Abstract
Gut bacterial toxins are thought to contribute to the development of colorectal cancer (CRC). This study examines the presence of specific gut bacterial toxin genes in stool samples from individuals with colorectal neoplasia (adenomas and/or CRC). The presence of bacterial genes encoding genotoxic or pro-inflammatory factors (pks, tcpC, gelE, cnf-1, AMmurB, and usp) was established by PCR of stool samples from individuals from mainland US (n = 30; controls = 10, adenoma = 10, CRC = 10) and from Puerto Rico (PR) (n = 33; controls = 13; adenomas = 8; CRC = 12). Logistic regression models and multinomial logistic regression models were used to estimate the magnitude of association. Distinct bacterial gene profiles were observed in each sample cohort. In individuals with CRC, AMmurB was detected more frequently in samples from the US and gelE in samples from PR. In samples from PR, individuals with ≥2 gut bacterial toxin genes in stool had higher odds of having colorectal neoplasia (OR = 11.0, 95%: CI 1.0–637.1): however, no significant association between bacterial genes and colorectal neoplasia was observed in the US cohort. Further analyses are warranted in a larger cohort to validate these preliminary findings, but these encouraging results highlight the importance of developing bacterial markers as tools for CRC diagnosis or risk stratification.
1. Introduction
Sporadic, non-hereditary colorectal cancer (CRC) is a complex and multifactorial disease involving genetic, environmental, and lifestyle risk factors. CRC survival is largely dependent on prevention and early detection [1,2]. Currently, routine CRC screening and removal of adenomas (pre-cancerous lesions) are the primary means for prevention; however, the fact that 61% of CRC patients are diagnosed at more advanced, less treatable stages emphasizes the need for risk-stratified CRC prevention strategies that incorporate the individual’s modifiable and non-modifiable risk factors [2].
One of the most currently studied environmental risk factors associated with CRC is the gut microbiota [3,4]. Compared to healthy subjects, individuals with CRC have been reported to have a distinct gut microbiota composition enriched in gram-negative bacteria [5,6], which may include opportunistic pathogens such as Escherichia and Campylobacter that may harbor toxins that induce DNA damage, genomic instability, inflammation, and aberrant cell signaling and other hallmarks of cancer [7,8,9,10,11]. Accumulating evidence supports the notion that a subset of the gut microbiota can promote CRC development through chronic inflammation and genotoxicity, among other possible pathways [12,13,14,15,16,17,18,19,20]. However, the precise mechanisms by which the gut microbiota exerts its CRC-promoting effects are still not fully understood.
Although numerous studies suggest the involvement of individual gut bacterial species in the etiology of CRC, causality is yet to be established [3,10,14,21,22]. Recent studies have reported that genotoxin-producing E. coli strains are more prevalent in CRC [8,23,24] and that CRC tissues have more mucosa-associated E. coli than seemingly healthy tissues [25]. These findings support the idea that bacteria with genotoxic and/or pro-inflammatory toxins are not only more abundant in CRC, but that they are also in close proximity to the colonic epithelium where they can exert their pro-carcinogenic effects.
In this study, we have assembled a panel of genes that includes six specific, genotoxic and/or pro-inflammatory gut bacterial genes that encode toxins with known pathogenic mechanisms, that may contribute to colorectal carcinogenesis (Table 1). Previously, our group reported a method for the detection of six gut bacterial toxin genes in DNA isolated from clinical stool samples [26]. In the present study, we report the association between the presence of these six gut bacterial toxin genes in stool and colorectal neoplasia using samples from two different geographical locations (mainland US and PR).

Table 1.
A list of six bacterial genes in this study and their known pathogenic mechanism.
2. Materials and Methods
2.1. Stool Sample Collection
Human stool samples were obtained from the Early Detection Research Network (EDRN; https://edrn.nci.nih.gov/) and the Puerto Rico Familial Colorectal Cancer Registry (PURIFICAR; http://purificar.rcm.upr.edu/ index_eng.html). The EDRN, an initiative of the National Cancer Institute (NCI), brings together dozens of institutions to help accrue biospecimens, accelerate the translation of biomarker information into clinical applications, and to evaluate new diagnostic tests for cancer. PURIFICAR is an island-wide registry that recruits healthy individuals and those with colorectal neoplasia. All subjects recruited by PURIFICAR complete the Colon Cancer Family Registry risk factor questionnaire, which collects sociodemographic and clinical information including: medical history, body mass index (BMI), lifestyle, family history of cancer, and demographic information, among others. PURIFICAR was approved by the University of Puerto Rico Institutional Review Board (approval number: A2210207).
The EDRN kindly provided 30 age- and gender-matched stool samples from individuals residing in the mainland US (controls = 10; adenoma = 10; and CRC = 10). Samples from individuals living in PR were obtained through PURIFICAR (n = 33; controls = 13; adenoma = 8; and CRC = 12), an island-wide population-based registry that collects biospecimens (blood, colorectal tissue, and stool) from both cases (individuals with colorectal neoplasia) and controls (healthy individuals without prior history of colorectal neoplasia). Only individuals with pathologic confirmation of adenomas and CRC were included in this study. Individuals diagnosed with Crohn’s disease or ulcerative colitis, that have undergone previous subtotal or total colectomies, or have had antibiotic treatment at any time in the three months previous to recruitment were excluded. All stool samples obtained through PURIFICAR were collected during a six-month period and stored at −80 °C.
2.2. Bacterial DNA Extraction
Bacterial DNA was extracted from human stool samples (200 mg/per sample) using the QIAamp® DNA Stool Mini Kit (QIAGEN) according the manufacturer’s instructions. DNA extractions were performed within a one-month period since stool samples were received at the laboratory. A total of 5 μL of DNA extract was used as template for subsequent PCR analyses. DNA concentrations were determined using a Nanodrop (ThermoFisher Scientific) and total bacterial DNA was quantified using the formula: μg of DNA = A260 × 0.05 × Vol, where A260 is the absorbance at 260 nm and Vol is the total volume of eluted DNA in microliters.
2.3. PCR Profiling of Specific Bacterial Toxin Genes
The detection and quantification of the six gut bacterial toxin genes in our panel was carried out by PCR analyses as previously described [26]. DNA extracts from bacterial isolates known to contain the genes of interest were used as positive controls. The strains used as positive controls for the pks, tcpC, and cnf qPCRs were selected from a collection of E. coli clinical isolates that were part of a nosocomial infection surveillance study [34]. The E. faecalis strain H32, an isolate previously known to contain gelE, was kindly donated by Dr. Luis Ríos-Hernández from University of Puerto Rico-Mayagüez. A. muciniphila genomic DNA was purchased from the American Type Culture Collection.
Gut bacterial toxin gene primer sequences, annealing temperatures, and expected PCR products are summarized in Table 2 and Table 3. Due to the limited amount of stool sample from individuals from the mainland US, only end-point PCR was used to detect the bacterial toxin genes. Briefly, an initial denaturation step of 1 min at 94 °C was performed followed 30 s at 94 °C, 30 s at the corresponding annealing temperature, and 3 min at 68 °C. All reactions were finalized with a final extension step of 10 min at 72 °C. pks, tcpC, gelE, cnf-1, AMmurB, and usp amplicons were sequenced to ensure primer specificity. Samples positive for any of the gut bacterial toxin genes in our panel were further analyzed to by qPCR quantify the corresponding gene copy number.

Table 2.
Gut bacterial toxin gene primer sequences, annealing temperatures, and expected amplicon size for end-point PCR analyses.

Table 3.
Gut bacterial toxin gene primer sequences, annealing temperatures, and expected amplicon sizes for quantitative real-time PCR analyses.
Stool samples from individuals living in PR served as a validation set. The gut bacterial toxin genes in these samples were detected by qPCR. Detection of pks, tcpC, gelE, cnf-1, AMmurB, and usp in stool was performed in triplicate by qPCR analysis using the QuantiTect SYBR Green PCR kit (QIAGEN). Briefly, qPCR reactions required a 15 min incubation at 95 °C, followed by 50 cycles (for quantifying gelE) or 30 cycles (for the other genes in the panel) of 30 s at 94 °C, 30 s at the corresponding annealing temperature, and 3 min at 68 °C. All reactions had a final extension step of 10 min at 72 °C to ensure maximum detection. The presence of a single DNA amplicon at the end of the qPCR cycle was ascertained by a melting curve with a single-phase transition (Figure A1). Two independent qPCR assays were performed independently. Only the samples that tested positive in both independent measurements were counted as true positives (86% of all positives).
Standard curves using DNA extracts from bacterial isolates known to contain the bacterial toxin genes (Figure A2) were generated for the quantification of the gene copy numbers in our panel. All qPCRs reactions were performed as described in the section above. Gene copy numbers were calculated in stool samples for the bacterial toxin genes of interest. To calculate the number of gene copies in each sample, the DNA copy number conversion is determined from a measurement of absorbance at 260 nm, according to the following relationship:
where A260 is the absorbance of the sample at 260 nm and GW is the weight of the entire genome of the organism harboring the gene (5.1 × 10−15 g for E. coli, 3.6 × 10−15 g for E. faecalis, and, 2.7 × 10 × 10−15 g for A. muciniphila). The E. coli genome weight is used to calculate pks, tcpc, cnf-1, and usp gene copy numbers. E. faecalis and A. muciniphila genome weights are used to calculate gelE and AMmurB copy numbers, respectively.
gene copies = A260 × (50 × 10−9 gDNA/μL)/(GW)(1)
2.4. Statistical Analysis
Logistic regression models were used to estimate the magnitude of association (Odds ratio with 95% confidence interval, OR with 95% CI) between colorectal neoplasia (CRC and polyps) and bacterial genes. In addition, multinomial (polytomous) logistic regression models were fitted to estimate the OR’s with 95% CI for CRC (outcome 1) and adenomas (outcome 2) compared with controls. The multinomial logistic regression was used to predict the probabilities of the different possible outcomes (CRC or adenomas) of a categorically distributed dependent variable given a set of independent variables (bacterial genes). All data was analyzed using Stata for Windows release 14.0 (Stata Corporation, College Station, TX, USA).
3. Results
3.1. Detection of Genotoxic or Pro-Inflammatory Bacterial Genes by PCR
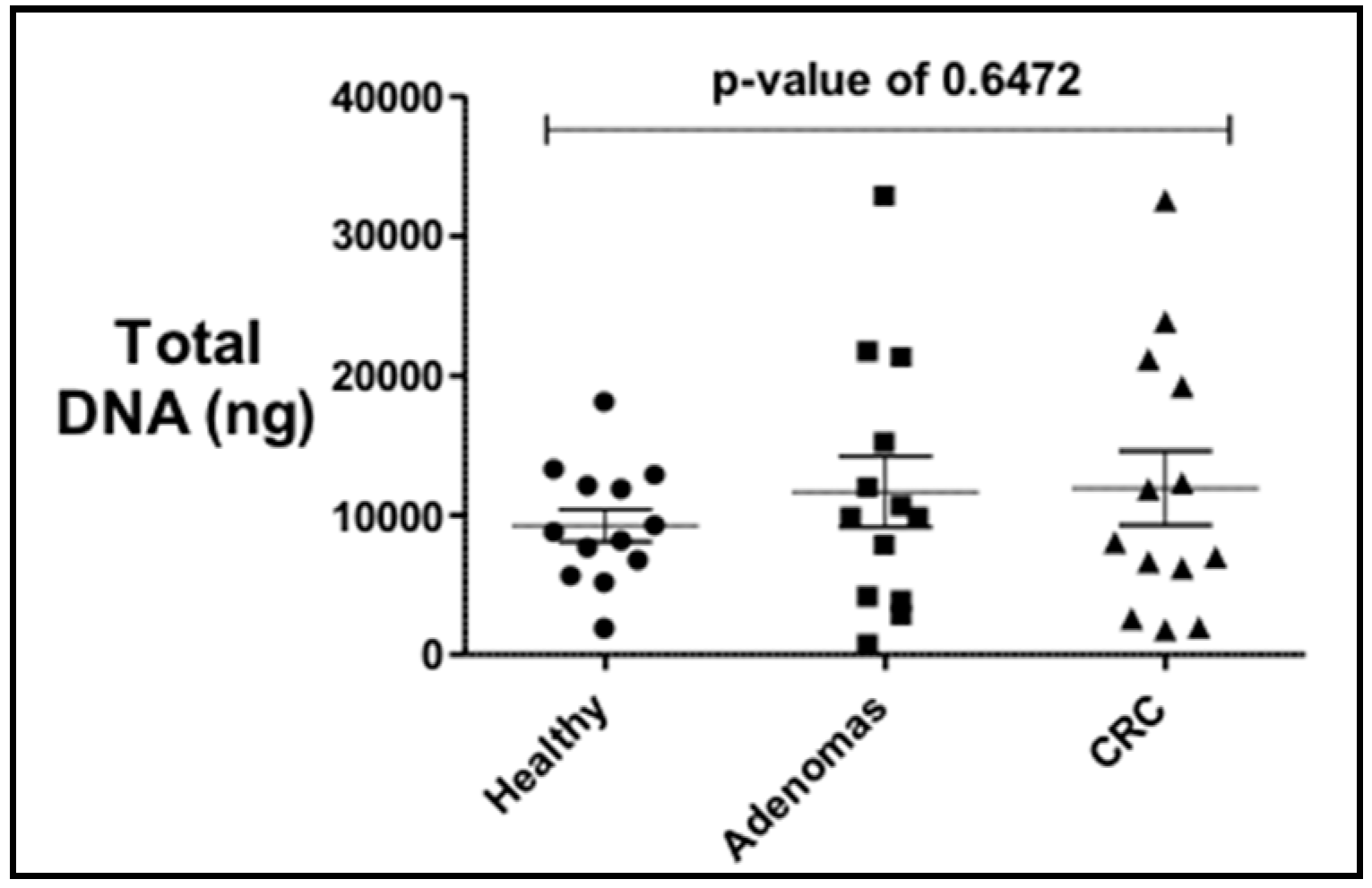
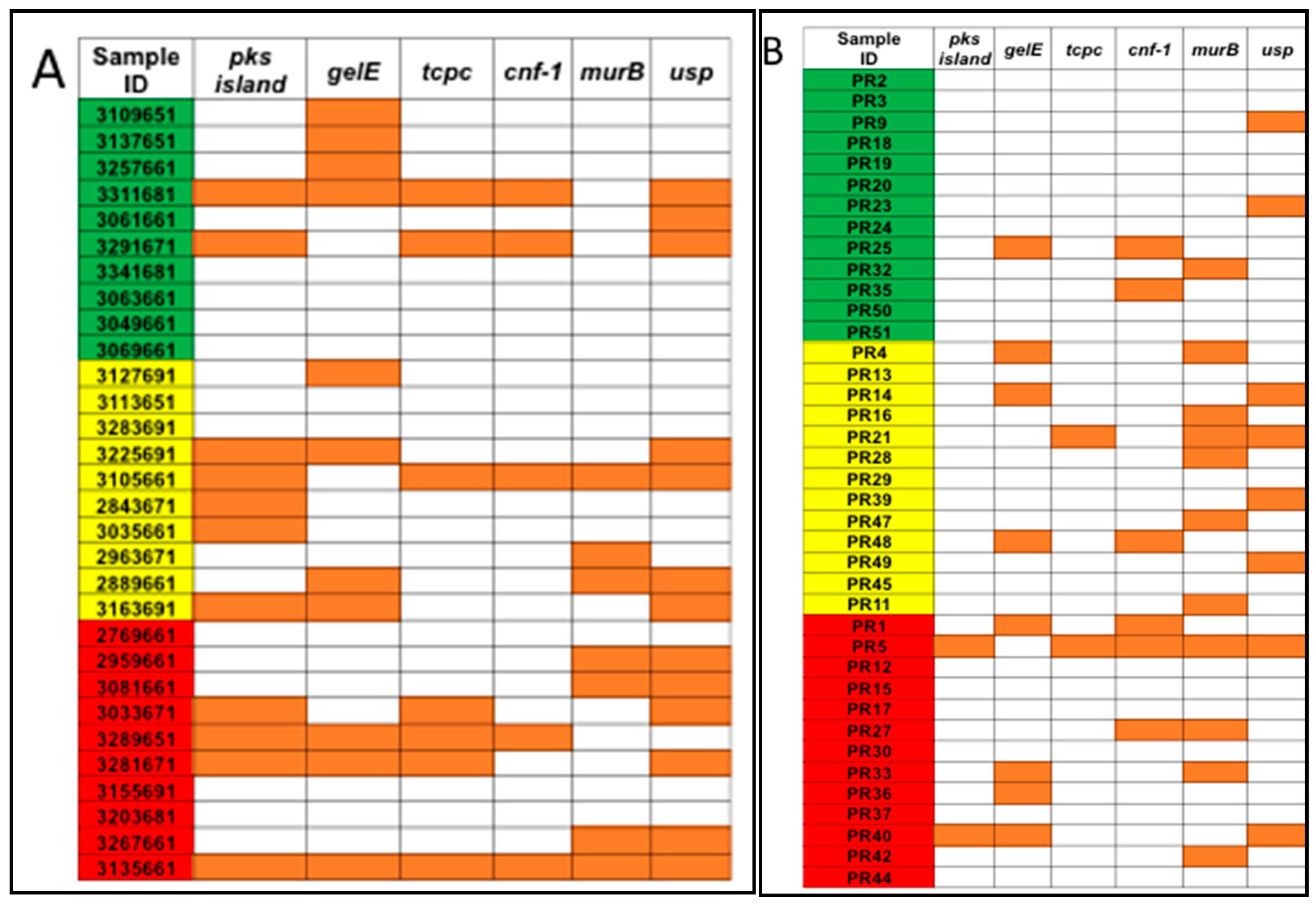
The total bacterial DNA extracted ranged between 0.5 μg and 32 μg per 200 mg per stool sample. No statistical difference in the average total bacterial DNA concentrations between controls (9.2 μg), adenoma (11.6 μg), and CRC groups (11.9 μg) and the stool sample subgroups (US and PR) was observed (Figure 1). PCR analyses of stool samples from both mainland US and PR showed a higher frequency of genotoxic or pro-inflammatory toxin genes detected in samples from individuals with adenoma and CRC compared to controls (Figure 2). Also, differences were observed in the gut bacterial toxin gene profiles between samples from these two subgroups. In samples from the US, all of the gut bacterial toxin genes in our panel, except AMmurB, were detected in samples from healthy controls (Figure 2A). In the healthy group from PR, 4 of 6 of the gut bacterial toxin genes in our panel (pks, tcpC, cnf-1, and AMmurB) were detected (Figure 2B). In samples from PR with adenoma and CRC, AMmurB (50%) was more frequently detected in samples from individuals with adenomas and gelE (42%) in CRC samples. In the US cohort, pks (50%) was more frequently detected in samples from individuals with adenomas and usp (60%) in CRC stool samples.
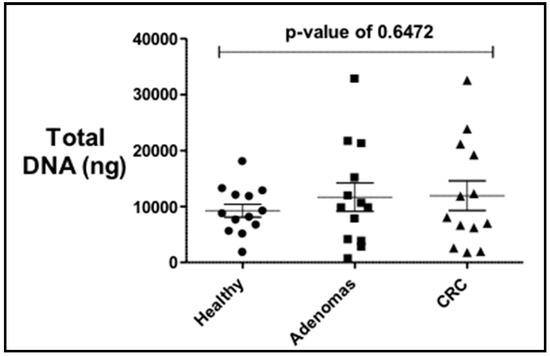
Figure 1.
Total bacterial DNA extracted from stool samples of controls, adenomas and colorectal cancer (CRC) individuals shows no statistical difference by diagnosis. There is a higher variability in the adenomas and CRC.
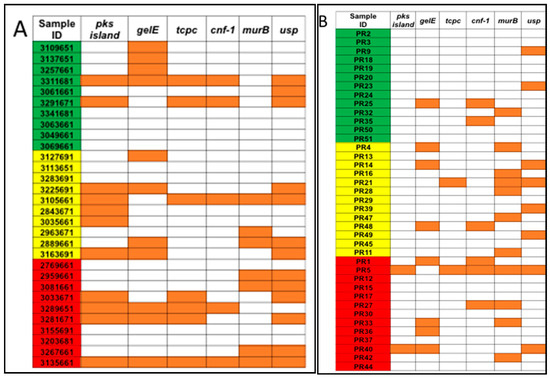
Figure 2.
Pro-inflammatory and/or genotoxic bacterial genes were detected for (A) US samples from the Early Detection Research Network (EDRN) and (B) Puerto Rico samples from the University of Puerto Rico Comprehensive Cancer Center (UPR CCC). Samples were divided by diagnosis as controls (green), adenomas (yellow), and CRC (red). Orange squares represent samples that were positive for the gene.
The presence of genotoxic and/or pro-inflammatory bacterial toxin genes was associated with colorectal neoplasia in both of the study groups (US and PR cohorts); however, the degree of the association and the bacterial toxin genes associated varied between the two populations. Among the US cohort (Table 4), the odds of having CRC among individuals with AMmurB gene in their stool were 14.5-times (OR = 14.5, 95% CI: 0.7–316.7) compared to controls. However, this association was marginally significant (p = 0.09). No statistically significant associations between the presence of the gut bacterial toxin genes and colorectal neoplasia (adenoma and CRC) were observed in stool samples from individuals from the US. In the PR cohort, the presence of gelE in stool was marginally associated with adenomas (OR = 8.6, 95% CI: 0.8–89.04) (Table 5). In addition, the presence of ≥2 bacterial toxin genes in stool was significantly associated with adenomas (OR = 24; 95% CI: 1.11–518.6) and colorectal neoplasia (OR = 11.3; 95% CI: 1.0–637.1) compared to controls.

Table 4.
Odds ratio (OR) estimation for the association between the presence of gut bacterial toxin genes in stool samples from individuals in the U.S. and colorectal neoplasia (n = 30).

Table 5.
Odds ratio (OR) estimation for the association between the presence of gut bacterial toxin genes in stool samples from individuals from Puerto Rico and colorectal neoplasia (n = 33).
3.2. Gene Copy Number Measurements
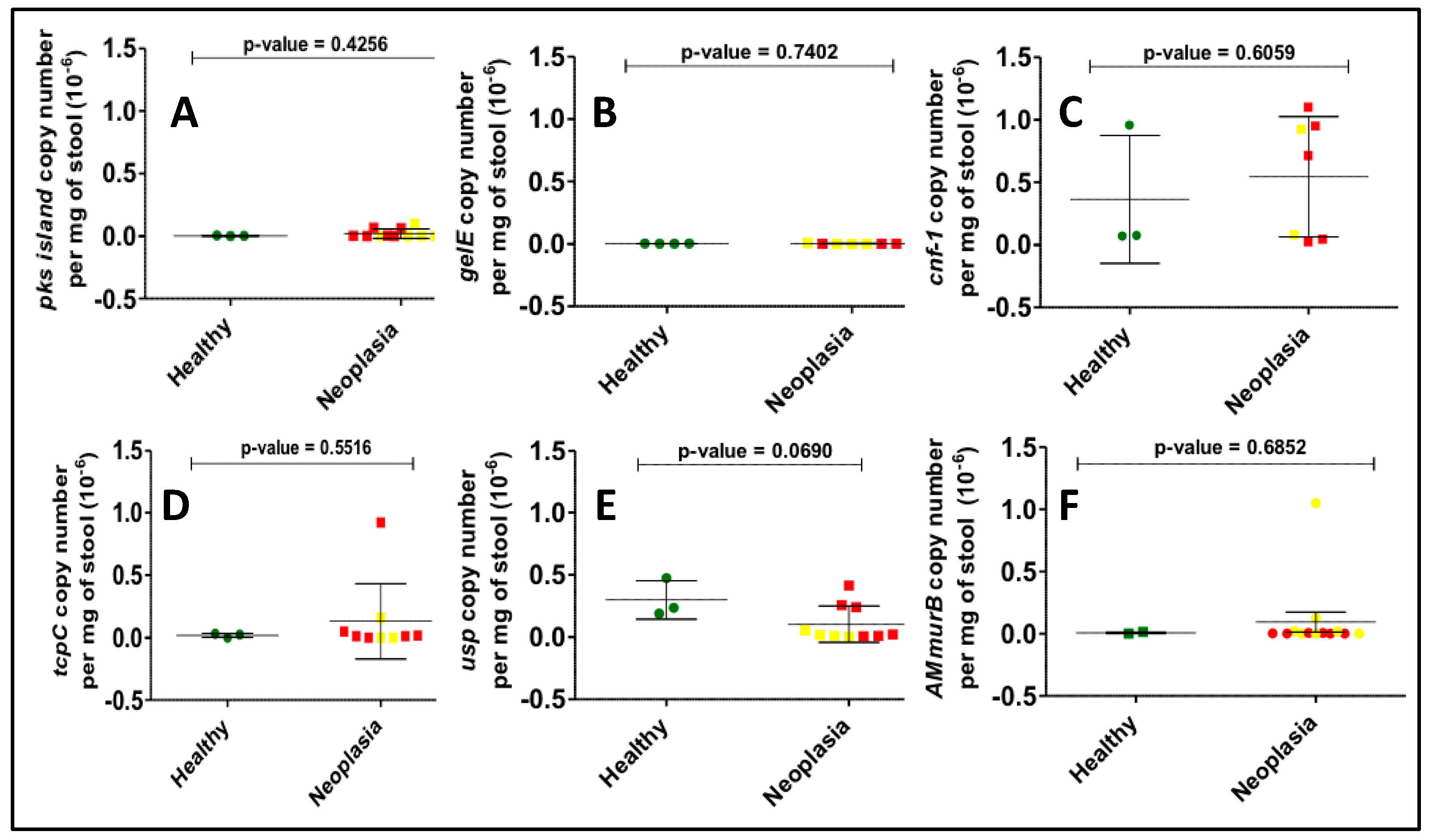
Bacterial toxin gene copy numbers were calculated in all samples positive for the bacterial genes in our panel. Although colorectal neoplasia samples were expected to have a higher bacterial gene copy number compared to the corresponding control samples, no differences were observed between the mean copy number of any of the bacterial genes studied (Figure 3). A higher degree of variability was observed for usp and pks gene copy numbers than for other genes.
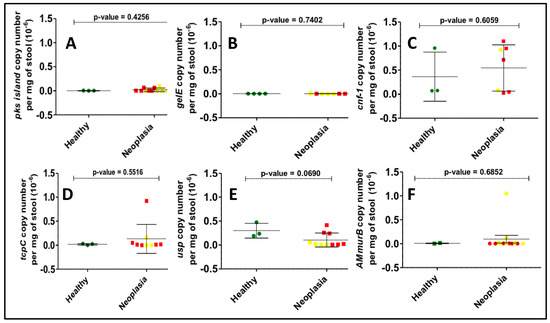
Figure 3.
Determination of gene copy number per mg of stool for all positives among the US samples by real-time quantitative PCR. Here we show the results for (A) pks island, (B) gelE, (C) cnf-1, (D) tcpC, (E) usp, (F) AMmurB showing no statistical differences between the healthy (green dots) and neoplasia groups, which include adenomas (yellow dots) and CRC (red dots) individuals.
4. Discussion
The gut microbiota has emerged as a major contributor to gastrointestinal carcinogenesis [35,36,37]. Despite new insights about the relationships between the gut microbiota and CRC, no methods have been developed that use microbial species or genes as markers for the purposes of screening or risk stratification [38]. In this case-control study, we have examined the association between the presence in stool of a subset of known pro-inflammatory and/or genotoxic bacterial genes and colorectal neoplasia, in samples from two geographically and ethnically distinct populations. This study, even with the limited number of stool samples, revealed a gut bacterial toxin gene profile that was different for each of the two populations (US and PR). Our analyses also revealed that individuals who are positive for multiple bacterial toxin genes have higher odds of developing colorectal neoplasia. Due to the small sample size, most of our observations are not statistically significant, but they reveal a trend that underscores the possibility of incorporating bacterial biomarkers into CRC screening protocols or as tools for risk stratification [38].
In this work, rather than report the detection and prevalence of pro-carcinogenic bacterial species, such as F. nucleatum and B. fragilis, we screened stool samples for the presence of six specific gut bacterial genes that encode toxins that have been previously shown to promote DNA damage and inflammation [26,28,29,30,32]. Two distinct bacterial toxin gene signatures were observed in stool samples from individuals living in the mainland US and PR. The sample size in these two cohorts (n = 30 in US and n = 33 in PR) is too small to draw any conclusions regarding how the geographical differences between these groups may influence the gut microbiome. However, dietary patterns are known to shape and influence the gut microbiota, their metabolism, and functional characteristics [39,40,41]. Studies have shown that the dietary patterns of Puerto Rican adults differ from those in the general US population [42]. In addition, host genetics have been reported to modulate the gut microbial composition [43,44,45] and Puerto Rican Hispanics have been shown to have a unique genomic composition [46], which could possibly contribute to the differences observed in their stool bacterial toxin gene profiles compared to the profiles from individuals from the mainland US. Nonetheless, additional studies with a larger number of samples are needed in order to examine the factors that contribute to the observed differences in the gut microbiota in these two populations.
In the samples from the mainland USA, the gene AMmurB was found to be associated with CRC. Although not encoding a bacterial toxin itself, this gene is a marker for the presence of Akkermansia muciniphila, a mucolytic bacterium whose link to CRC had been previously established [33,47]. It has been thought that the presence of this bacterium in CRC is due to the increased production of the glycoprotein mucin in diseased tissue [33]. This is not surprising since colorectal adenomas have been reported to contain increased levels of certain mucins [48]. Although not reaching statistical significance, the presence of pks island was associated with higher odds (OR = 2.7) of having adenomas or CRC. The pks island encodes a number of enzymes for the production of a genotoxic natural product colibactin, whose chemical structure and mechanism of action have not been elucidated, but its presence had been previously shown to correlate with CRC and tumor formation [8]. We also report the increased presence of tcpC and usp in CRC samples (OR > 2.7). The possible role of TcpC in cancer could be related with its known activity as an antagonist of Toll-like receptor 4, an activity that promotes aberrant tissue inflammation [29]. Finally the presence of usp in the GI tract had been documented, but its main previously known activity was the promotion of cell cycle arrest and DNA damage in the urinary tract [32]. Although there are a number of plausible mechanisms by which these bacterial genes could favor cancer promotion, we believe that the true clinical significance of these preliminary associations will emerge from validation studies with larger cohorts that more fully reflect the geographical and racial diversity of the US.
In the Puerto Rico samples, stronger associations were observed between the presence of the bacterial toxin genes in our panel and increased odds of having adenomas or CRC. As in the US samples, marginally significant associations were detected between the presence of pks, tcpC, or usp, and colorectal neoplasia. Having 2 or more of any bacterial toxin gene in our panel in stool showed a strong significant association with higher odds of having adenomas. The detection of such a strong significant association within this small sample group warrants further investigation in larger number of individuals and supports that this panel may have potential as a CRC risk stratification tool.
Taken together, our results provide a glimpse into the possible feasibility and clinical utility of a CRC risk stratification and/or screening strategies based on the detection of specific bacterial toxin genes in stool. Although our results need to be further validated in a larger number of patient samples and with more diverse patient populations, it is clear that distinct bacterial toxin gene signatures can be detected and quantified, without having to isolate bacterial clones or to reconstruct species composition. Further research is warranted to evaluate the clinical utility of this PCR-based method in CRC risk stratification and/or screening, and to further explore the mechanisms by which these genes could act as functional effectors in colorectal carcinogenesis.
Author Contributions
R.G.-M., M.G.-P., M.C.-C., and A.B.-O. designed the study. M.G.-P., M.C.-C., and A.B.-O. obtained the funding necessary for the study. R.G.-M. and M.G.-P. performed the experiments, and together with M.C.-C. and A.B.-O., MSS performed data analyses and interpretation of the data. All authors contributed in the preparation of tables and figures, and in drafting the manuscript. Some of this work will be published as part of the doctoral thesis of R.G.-M.
Funding
This research was supported by National Cancer Institute (NCI) award number CA198963 and CA096297/CA096300, National Institute of General Medical Sciences award number R25GM061838, and National Institute on Minority Health and Health Disparities (NIMHD) and the National Institute of Allergy and Infectious Diseases (NIAID) award number MD007587. Additional support was provided by NIMHD award Number G12 MD007600 and National Center for Research Resources (NCRR) award number G12RR03051. Study data were managed using REDCap electronic data capture tools hosted at the Puerto Rico Clinical and Translational Research Consortium at the University of Puerto Rico Medical Sciences Campus.
Acknowledgments
The authors thank Luis Ríos-Hernández from UPR Mayagüez for the E. faecalis strains. The authors acknowledge the support of Dean Brenner and Ananda Sen from the University of Michigan and from the Early Detection Research Network (EDRN) and its Great Lakes/New England Clinical Validation Center for providing stool samples from individuals in the mainland US.
Consent of Participants
Informed consent was obtained for each of the human participants and the confidential nature of their participation is protected per IRB protocol A2210207.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A
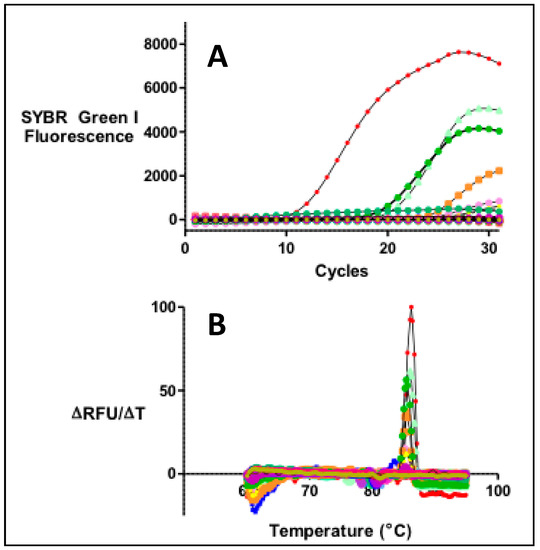
Figure A1.
(A) Real-time PCR reactions were performed for the quantitation of DNA in stool samples compared with a set of standards of known concentration for each of the genes, (B) The presence of a single PCR product was ascertained by monitoring a DNA melting profile immediately after the PCR reaction. The presence of a single peak for each sample is indicative of a single PCR product.
Figure A1.
(A) Real-time PCR reactions were performed for the quantitation of DNA in stool samples compared with a set of standards of known concentration for each of the genes, (B) The presence of a single PCR product was ascertained by monitoring a DNA melting profile immediately after the PCR reaction. The presence of a single peak for each sample is indicative of a single PCR product.
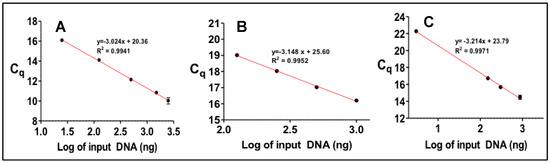
Figure A2.
For the quantitation of DNA by real-time PCR a number of standards of known concentration were analyzed. The results for each gene was expressed as a function of average number of cycles (Cq) vs. known DNA concentration. Results are shown for (A) pks island, (B) usp and (C) cnf-1.
Figure A2.
For the quantitation of DNA by real-time PCR a number of standards of known concentration were analyzed. The results for each gene was expressed as a function of average number of cycles (Cq) vs. known DNA concentration. Results are shown for (A) pks island, (B) usp and (C) cnf-1.
References
- Smith, B.D.; Smith, G.L.; Hurria, A.; Hortobagyi, G.N.; Buchholz, T.A. Future of Cancer Incidence in the United States: Burdens Upon an Aging, Changing Nation. JCO 2009, 27, 2758–2765. [Google Scholar] [CrossRef] [PubMed]
- Colorectal Cancer Facts & Figures 2017–2019; American Cancer Society: Atlanta, GA, USA, 2017.
- Tjalsma, H.; Boleij, A.; Marchesi, J.R.; Dutilh, B.E. A bacterial driver-passenger model for colorectal cancer: Beyond the usual suspects. Nat. Rev. Microbiol. 2012, 10, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Sears, C.L.; Garrett, W.S. Microbes, microbiota, and colon cancer. Cell Host Microbe 2014, 15, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Konstantinov, S.R.; Kuipers, E.J.; Peppelenbosch, M.P. Functional genomic analyses of the gut microbiota for CRC screening. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 741–745. [Google Scholar] [CrossRef] [PubMed]
- Sobhani, I.; Tap, J.; Roudot-Thoraval, F.; Roperch, J.P.; Letulle, S.; Langella, P.; Corthier, G.; Van Nhieu, J.T.; Furet, J.P. Microbial Dysbiosis in Colorectal Cancer (CRC) Patients. PLoS ONE 2011, 6, e16393. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Cai, G.; Qiu, Y.; Fei, N.; Zhang, M.; Pang, X.; Jia, W.; Cai, S.; Zhao, L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2011, 6, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Arthur, J.C.; Perez-Chanona, E.; Mühlbauer, M.; Tomkovich, S.; Uronis, J.M.; Fan, T.; Campbell, B.J.; Abujamel, T.; Dogan, B.; Rogers, A.B.; et al. Intestinal Inflammation Targets Cancer-Inducing Activity of the Microbiota. Science 2012, 338, 120–123. [Google Scholar] [CrossRef]
- Bezine, E.; Vignard, J.; Mirey, G. The Cytolethal Distending Toxin Effects on Mammalian Cells: A DNA Damage Perspective. Cells 2014, 3, 592–615. [Google Scholar] [CrossRef] [PubMed]
- Putze, J.; Hennequin, C.; Nougayrede, J.; Zhang, W.; Homburg, S.; Karch, H.; Bringer, M.; Fayolle, C.; Carniel, E.; Rabsch, W.; et al. Genetic Structure and Distribution of the Colibactin Genomic Island among Members of the Family Enterobacteriaceae. Infect. Immun. 2009, 77, 4696–4703. [Google Scholar] [CrossRef]
- Dejea, C.M.; Fathi, P.; Craig, J.M.; Boleij, A.; Taddese, R.; Geis, A.L.; Wu, X.; DeStefano Shields, C.E.; Hechenbleikner, E.M.; Huso, D.L.; et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 2018, 359, 592. [Google Scholar] [CrossRef]
- Brennan, C.A.; Garrett, W.S. Gut Microbiota, Inflammation, and Colorectal Cancer. Annu. Rev. Microbiol. 2016, 70, 395–411. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kundu, P.; Seow, S.W.; Texeira de Matos, C.; Aronsson, L.; Chin, K.C.; Kärre, K.; Pettersson, S.; Greicius, G. Gut microbiota accelerate tumor growth via c-jun and STAT3 phosphorylation in APC Min/+ mice. Carcinogenesis 2012, 33, 1231–1238. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Rhee, K.; Albesiano, E.; Rabizadeh, S.; Wu, X.; Yen, H.; Huso, D.L.; Brancati, F.L.; Wick, E.; McAllister, F.; et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 2009, 15, 1016–1022. [Google Scholar] [CrossRef] [PubMed]
- Arthur, J.C.; Gharaibeh, R.Z.; Mühlbauer, M.; Perez-Chanona, E.; Uronis, J.M.; McCafferty, J.; Fodor, A.A.; Jobin, C. Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nat. Commun. 2014, 5, 4724. [Google Scholar] [CrossRef] [PubMed]
- Schwabe, R.F.; Jobin, C. The microbiome and cancer. Nat. Rev. Cancer 2013, 13, 800–812. [Google Scholar] [CrossRef] [PubMed]
- Grivennikov, S.I.; Wang, K.; Mucida, D.; Stewart, C.A.; Schnabl, B.; Jauch, D.; Taniguchi, K.; Yu, G.; Osterreicher, C.H.; Hung, K.E.; et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature 2012, 491, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Vannucci, L.; Stepankova, R.; Kozakova, H.; Fiserova, A.; Rossmann, P.; Tlaskalova-Hogenova, H. Colorectal carcinogenesis in germ-free and conventionally reared rats: Different intestinal environments affect the systemic immunity. Int. J. Oncol. 2008, 32, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.Y.; Shaw, M.H.; Redondo, G.; Núñez, G. The Innate Immune Receptor Nod1 Protects the Intestine from Inflammation-Induced Tumorigenesis. Cancer Res. 2008, 68, 10060. [Google Scholar] [CrossRef]
- Dove, W.F.; Clipson, L.; Gould, K.A.; Luongo, C.; Marshall, D.J.; Moser, A.R.; Newton, M.A.; Jacoby, R.F. Intestinal Neoplasia in the ApcMin Mouse: Independence from the Microbial and Natural Killer (beige Locus) Status. Cancer Res. 1997, 57, 812. [Google Scholar] [PubMed]
- Kostic, A.D.; Gevers, D.; Pedamallu, C.S.; Michaud, M.; Duke, F.; Earl, A.M.; Ojesina, A.I.; Jung, J.; Bass, A.J.; Tabernero, J.; et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2011, 22, 292–298. [Google Scholar] [CrossRef]
- Kostic, A.D.; Chun, E.; Robertson, L.; Glickman, J.N.; Gallini, C.A.; Michaud, M.; Clancy, T.E.; Chung, D.C.; Lochhead, P.; Hold, G.L.; et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor immune microenvironment. Cell Host Microbe 2013, 14, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Buc, E.; Dubois, D.; Sauvanet, P.; Raisch, J.; Delmas, J.; Darfeuille-Michaud, A.; Pezet, D.; Bonnet, R. High Prevalence of Mucosa-Associated E. coli Producing Cyclomodulin and Genotoxin in Colon Cancer. PLoS ONE 2013, 8, e56964. [Google Scholar] [CrossRef] [PubMed]
- Prorok-Hamon, M.; Friswell, M.K.; Alswied, A.; Roberts, C.L.; Song, F.; Flanagan, P.K.; Knight, P.; Codling, C.; Marchesi, J.R.; Winstanley, C.; et al. Colonic mucosa-associated diffusely adherent afaC+ Escherichia coli expressing lpfA and pks are increased in inflammatory bowel disease and colon cancer. Gut 2013, 63, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Martin, H.M.; Campbell, B.J.; Hart, C.A.; Mpofu, C.; Nayar, M.; Singh, R.; Englyst, H.; Williams, H.F.; Rhodes, J.M. Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer 1. Gastroenterology 2004, 127, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Moreno, R.; Robledo, I.E.; Baerga-Ortiz, A. Direct Detection and Quantification of Bacterial Genes Associated with Inflammation in DNA Isolated from Stool. Adv. Microbiol. 2014, 4, 1065–1075. [Google Scholar] [CrossRef]
- Nougayrede, J.P.; Homburg, S.; Taieb, F.; Boury, M.; Brzuszkiewicz, E.; Gottschalk, G.; Buchrieser, C.; Hacker, J.; Dobrindt, U.; Oswald, E. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 2006, 313, 848–851. [Google Scholar] [CrossRef] [PubMed]
- Cuevas-Ramos, G.; Petit, C.R.; Marcq, I.; Boury, M.; Oswald, E.; Nougayrède, J. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc. Natl. Acad. Sci. USA 2010, 107, 11537–11542. [Google Scholar] [CrossRef]
- Snyder, G.A.; Cirl, C.; Jiang, J.; Chen, K.; Waldhuber, A.; Smith, P.; Römmler, F.; Snyder, N.; Fresquez, T.; Dürr, S.; et al. Molecular mechanisms for the subversion of MyD88 signaling by TcpC from virulent uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 2013, 110, 6985–6990. [Google Scholar] [CrossRef]
- Sifri, C.D.; Mylonakis, E.; Singh, K.V.; Qin, X.; Garsin, D.A.; Murray, B.E.; Ausubel, F.M.; Calderwood, S.B. Virulence Effect of Enterococcus faecalis Protease Genes and the Quorum-Sensing Locus fsr in Caenorhabditis elegans and Mice. Infect. Immun. 2002, 70, 5647–5650. [Google Scholar] [CrossRef]
- Nougayrède, J.; Taieb, F.; Rycke, J.D.; Oswald, E. Cyclomodulins: Bacterial effectors that modulate the eukaryotic cell cycle. Trends Microbiol. 2005, 13, 103–110. [Google Scholar] [CrossRef]
- Nipic, D.; Podlesek, Z.; Budic, M.; Crnigoj, M.; Zgur-Bertok, D. Escherichia coli uropathogenic-specific protein, Usp, is a bacteriocin-like genotoxin. J. Infect. Dis. 2013, 208, 1545–1552. [Google Scholar] [CrossRef] [PubMed]
- Weir, T.L.; Manter, D.K.; Sheflin, A.M.; Barnett, B.A.; Heuberger, A.L.; Ryan, E.P. Stool Microbiome and Metabolome Differences between Colorectal Cancer Patients and Healthy Adults. PLoS ONE 2013, 8, e70803. [Google Scholar] [CrossRef] [PubMed]
- Robledo, I.E.; Aquino, E.E.; Vazquez, G.J. Detection of the KPC gene in Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii during a PCR-based nosocomial surveillance study in Puerto Rico. Antimicrob. Agents Chemother. 2011, 55, 2968–2970. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Soerjomataram, I.; Ervik, M.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [Internet]; International Agency for Research on Cancer: Lyon, France, 2013. [Google Scholar]
- Guinane, C.M.; Cotter, P.D. Role of the gut microbiota in health and chronic gastrointestinal disease: Understanding a hidden metabolic organ. Ther. Adv. Gastroenterol. 2013, 6, 295–308. [Google Scholar] [CrossRef] [PubMed]
- Plottel, C.S.; Blaser, M.J. Microbiome and Malignancy. Cell Host Microbe 2011, 10, 324–335. [Google Scholar] [CrossRef] [PubMed]
- Stary, L.; Mezerova, K.; Skalicky, P.; Zboril, P.; Raclavsky, V. Are we any closer to screening for colorectal cancer using microbial markers? A critical review. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub. 2017, 161, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.W.; Sanderson, J.D.; Churcher, C.; Parkes, G.C.; Hudspith, B.N.; Rayment, N.; Brostoff, J.; Parkhill, J.; Dougan, G.; Petrovska, L. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011, 11, 7. [Google Scholar] [CrossRef]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking Long-Term Dietary Patterns with Gut Microbial Enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef]
- Duncan, S.H.; Louis, P.; Flint, H.J. Cultivable bacterial diversity from the human colon. Lett. Appl. Microbiol. 2007, 44, 343–350. [Google Scholar] [CrossRef]
- Tucker, K.L.; Bianchi, L.A.; Maras, J.; Bermudez, O.I. Adaptation of a food frequency questionnaire to assess diets of Puerto Rican and non-Hispanic adults. Am. J. Epidemiol. 1998, 148, 507–518. [Google Scholar] [CrossRef]
- Goodrich, J.; Waters, J.; Poole, A.; Sutter, J.; Koren, O.; Blekhman, R.; Beaumont, M.; Van Treuren, W.; Knight, R.; Bell, J.; et al. Human Genetics Shape the Gut Microbiome. Cell 2014, 159, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Blekhman, R.; Goodrich, J.K.; Huang, K.; Sun, Q.; Bukowski, R.; Bell, J.T.; Spector, T.D.; Keinan, A.; Ley, R.E.; Gevers, D.; et al. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 2015, 16, 191. [Google Scholar] [CrossRef] [PubMed]
- Spor, A.; Koren, O.; Ley, R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 2011, 9, 279. [Google Scholar] [CrossRef] [PubMed]
- Via, M.; Gignoux, C.R.; Roth, L.A.; Fejerman, L.; Galanter, J.; Choudhry, S.; Toro-Labrador, G.; Viera-Vera, J.; Oleksyk, T.K.; Beckman, K.; et al. History Shaped the Geographic Distribution of Genomic Admixture on the Island of Puerto Rico. PLoS ONE 2011, 6, e16513. [Google Scholar] [CrossRef] [PubMed]
- Dingemanse, C.; Belzer, C.; van Hijum, S.A.F.T.; Günthel, M.; Salvatori, D.; Dunnen, J.T.D.; Kuijper, E.J.; Devilee, P.; de Vos, W.M.; van Ommen, G.B.; et al. Akkermansia muciniphila and Helicobacter typhlonius modulate intestinal tumor development in mice. Carcinogenesis 2015, 36, 1388–1396. [Google Scholar] [CrossRef] [PubMed]
- Byrd, J.C.; Bresalier, R.S. Mucins and mucin binding proteins in colorectal cancer. Cancer Metastasis Rev. 2004, 23, 77–99. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).