. [Google Scholar] [CrossRef]
- Niijima, A. The afferent discharges from sensors for interleukin 1 beta in the hepatoportal system in the anesthetized rat. J. Auton. Nerv. Syst. 1996, 61, 287–291. [Google Scholar] [CrossRef]
- Howland, R.H. Vagus Nerve Stimulation. Curr. Behav. Neurosci. Rep. 2014, 1, 64–73. [Google Scholar] [CrossRef]
- Wang, X.; Yang, Z.; Xue, B.; Shi, H. Activation of the Cholinergic Antiinflammatory Pathway Ameliorates Obesity-Induced Inflammation and Insulin Resistance. Endocrinology 2011, 152, 836–846. [Google Scholar] [CrossRef]
- Borovikova, L.V.; Ivanova, S.; Zhang, M.; Yang, H.; Botchkina, G.I.; Watkins, L.R.; Wang, H.; Abumrad, N.; Eaton, J.W.; Tracey, K.J. Vagus nerve stimulation attenuates the systemic inflammatory response to endotoxin. Nature 2000, 405, 458–462. [Google Scholar] [CrossRef]
- Shah, E.; Rezaie, A.; Riddle, M.; Pimentel, M. Psychological disorders in gastrointestinal disease: Epiphenomenon, cause or consequence? Ann. Gastroenterol. 2014, 27, 224–230. [Google Scholar] [PubMed]
- Mu, Q.; Kirby, J.; Reilly, C.M.; Luo, X.M. Leaky Gut as a Danger Signal for Autoimmune Diseases. Front. Immunol. 2017, 8, 598. [Google Scholar] [CrossRef] [PubMed]
- Kozakova, H.; Schwarzer, M.; Tuckova, L.; Srutkova, D.; Czarnowska, E.; Rosiak, I.; Hudcovic, T.; Schabussova, I.; Hermanova, P.; Zakostelska, Z.; et al. Colonization of germ-free mice with a mixture of three lactobacillus strains enhances the integrity of gut mucosa and ameliorates allergic sensitization. Cell. Mol. Immunol. 2015, 1–12. [Google Scholar] [CrossRef]
- Kuethe, J.W.; Armocida, S.M.; Midura, E.F.; Rice, T.C.; Hildeman, D.A.; Healy, D.P.; Caldwell, C.C. Fecal Microbiota Transplant Restores Mucosal Integrity in a Murine Model of Burn Injury. Shock (Augusta Ga.) 2016, 45, 647–652. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Stärkel, P.; Turner, J.R.; Ho, S.B.; Schnabl, B. Dysbiosis-induced intestinal inflammation activates tumor necrosis factor receptor I and mediates alcoholic liver disease in mice. Hepatology 2015, 61, 883–894. [Google Scholar] [CrossRef] [PubMed]
- Al-Sadi, R. Mechanism of cytokine modulation of epithelial tight junction barrier. Front. Biosci. 2009, 14, 2765. [Google Scholar] [CrossRef]
- Telesford, K.M.; Yan, W.; Ochoa-Reparaz, J.; Pant, A.; Kircher, C.; Christy, M.A.; Begum-Haque, S.; Kasper, D.L.; Kasper, L.H. A commensal symbiotic factor derived from Bacteroides fragilis promotes human CD39(+)Foxp3(+) T cells and Treg function. Gut Microbes 2015, 6, 234–242. [Google Scholar] [CrossRef]
- Mazmanian, S.K.; Round, J.L.; Kasper, D.L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 2008, 453, 620–625. [Google Scholar] [CrossRef]
- Johnson, J.L.; Jones, M.B.; Cobb, B.A. Bacterial capsular polysaccharide prevents the onset of asthma through T-cell activation. Glycobiology 2015, 25, 368–375. [Google Scholar] [CrossRef]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; González, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef]
- Obrenovich, M.E.M. Leaky Gut, Leaky Brain? Microorganisms 2018, 6, 107. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Wu, X.; Block, M.L.; Liu, Y.; Breese, G.R.; Hong, J.-S.; Knapp, D.J.; Crews, F.T. Systemic LPS Causes Chronic Neuroinflammation and Progressive Neurodegeneration. Glia 2007, 55, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Valentin-Torres, A.; Savarin, C.; Hinton, D.R.; Phares, T.W.; Bergmann, C.C.; Stohlman, S.A. Sustained TNF production by central nervous system infiltrating macrophages promotes progressive autoimmune encephalomyelitis. J. Neuroinflamm. 2016, 13, 46. [Google Scholar] [CrossRef]
- Hofman, F.M.; Hinton, D.R.; Johnson, K.; Merrill, J.E. Tumor necrosis factor identified in multiple sclerosis brain. J. Exp. Med. 1989, 170, 607–612. [Google Scholar] [CrossRef]
- Braniste, V.; Al-Asmakh, M.; Kowal, C.; Anuar, F.; Abbaspour, A.; Tóth, M.; Korecka, A.; Bakocevic, N.; Ng, L.G.; Kundu, P.; et al. The gut microbiota influences blood-brain barrier permeability in mice. Sci Transl. Med. 2014, 6, 263ra158. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-B.; Wang, P.-Y.; Wang, X.; Wan, Y.-L.; Liu, Y.-C. Butyrate Enhances Intestinal Epithelial Barrier Function via Up-Regulation of Tight Junction Protein Claudin-1 Transcription. Dig. Dis. Sci. 2012, 57, 3126–3135. [Google Scholar] [CrossRef]
- Erny, D.; Hrabě de Angelis, A.L.; Jaitin, D.; Wieghofer, P.; Staszewski, O.; David, E.; Keren-Shaul, H.; Mahlakoiv, T.; Jakobshagen, K.; Buch, T.; et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 2015, 18, 965–977. [Google Scholar] [CrossRef]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 1–8. [Google Scholar] [CrossRef]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; deRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef]
- Lukić, I.; Getselter, D.; Koren, O.; Elliott, E. Role of Tryptophan in Microbiota-Induced Depressive-Like Behavior: Evidence from Tryptophan Depletion Study. Front. Behav. Neurosci. 2019, 13. [Google Scholar] [CrossRef]
- Yano, J.M.; Yu, K.; Donaldson, G.P.; Shastri, G.G.; Ann, P.; Ma, L.; Nagler, C.R.; Ismagilov, R.F.; Mazmanian, S.K.; Hsiao, E.Y. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 2015, 161, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.G.T.; Bottiglieri, T.; Snead, O.C. GABA, γ-hydroxybutyric acid, and neurological disease. Ann. Neurol. 2003, 54, S3–S12. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Miao, K.; Niyaphorn, S.; Qu, X. Production of Gamma-Aminobutyric Acid from Lactic Acid Bacteria: A Systematic Review. Int. J. Mol. Sci. 2020, 21, 995. [Google Scholar] [CrossRef] [PubMed]
- Dhakal, R.; Bajpai, V.K.; Baek, K.-H. Production of gaba (γ-aminobutyric acid) by microorganisms: A review. Braz. J. Microbiol. 2012, 43, 1230–1241. [Google Scholar] [CrossRef] [PubMed]
- Janik, R.; Thomason, L.A.M.; Stanisz, A.M.; Forsythe, P.; Bienenstock, J.; Stanisz, G.J. Magnetic resonance spectroscopy reveals oral Lactobacillus promotion of increases in brain GABA, N-acetyl aspartate and glutamate. NeuroImage 2016, 125, 988–995. [Google Scholar] [CrossRef] [PubMed]
- Bravo, J.A.; Forsythe, P.; Chew, M.V.; Escaravage, E.; Savignac, H.M.; Dinan, T.G.; Bienenstock, J.; Cryan, J.F. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc. Natl. Acad. Sci. USA 2011, 108, 16050–16055. [Google Scholar] [CrossRef]
- Dinan, T.G.; Stilling, R.M.; Stanton, C.; Cryan, J.F. Collective unconscious: How gut microbes shape human behavior. J. Psychiatr. Res. 2015, 63, 1–9. [Google Scholar] [CrossRef]
- O’Mahony, S.M.; Clarke, G.; Borre, Y.E.; Dinan, T.G.; Cryan, J.F. Serotonin, tryptophan metabolism and the brain-gut-microbiome axis. Behav. Brain Res. 2015, 277, 32–48. [Google Scholar] [CrossRef]
- De Stefano, N.; Giorgio, A. GABA: A new imaging biomarker of neurodegeneration in multiple sclerosis? Brain 2015, 138, 2467–2468. [Google Scholar] [CrossRef][Green Version]
- Cao, G.; Edden, R.A.E.; Gao, F.; Li, H.; Gong, T.; Chen, W.; Liu, X.; Wang, G.; Zhao, B. Reduced GABA levels correlate with cognitive impairment in patients with relapsing-remitting multiple sclerosis. Eur. Radiol. 2018, 28, 1140–1148. [Google Scholar] [CrossRef]
- Bhat, R.; Axtell, R.; Mitra, A.; Miranda, M.; Lock, C.; Tsien, R.W.; Steinman, L. Inhibitory role for GABA in autoimmune inflammation. Proc. Natl. Acad. Sci. USA 2010, 107, 2580–2585. [Google Scholar] [CrossRef] [PubMed]
- Bjurstöm, H.; Wang, J.; Ericsson, I.; Bengtsson, M.; Liu, Y.; Kumar-Mendu, S.; Issazadeh-Navikas, S.; Birnir, B. GABA, a natural immunomodulator of T lymphocytes. J. Neuroimmunol. 2008, 205, 44–50. [Google Scholar] [CrossRef]
- Farooqi, N.; Gran, B.; Constantinescu, C.S. Are current disease-modifying therapeutics in multiple sclerosis justified on the basis of studies in experimental autoimmune encephalomyelitis? J. Neurochem. 2010, 115, 829–844. [Google Scholar] [CrossRef] [PubMed]
- Burrows, D.J.; McGown, A.; Jain, S.A.; De Felice, M.; Ramesh, T.M.; Sharrack, B.; Majid, A. Animal models of multiple sclerosis: From rodents to zebrafish. Mult. Scler. 2019, 25, 306–324. [Google Scholar] [CrossRef] [PubMed]
- Hohlfeld, R. Multiple sclerosis: Human model for EAE? Eur. J. Immunol. 2009, 39, 2036–2039. [Google Scholar] [CrossRef]
- Ochoa-Reparaz, J.; Mielcarz, D.W.; Begum-Haque, S.; Kasper, L.H. Gut, bugs, and brain: Role of commensal bacteria in the control of central nervous system disease. Ann. Neurol. 2011, 69, 240–247. [Google Scholar] [CrossRef]
- Yokote, H.; Miyake, S.; Croxford, J.L.; Oki, S.; Mizusawa, H.; Yamamura, T. NKT cell-dependent amelioration of a mouse model of multiple sclerosis by altering gut flora. Am. J. Pathol. 2008, 173, 1714–1723. [Google Scholar] [CrossRef]
- Ochoa-Repáraz, J.; Mielcarz, D.W.; Ditrio, L.E.; Burroughs, A.R.; Foureau, D.M.; Haque-Begum, S.; Kasper, L.H. Role of gut commensal microflora in the development of experimental autoimmune encephalomyelitis. J. Immunol. 2009, 183, 6041–6050. [Google Scholar] [CrossRef]
- Lee, Y.K.; Menezes, J.S.; Umesaki, Y.; Mazmanian, S.K. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4615–4622. [Google Scholar] [CrossRef]
- Berer, K.; Mues, M.; Koutrolos, M.; Rasbi, Z.A.; Boziki, M.; Johner, C.; Wekerle, H.; Krishnamoorthy, G. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature 2011, 479, 538–541. [Google Scholar] [CrossRef]
- Ochoa-Reparaz, J.; Mielcarz, D.W.; Ditrio, L.E.; Burroughs, A.R.; Begum-Haque, S.; Dasgupta, S.; Kasper, D.L.; Kasper, L.H. Central nervous system demyelinating disease protection by the human commensal Bacteroides fragilis depends on polysaccharide A expression. J. Immunol. 2010, 185, 4101–4108. [Google Scholar] [CrossRef] [PubMed]
- Berer, K.; Gerdes, L.A.; Cekanaviciute, E.; Jia, X.; Xiao, L.; Xia, Z.; Liu, C.; Klotz, L.; Stauffer, U.; Baranzini, S.E.; et al. Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc. Natl. Acad. Sci. USA 2017, 114, 10719–10724. [Google Scholar] [CrossRef] [PubMed]
- Cekanaviciute, E.; Yoo, B.B.; Runia, T.F.; Debelius, J.W.; Singh, S.; Nelson, C.A.; Kanner, R.; Bencosme, Y.; Lee, Y.K.; Hauser, S.L.; et al. Gut bacteria from multiple sclerosis patients modulate human t cells and exacerbate symptoms in mouse models. Proc. Natl. Acad. Sci. USA 2017, 114, 10713–10718. [Google Scholar] [CrossRef] [PubMed]
- Shahi, S.K.; Freedman, S.N.; Murra, A.C.; Zarei, K.; Sompallae, R.; Gibson-Corley, K.N.; Karandikar, N.J.; Murray, J.A.; Mangalam, A.K. Prevotella histicola, A Human Gut Commensal, Is as Potent as COPAXONE® in an Animal Model of Multiple Sclerosis. Front. Immunol. 2019, 10, 462. [Google Scholar] [CrossRef]
- Fletcher, J.M.; Lonergan, R.; Costelloe, L.; Kinsella, K.; Moran, B.; O’Farrelly, C.; Tubridy, N.; Mills, K.H.G. CD39+Foxp3+ Regulatory T Cells Suppress Pathogenic Th17 Cells and Are Impaired in Multiple Sclerosis. J. Immunol. 2009, 183, 7602–7610. [Google Scholar] [CrossRef]
- Ciccocioppo, F.; Lanuti, P.; Pierdomenico, L.; Simeone, P.; Bologna, G.; Ercolino, E.; Buttari, F.; Fantozzi, R.; Thomas, A.; Onofrj, M.; et al. The Characterization of Regulatory T-Cell Profiles in Alzheimer’s Disease and Multiple Sclerosis. Sci. Rep. 2019, 9, 8788. [Google Scholar] [CrossRef]
- Thompson, A.J.; Baranzini, S.E.; Geurts, J.; Hemmer, B.; Ciccarelli, O. Multiple sclerosis. Lancet 2018, 391, 1622–1636. [Google Scholar] [CrossRef]
- Mestre, L.; Carrillo-Salinas, F.J.; Mecha, M.; Feliú, A.; Espejo, C.; Álvarez-Cermeño, J.C.; Villar, L.M.; Guaza, C. Manipulation of Gut Microbiota Influences Immune Responses, Axon Preservation, and Motor Disability in a Model of Progressive Multiple Sclerosis. Front. Immunol. 2019, 10, 1374. [Google Scholar] [CrossRef]
- Kap, Y.S.; Bus-Spoor, C.; van Driel, N.; Dubbelaar, M.L.; Grit, C.; Kooistra, S.M.; Fagrouch, Z.C.; Verschoor, E.J.; Bauer, J.; Eggen, B.J.L.; et al. Targeted Diet Modification Reduces Multiple Sclerosis-like Disease in Adult Marmoset Monkeys from an Outbred Colony. J. Immunol. 2018, 201, 3229–3243. [Google Scholar] [CrossRef]
- Colpitts, S.L.; Kasper, E.J.; Keever, A.; Liljenberg, C.; Kirby, T.; Magori, K.; Kasper, L.H.; Ochoa-Reparaz, J. A bidirectional association between the gut microbiota and CNS disease in a biphasic murine model of multiple sclerosis. Gut Microbes 2017, 8, 561–573. [Google Scholar] [CrossRef]
- Ochoa-Repáraz, J.; Kasper, L.H. Gut microbiome and the risk factors in central nervous system autoimmunity. FEBS Lett. 2014, 588, 4214–4222. [Google Scholar] [CrossRef] [PubMed]
- Makkawi, S.; Camara-Lemarroy, C.; Metz, L. Fecal microbiota transplantation associated with 10 years of stability in a patient with SPMS. Neurol. Neuroimmunol. Neuroinflamm. 2018, 5, e459. [Google Scholar] [CrossRef] [PubMed]
- Miyake, S.; Kim, S.; Suda, W.; Oshima, K.; Nakamura, M.; Matsuoka, T.; Chihara, N.; Tomita, A.; Sato, W.; Kim, S.-W.; et al. Dysbiosis in the Gut Microbiota of Patients with Multiple Sclerosis, with a Striking Depletion of Species Belonging to Clostridia XIVa and IV Clusters. PLoS ONE 2015, 10, e0137429. [Google Scholar] [CrossRef] [PubMed]
- Haghi, F.; Goli, E.; Mirzaei, B.; Zeighami, H. The association between fecal enterotoxigenic B. fragilis with colorectal cancer. BMC Cancer 2019, 19, 879. [Google Scholar] [CrossRef] [PubMed]
- Collins, M.D.; Lawson, P.A.; Willems, A.; Cordoba, J.J.; Fernandez-Garayzabal, J.; Garcia, P.; Cai, J.; Hippe, H.; Farrow, J.A. The phylogeny of the genus Clostridium: Proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 1994, 44, 812–826. [Google Scholar] [CrossRef] [PubMed]
- Shen, A. Clostridium difficile Toxins: Mediators of Inflammation. JIN 2012, 4, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chia, N.; Kalari, K.R.; Yao, J.Z.; Novotna, M.; Paz Soldan, M.M.; Luckey, D.H.; Marietta, E.V.; Jeraldo, P.R.; Chen, X.; et al. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci. Rep. 2016, 6, 28484. [Google Scholar] [CrossRef] [PubMed]
- Jangi, S.; Gandhi, R.; Cox, L.M.; Li, N.; von Glehn, F.; Yan, R.; Patel, B.; Mazzola, M.A.; Liu, S.; Glanz, B.L.; et al. Alterations of the human gut microbiome in multiple sclerosis. Nat. Commun. 2016, 7, 12015. [Google Scholar] [CrossRef]
- Hinds, J.P.; Eidelman, B.H.; Wald, A. Prevalence of bowel dysfunction in multiple sclerosis. Gastroenterology 1990, 98, 1538–1542. [Google Scholar] [CrossRef]
- Quigley, E.M.M.; Spiller, R.C. Constipation and the Microbiome: Lumen Versus Mucosa! Gastroenterology 2016, 150, 300–303. [Google Scholar] [CrossRef]
- Ventura, R.E.; Antezana, A.O.; Bacon, T.; Kister, I. Hispanic Americans and African Americans with multiple sclerosis have more severe disease course than Caucasian Americans. Mult. Scler. 2017, 23, 1554–1557. [Google Scholar] [CrossRef] [PubMed]
- Sand, I.K.; Zhu, Y.; Ntranos, A.; Clemente, J.C.; Cekanaviciute, E.; Brandstadter, R.; Crabtree-Hartman, E.; Singh, S.; Bencosme, Y.; Debelius, J.; et al. Disease-modifying therapies alter gut microbial composition in MS. Neurol. Neuroimmunol. Neuroinflamm. 2019, 6, e517. [Google Scholar] [CrossRef]
- Rumah, K.R.; Vartanian, T.K.; Fischetti, V.A. Oral Multiple Sclerosis Drugs Inhibit the In vitro Growth of Epsilon Toxin Producing Gut Bacterium, Clostridium perfringens. Front. Cell. Infect. Microbiol. 2017, 7, 11. [Google Scholar] [CrossRef] [PubMed]
- Cree, B.A.C.; Spencer, C.M.; Varrin-Doyer, M.; Baranzini, S.E.; Zamvil, S.S. Gut microbiome analysis in neuromyelitis optica reveals overabundance of Clostridium perfringens. Ann. Neurol. 2016, 80, 443–447. [Google Scholar] [CrossRef] [PubMed]
- Wagley, S.; Bokori-Brown, M.; Morcrette, H.; Malaspina, A.; D’Arcy, C.; Gnanapavan, S.; Lewis, N.; Popoff, M.R.; Raciborska, D.; Nicholas, R.; et al. Evidence of Clostridium perfringens epsilon toxin associated with multiple sclerosis. Mult. Scler. 2019, 25, 653–660. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.M.; Koskella, B.; Lin, H.C. Phage therapy: An alternative to antibiotics in the age of multi-drug resistance. World, J. Gastrointest. Pharmacol Ther. 2017, 8, 162–173. [Google Scholar] [CrossRef]
- Glenn, J.D.; Mowry, E.M. Emerging Concepts on the Gut Microbiome and Multiple Sclerosis. J. Interferon Cytokine Res. 2016, 36, 347–357. [Google Scholar] [CrossRef]
- Burgess, J.N.; Pant, A.B.; Kasper, L.H.; Colpitts Brass, S. CD4+ T cells from multiple sclerosis patients respond to a commensal-derived antigen. Ann. Clin. Transl. Neurol. 2017, 4, 825–829. [Google Scholar] [CrossRef]
- He, B.; Hoang, T.K.; Tian, X.; Taylor, C.M.; Blanchard, E.; Luo, M.; Bhattacharjee, M.B.; Freeborn, J.; Park, S.; Couturier, J.; et al. Lactobacillus reuteri Reduces the Severity of Experimental Autoimmune Encephalomyelitis in Mice by Modulating Gut Microbiota. Front. Immunol. 2019, 10, 385. [Google Scholar] [CrossRef]
- Yamashita, M.; Ukibe, K.; Matsubara, Y.; Hosoya, T.; Sakai, F.; Kon, S.; Arima, Y.; Murakami, M.; Nakagawa, H.; Miyazaki, T. Lactobacillus helveticus SBT2171 Attenuates Experimental Autoimmune Encephalomyelitis in Mice. Front. Microbiol. 2018, 8, 2596. [Google Scholar] [CrossRef]
- Calvo-Barreiro, L.; Eixarch, H.; Ponce-Alonso, M.; Castillo, M.; Lebrón-Galán, R.; Mestre, L.; Guaza, C.; Clemente, D.; del Campo, R.; Montalban, X.; et al. A Commercial Probiotic Induces Tolerogenic and Reduces Pathogenic Responses in Experimental Autoimmune Encephalomyelitis. Cells 2020, 9, 906. [Google Scholar] [CrossRef]
- Motta, J.-P.; Magne, L.; Descamps, D.; Rolland, C.; Squarzoni-Dale, C.; Rousset, P.; Martin, L.; Cenac, N.; Balloy, V.; Huerre, M.; et al. Modifying the protease, antiprotease pattern by elafin overexpression protects mice from colitis. Gastroenterology 2011, 140, 1272–1282. [Google Scholar] [CrossRef] [PubMed]
- Steidler, L.; Hans, W.; Schotte, L.; Neirynck, S.; Obermeier, F.; Falk, W.; Fiers, W.; Remaut, E. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science 2000, 289, 1352–1355. [Google Scholar] [CrossRef]
- Han, W.; Mercenier, A.; Ait-Belgnaoui, A.; Pavan, S.; Lamine, F.; van Swam, I.I.; Kleerebezem, M.; Salvador-Cartier, C.; Hisbergues, M.; Bueno, L.; et al. Improvement of an experimental colitis in rats by lactic acid bacteria producing superoxide dismutase. Inflamm. Bowel. Dis. 2006, 12, 1044–1052. [Google Scholar] [CrossRef] [PubMed]
- Stordeur, P.; Goldman, M. Interleukin-10 as a regulatory cytokine induced by cellular stress: Molecular aspects. Int. Rev. Immunol. 1998, 16, 501–522. [Google Scholar] [CrossRef] [PubMed]
- Robert, S.; Gysemans, C.; Takiishi, T.; Korf, H.; Spagnuolo, I.; Sebastiani, G.; Huynegem, K.V.; Steidler, L.; Caluwaerts, S.; Demetter, P.; et al. Oral Delivery of Glutamic Acid Decarboxylase (GAD)-65 and IL10 by Lactococcus lactis Reverses Diabetes in Recent-Onset NOD Mice. Diabetes 2014, 63, 2876–2887. [Google Scholar] [CrossRef] [PubMed]
- Carroll, I.M.; Andrus, J.M.; Bruno-Bárcena, J.M.; Klaenhammer, T.R.; Hassan, H.M.; Threadgill, D.S. Anti-inflammatory properties of Lactobacillus gasseri expressing manganese superoxide dismutase using the interleukin 10-deficient mouse model of colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 293, G729–G738. [Google Scholar] [CrossRef]
- Watterlot, L.; Rochat, T.; Sokol, H.; Cherbuy, C.; Bouloufa, I.; Lefèvre, F.; Gratadoux, J.-J.; Honvo-Hueto, E.; Chilmonczyk, S.; Blugeon, S.; et al. Intragastric administration of a superoxide dismutase-producing recombinant Lactobacillus casei BL23 strain attenuates DSS colitis in mice. Int. J. Food Microbiol. 2010, 144, 35–41. [Google Scholar] [CrossRef]
- Maddaloni, M.; Kochetkova, I.; Hoffman, C.; Pascual, D.W. Delivery of IL-35 by Lactococcus lactis Ameliorates Collagen-Induced Arthritis in Mice. Front. Immunol. 2018, 9, 2691. [Google Scholar] [CrossRef]
- Verma, A.; Xu, K.; Du, T.; Zhu, P.; Liang, Z.; Liao, S.; Zhang, J.; Raizada, M.K.; Grant, M.B.; Li, Q. Expression of Human ACE2 in Lactobacillus and Beneficial Effects in Diabetic Retinopathy in Mice. Mol. Ther. Methods Clin. Dev. 2019, 14, 161–170. [Google Scholar] [CrossRef]
- Gomes-Santos, A.C.; de Oliveira, R.P.; Moreira, T.G.; Castro-Junior, A.B.; Horta, B.C.; Lemos, L.; de Almeida, L.A.; Rezende, R.M.; Cara, D.C.; Oliveira, S.C.; et al. Hsp65-Producing Lactococcus lactis Prevents Inflammatory Intestinal Disease in Mice by IL-10- and TLR2-Dependent Pathways. Front. Immunol. 2017, 8, 30. [Google Scholar] [CrossRef] [PubMed]
- Rezende, R.M.; Oliveira, R.P.; Medeiros, S.R.; Gomes-Santos, A.C.; Alves, A.C.; Loli, F.G.; Guimarães, M.A.F.; Amaral, S.S.; da Cunha, A.P.; Weiner, H.L.; et al. Hsp65-producing Lactococcus lactis prevents experimental autoimmune encephalomyelitis in mice by inducing CD4+LAP+ regulatory T cells. J. Autoimmun. 2013, 40, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Motta, J.-P.; Bermúdez-Humarán, L.G.; Deraison, C.; Martin, L.; Rolland, C.; Rousset, P.; Boue, J.; Dietrich, G.; Chapman, K.; Kharrat, P.; et al. Food-Grade Bacteria Expressing Elafin Protect Against Inflammation and Restore Colon Homeostasis. Sci. Transl. Med. 2012, 4, 158ra144. [Google Scholar] [CrossRef] [PubMed]
- Morshedi, M.; Hashemi, R.; Moazzen, S.; Sahebkar, A.; Hosseinifard, E.-S. Immunomodulatory and anti-inflammatory effects of probiotics in multiple sclerosis: A systematic review. J. Neuroinflamm. 2019, 16, 231. [Google Scholar] [CrossRef] [PubMed]
- Gareau, M.G.; Sherman, P.M.; Walker, W.A. Probiotics and the gut microbiota in intestinal health and disease. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 503–514. [Google Scholar] [CrossRef]
- Bermúdez-Humarán, L.G.; Kharrat, P.; Chatel, J.-M.; Langella, P. Lactococci and lactobacilli as mucosal delivery vectors for therapeutic proteins and DNA vaccines. Microb. Cell Fact. 2011, 10, S4. [Google Scholar] [CrossRef] [PubMed]
- Bermúdez-Humarán, L.G.; Aubry, C.; Motta, J.-P.; Deraison, C.; Steidler, L.; Vergnolle, N.; Chatel, J.-M.; Langella, P. Engineering lactococci and lactobacilli for human health. Curr. Opin. Microbiol. 2013, 16, 278–283. [Google Scholar] [CrossRef]
- Guo, T.; Xin, Y.; Zhang, Y.; Gu, X.; Kong, J. A rapid and versatile tool for genomic engineering in Lactococcus lactis. Microb. Cell Fact. 2019, 18, 22. [Google Scholar] [CrossRef]
- Durrer, K.E.; Allen, M.S.; von Herbing, I.H. Genetically engineered probiotic for the treatment of phenylketonuria (PKU); assessment of a novel treatment in vitro and in the PAHenu2 mouse model of PKU. PLoS ONE 2017, 12, e0176286. [Google Scholar] [CrossRef]
- Kimoto, H.; Nomura, M.; Kobayashi, M.; Mizumachi, K.; Okamoto, T. Survival of lactococci during passage through mouse digestive tract. Can. J. Microbiol. 2003, 49, 707–711. [Google Scholar] [CrossRef]
- Pavan, S.; Desreumaux, P.; Mercenier, A. Use of mouse models to evaluate the persistence, safety, and immune modulation capacities of lactic acid bacteria. Clin. Diagn. Lab. Immunol. 2003, 10, 696–701. [Google Scholar] [CrossRef] [PubMed]
- Vesa, T.; Pochart, P.; Marteau, P. Pharmacokinetics of Lactobacillus plantarum NCIMB 8826, Lactobacillus fermentum KLD, and Lactococcus lactis MG 1363 in the human gastrointestinal tract. Aliment. Pharmacol. Ther. 2000, 14, 823–828. [Google Scholar] [CrossRef]
- Seegers, J.F.M.L. Lactobacilli as live vaccine delivery vectors: Progress and prospects. Trends Biotechnol. 2002, 20, 508–515. [Google Scholar] [CrossRef]
- Song, A.A.-L.; In, L.L.A.; Lim, S.H.E.; Rahim, R.A. A review on Lactococcus lactis: From food to factory. Microb. Cell Fact. 2017, 16, 55. [Google Scholar] [CrossRef] [PubMed]
- de Ruyter, P.G.; Kuipers, O.P.; de Vos, W.M. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 1996, 62, 3662–3667. [Google Scholar] [CrossRef] [PubMed]
- Mierau, I.; Kleerebezem, M. 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl. Microbiol. Biotechnol. 2005, 68, 705–717. [Google Scholar] [CrossRef] [PubMed]
- Linares, D.M.; Perez, M.; Ladero, V.; del Rio, B.; Redruello, B.; Martin, M.C.; Fernandez, M.; Alvarez, M.A. An agmatine-inducible system for the expression of recombinant proteins in Enterococcus faecalis. Microb. Cell Fact. 2014, 13, 169. [Google Scholar] [CrossRef]
- Linares, D.M.; Alvarez-Sieiro, P.; del Rio, B.; Ladero, V.; Redruello, B.; Martin, M.C.; Fernandez, M.; Alvarez, M.A. Implementation of the agmatine-controlled expression system for inducible gene expression in Lactococcus lactis. Microb. Cell Fact. 2015, 14, 208. [Google Scholar] [CrossRef]
- Kloosterman, T.G.; van der Kooi-Pol, M.M.; Bijlsma, J.J.E.; Kuipers, O.P. The novel transcriptional regulator SczA mediates protection against Zn 2+ stress by activation of the Zn 2+-resistance gene czcD in Streptococcus pneumoniae. Mol. Microbiol. 2007, 65, 1049–1063. [Google Scholar] [CrossRef]
- Mu, D.; Montalban-Lopez, M.; Masuda, Y.; Kuipers, O.P. Zirex: A Novel Zinc-Regulated Expression System for Lactococcus lactis. Appl. Environ. Microbiol. 2013, 79, 4503–4508. [Google Scholar] [CrossRef]
- Llull, D.; Poquet, I. New Expression System Tightly Controlled by Zinc Availability in Lactococcus lactis. AEM 2004, 70, 5398–5406. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, A.; Jamet, E.; Commissaire, J.; Renault, P.; Langella, P.; Azevedo, V. A xylose-inducible expression system for Lactococcus lactis. FEMS Microbiol. Lett. 2004, 239, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Benbouziane, B.; Ribelles, P.; Aubry, C.; Martin, R.; Kharrat, P.; Riazi, A.; Langella, P.; Bermúdez-Humarán, L.G. Development of a Stress-Inducible Controlled Expression (SICE) system in Lactococcus lactis for the production and delivery of therapeutic molecules at mucosal surfaces. J. Biotechnol. 2013, 168, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Madsen, S.M.; Arnau, J.; Vrang, A.; Givskov, M.; Israelsen, H. Molecular characterization of the pH-inducible and growth phase-dependent promoter P170 of Lactococcus lactis. Mol. Microbiol. 1999, 32, 75–87. [Google Scholar] [CrossRef]
- van der Vossen, J.M.; van der Lelie, D.; Venema, G. Isolation and characterization of Streptococcus cremoris Wg2-specific promoters. Appl. Environ. Microbiol. 1987, 53, 2452–2457. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Liu, F.; Xu, H.; Bai, Y.; Zhang, X.; Saris, P.E.J.; Qiao, M. Isolation of strong constitutive promoters from Lactococcus lactis subsp. lactis N8. FEMS Microbiol. Lett. 2015, 362. [Google Scholar] [CrossRef] [PubMed]
- Jensen, P.R.; Hammer, K. The Sequence of Spacers between the Consensus Sequences Modulates the Strength of Prokaryotic Promoters. Appl. Environ. Microbiol. 1998, 64, 82–87. [Google Scholar] [CrossRef]
- Wells, J. Mucosal vaccination and therapy with genetically modified lactic acid bacteria. Annu. Rev. Food Sci. Technol. 2011, 2, 423–445. [Google Scholar] [CrossRef]
- Bahey-El-Din, M. Lactococcus lactis-based vaccines from laboratory bench to human use: An overview. Vaccine 2012, 30, 685–690. [Google Scholar] [CrossRef]
- Berlec, A.; Malovrh, T.; Zadravec, P.; Steyer, A.; Ravnikar, M.; Sabotič, J.; Poljšak-Prijatelj, M.; Štrukelj, B. Expression of a hepatitis A virus antigen in Lactococcus lactis and Escherichia coli and evaluation of its immunogenicity. Appl. Microbiol. Biotechnol. 2013, 97, 4333–4342. [Google Scholar] [CrossRef]
- van Asseldonk, M.; Rutten, G.; Oteman, M.; Siezen, R.J.; de Vos, W.M.; Simons, G. Cloning of usp45, a gene encoding a secreted protein from Lactococcus lactis subsp. lactis MG1363. Gene 1990, 95, 155–160. [Google Scholar] [CrossRef]
- Poquet, I.; Ehrlich, S.D.; Gruss, A. An Export-Specific Reporter Designed for Gram-Positive Bacteria: Application to Lactococcus lactis. J. Bacteriol. 1998, 180, 1904–1912. [Google Scholar] [CrossRef]
- Ravn, P.; Arnau, J.; Madsen, S.M.; Vrang, A.; Israelsen, H. The development of TnNuc and its use for the isolation of novel secretion signals in Lactococcus lactis. Gene 2000, 242, 347–356. [Google Scholar] [CrossRef]
- Perez-Martinez, G.; Kok, J.; Venema, G.; van Dijl, J.M.; Smith, H.; Bron, S. Protein export elements from Lactococcus lactis. Mol. Gen. Genet. 1992, 234, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Ravn, P.; Arnau, J.; Madsen, S.M.; Vrang, A.; Israelsen, H. Optimization of signal peptide SP310 for heterologous protein production in Lactococcus lactis. Microbiology 2003, 149, 2193–2201. [Google Scholar] [CrossRef]
- Ng, D.T.W.; Sarkar, C.A. Engineering Signal Peptides for Enhanced Protein Secretion from Lactococcus lactis. Appl. Environ. Microbiol. 2013, 79, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Nouaille, S.; Morello, E.; Cortez-Peres, N.; Le Loir, Y.; Commissaire, J.; Gratadoux, J.J.; Poumerol, E.; Gruss, A.; Langella, P. Complementation of the Lactococcus lactis Secretion Machinery with Bacillus subtilis SecDF Improves Secretion of Staphylococcal Nuclease. AEM 2006, 72, 2272–2279. [Google Scholar] [CrossRef] [PubMed]
- Bolotin, A.; Wincker, P.; Mauger, S.; Jaillon, O.; Malarme, K.; Weissenbach, J.; Ehrlich, S.D.; Sorokin, A. The Complete Genome Sequence of the Lactic Acid Bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 2001, 11, 731–753. [Google Scholar] [CrossRef]
- Frees, D.; Savijoki, K.; Varmanen, P.; Ingmer, H. Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, Gram-positive bacteria. Mol. Microbiol. 2007, 63, 1285–1295. [Google Scholar] [CrossRef]
- Cortes-Perez, N.G.; Poquet, I.; Oliveira, M.; Gratadoux, J.J.; Madsen, S.M.; Miyoshi, A.; Corthier, G.; Azevedo, V.; Langella, P.; Bermúdez-Humarán, L.G. Construction and characterization of a Lactococcus lactis strain deficient in intracellular ClpP and extracellular HtrA proteases. Microbiology 2006, 152, 2611–2618. [Google Scholar] [CrossRef]
- Miyoshi, A.; Poquet, I.; Azevedo, V.; Commissaire, J.; Bermudez-Humaran, L.; Domakova, E.; Le Loir, Y.; Oliveira, S.C.; Gruss, A.; Langella, P. Controlled Production of Stable Heterologous Proteins in Lactococcus lactis. AEM 2002, 68, 3141–3146. [Google Scholar] [CrossRef] [PubMed]
- Morello, E.; Nouaille, S.; Cortes-Perez, N.G.; Blugeon, S.; Medina, L.F.C.; Azevedo, V.; Gratadoux, J.J.; Bermúdez-Humarán, L.G.; Le Loir, Y.; Langella, P. Inactivation of the ybdD Gene in Lactococcus lactis Increases the Amounts of Exported Proteins. Appl. Environ. Microbiol. 2012, 78, 7148–7151. [Google Scholar] [CrossRef] [PubMed]
- Morello, E.; Bermúdez-Humarán, L.G.; Llull, D.; Solé, V.; Miraglio, N.; Langella, P.; Poquet, I. Lactococcus lactis, an efficient cell factory for recombinant protein production and secretion. J. Mol. Microbiol. Biotechnol. 2008, 14, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Zadravec, P.; Mavrič, A.; Bogovič Matijašić, B.; Štrukelj, B.; Berlec, A. Engineering BmpA as a carrier for surface display of IgG-binding domain on Lactococcus lactis. Protein Eng. Des. Sel. 2014, 27, 21–27. [Google Scholar] [CrossRef]
- Dieye, Y.; Usai, S.; Clier, F.; Gruss, A.; Piard, J.-C. Design of a Protein-Targeting System for Lactic Acid Bacteria. J. Bacteriol. 2001, 183, 4157–4166. [Google Scholar] [CrossRef]
- Ton-That, H.; Marraffini, L.A.; Schneewind, O. Protein sorting to the cell wall envelope of Gram-positive bacteria. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2004, 1694, 269–278. [Google Scholar] [CrossRef]
- Steen, A.; Buist, G.; Leenhouts, K.J.; El Khattabi, M.; Grijpstra, F.; Zomer, A.L.; Venema, G.; Kuipers, O.P.; Kok, J. Cell wall attachment of a widely distributed peptidoglycan binding domain is hindered by cell wall constituents. J. Biol. Chem. 2003, 278, 23874–23881. [Google Scholar] [CrossRef]
- Platteeuw, C.; van Alen-Boerrigter, I.; van Schalkwijk, S.; de Vos, W.M. Food-grade cloning and expression system for Lactococcus lactis. Appl. Environ. Microbiol. 1996, 62, 1008–1013. [Google Scholar] [CrossRef]
- Plavec, T.V.; Berlec, A. Engineering of lactic acid bacteria for delivery of therapeutic proteins and peptides. Appl. Microbiol. Biotechnol. 2019, 103, 2053–2066. [Google Scholar] [CrossRef]
- Davies, F.L.; Gasson, M.J. Reviews of the progress of dairy science: Genetics of lactic acid bacteria. J. Dairy Res. 1981, 48, 363–376. [Google Scholar] [CrossRef]
- McKay, L.L. Functional properties of plasmids in lactic streptococci. Antonie Leeuwenhoek 1983, 49, 259–274. [Google Scholar] [CrossRef]
- Leenhouts, K.J.; Kok, J.; Venema, G. Campbell-like integration of heterologous plasmid DNA into the chromosome of Lactococcus lactis subsp. lactis. Appl. Environ. Microbiol. 1989, 55, 394–400. [Google Scholar] [CrossRef] [PubMed]
- Leenhouts, K.J.; Kok, J.; Venema, G. Stability of Integrated Plasmids in the Chromosome of Lactococcus lactis. Appl. Environ. Microbiol. 1990, 56, 2726–2735. [Google Scholar] [CrossRef] [PubMed]
- Maguin, E.; Duwat, P.; Hege, T.; Ehrlich, D.; Gruss, A. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 1992, 174, 5633–5638. [Google Scholar] [CrossRef] [PubMed]
- Leenhouts, K.; Venema, G.; Kok, J. A lactococcal pWV01-based integration toolbox for bacteria. Methods Cell Sci. 1998, 20, 35–50. [Google Scholar] [CrossRef]
- van de Guchte, M.; Daly, C.; Fitzgerald, G.F.; Arendt, E.K. Identification of int and attP on the genome of lactococcal bacteriophage Tuc2009 and their use for site-specific plasmid integration in the chromosome of Tuc2009-resistant Lactococcus lactis MG1363. Appl. Environ. Microbiol. 1994, 60, 2324–2329. [Google Scholar] [CrossRef]
- Brøndsted, L.; Hammer, K. Use of the Integration Elements Encoded by the Temperate Lactococcal Bacteriophage TP901-1 To Obtain Chromosomal Single-Copy Transcriptional Fusions in Lactococcus lactis. Appl. Environ. Microbiol. 1999, 65, 752–758. [Google Scholar] [CrossRef]
- Henrich, B.; Klein, J.R.; Weber, B.; Delorme, C.; Renault, P.; Wegmann, U. Food-Grade Delivery System for Controlled Gene Expression in Lactococcus lactis. Appl. Environ. Microbiol. 2002, 68, 5429–5436. [Google Scholar] [CrossRef]
- Pinto, J.P.C.; Zeyniyev, A.; Karsens, H.; Trip, H.; Lolkema, J.S.; Kuipers, O.P.; Kok, J. pSEUDO, a Genetic Integration Standard for Lactococcus lactis. Appl. Environ. Microbiol. 2011, 77, 6687–6690. [Google Scholar] [CrossRef]
- Solem, C.; Defoor, E.; Jensen, P.R.; Martinussen, J. Plasmid pCS1966, a new selection/counterselection tool for lactic acid bacterium strain construction based on the oroP gene, encoding an orotate transporter from Lactococcus lactis. Appl. Environ. Microbiol. 2008, 74, 4772–4775. [Google Scholar] [CrossRef]
- Blancato, V.S.; Magni, C. A chimeric vector for efficient chromosomal modification in Enterococcus faecalis and other lactic acid bacteria. Lett. Appl. Microbiol. 2010, 50, 542–546. [Google Scholar] [CrossRef] [PubMed]
- Martinussen, J.; Hammer, K. Cloning and characterization of upp, a gene encoding uracil phosphoribosyltransferase from Lactococcus lactis. J. Bacteriol 1994, 176, 6457–6463. [Google Scholar] [CrossRef]
- Xin, Y.; Guo, T.; Mu, Y.; Kong, J. Development of a counterselectable seamless mutagenesis system in lactic acid bacteria. Microb. Cell Fact. 2017, 16, 116. [Google Scholar] [CrossRef]
- Selle, K.; Barrangou, R. Harnessing CRISPR-Cas systems for bacterial genome editing. Trends Microbiol. 2015, 23, 225–232. [Google Scholar] [CrossRef] [PubMed]
- van der Els, S.; James, J.K.; Kleerebezem, M.; Bron, P.A. Versatile Cas9-Driven Subpopulation Selection Toolbox for Lactococcus lactis. Appl. Environ. Microbiol. 2018, 84. [Google Scholar] [CrossRef]
- van Pijkeren, J.-P.; Britton, R.A. High efficiency recombineering in lactic acid bacteria. Nucleic Acids Res. 2012, 40, e76. [Google Scholar] [CrossRef] [PubMed]
| Injectable Medications | |||
|---|---|---|---|
| Therapeutic | Target | Most Common Side Effects * | Proposed Mechanism of Action |
| Interferon beta-1a | Inflammation | Headache, flu-like symptoms, injection site pain, inflammation. | Anti-inflammatory effects |
| Interferon beta-1b | Inflammation | Flu-like symptoms, headache, injection site reactions, injection site skin breakdown, low white blood cell count. | |
| Glatiramer acetate | Inflammation | Injection site reactions, flushing, shortness of breath, rash, chest pain. | Immunomodulation |
| Oral Treatments | |||
| Teriflunomide | Inflammation | Headache, hair thinning, diarrhea, nausea, abnormal liver tests. | Controls proliferation of auto-reactive cells. |
| Fingolimod | Inflammation | Headache, flu-like symptoms, diarrhea, back pain, abnormal liver tests, sinusitis, abdominal pain, pain in extremities, cough. It can slow heart down. | Sphingosine 1-phosphate receptor modulator: Blocks lymphocyte egress from lymph nodes. |
| Cladribine | Inflammation | Upper respiratory infection, headache, low white blood cell counts. | Immunosuppressive effects on lymphocytes (T and B cells). |
| Dimethyl fumarate | Inflammation | Flushing, gastrointestinal issues. | Immunomodulatory and antioxidative effects. |
| Intravenous Infusion Treatments | |||
| Alemtuzumab | Inflammation | Rash, headache, fever, nasal congestion, nausea, urinary tract infection, fatigue, insomnia, upper respiratory tract infection, herpes viral infections, hives, itching, thyroid gland disorders, fungal infection, pain in joints, extremities and back, diarrhea, vomiting, flushing. Infusion reactions common. | Humanized anti-CD52 monoclonal antibody—depletes CD52 + lymphocytes. |
| Ocrelizumab | Inflammation | Infusion reactions; increased risk of infections; possible increase in malignancies, including breast cancer. | Humanized anti-CD20 monoclonal antibody: Targets CD20 + B cells |
| Natalizumab | Inflammation | Headache, fatigue, joint pain, chest discomfort, urinary tract infection, lower respiratory tract infection, gastroenteritis, vaginitis, depression, pain in extremity, abdominal discomfort, diarrhea, and rash. It increases Risk of progressive multifocal leukoencephalopathy (PML), a deadly opportunistic viral infection of the brain. | Anti-α4β1-integrin monoclonal antibody. Blocks T cell migration to CNS |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kohl, H.M.; Castillo, A.R.; Ochoa-Repáraz, J. The Microbiome as a Therapeutic Target for Multiple Sclerosis: Can Genetically Engineered Probiotics Treat the Disease? Diseases 2020, 8, 33. https://doi.org/10.3390/diseases8030033
Kohl HM, Castillo AR, Ochoa-Repáraz J. The Microbiome as a Therapeutic Target for Multiple Sclerosis: Can Genetically Engineered Probiotics Treat the Disease? Diseases. 2020; 8(3):33. https://doi.org/10.3390/diseases8030033
Chicago/Turabian StyleKohl, Hannah M., Andrea R. Castillo, and Javier Ochoa-Repáraz. 2020. "The Microbiome as a Therapeutic Target for Multiple Sclerosis: Can Genetically Engineered Probiotics Treat the Disease?" Diseases 8, no. 3: 33. https://doi.org/10.3390/diseases8030033
APA StyleKohl, H. M., Castillo, A. R., & Ochoa-Repáraz, J. (2020). The Microbiome as a Therapeutic Target for Multiple Sclerosis: Can Genetically Engineered Probiotics Treat the Disease? Diseases, 8(3), 33. https://doi.org/10.3390/diseases8030033






