Milk’s Role as an Epigenetic Regulator in Health and Disease
Abstract
:1. Introduction
2. Extracellular Vesicles: Signalosomes for Intercellular Communication
3. Milk Exosomes: Long-Distance Transmitters of Lactation-Specific miRNAs
3.1. Stability of Milk Exosomal miRNAs
3.2. Milk Exosome Uptake
3.3. Milk’s Exosomal miRNAs
4. Epigenetic Regulation of Lactation
5. DNMT-Targeting miRNAs of Milk: Activators of the Recipient’s Epigenome
6. Activation of Developmental Genes via DNA CpG Demethylation
6.1. FTO
6.2. NRF2
6.3. INS
6.4. IGF1
6.5. CAV1
6.6. FOXP3
6.7. NRA4
6.8. NFKBI
6.9. LCT
7. Appetite Control and Feeding Reward
8. Intestinal Growth
9. Adipogenesis
10. Myogenesis
11. Osteogenesis
12. Epidermal Differentiation
13. Milk-Mediated Epigenetic Signaling and Diseases of Civilization
13.1. Obesity
13.2. Type 2 Diabetes Mellitus
13.3. Cancer
13.4. Neurodegenerative Diseases
13.4.1. Alzheimer’s Disease
13.4.2. Parkinson’s Disease
14. Metformin
15. Enhancement of Dairy Milk Yield: A Potential Health Hazard
16. Future Prospects and Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
References
- Godfrey, K.M.; Costello, P.M.; Lillycrop, K.A. Development, epigenetics and metabolic programming. Nestle Nutr. Inst. Workshop Ser. 2016, 85, 71–80. [Google Scholar] [PubMed]
- Landecker, H. Food as exposure: Nutrinonal epigenetics and the new metabolism. Biosocieties 2011, 6, 167–194. [Google Scholar] [CrossRef] [PubMed]
- Vickers, M.H. Early life nutrition, epigenetics and programming of later life disease. Nutrients 2014, 6, 2165–2178. [Google Scholar] [CrossRef] [PubMed]
- Power, M.L.; Schulkin, J. Maternal regulation of offspring development in mammals is an ancient adaptation tied to lactation. Appl. Transl. Genom. 2013, 2, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Ozkan, H.; Tuzun, F.; Kumral, A.; Duman, N. Milk kinship hypothesis in light of epigenetic knowledge. Clin. Epigenet. 2012, 4, 14. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C.; John, S.M.; Schmitz, G. Milk is not just food but most likely a genetic transfection system activating mTORC1 signaling for postnatal growth. Nutr. J. 2013, 12, 103. [Google Scholar] [CrossRef] [PubMed]
- Mathers, J.C.; Strathdee, G.; Relton, C.L. Induction of epigenetic alterations by dietary and other environmental factors. Adv. Genet. 2010, 71, 3–39. [Google Scholar] [PubMed]
- Lillycrop, K.A.; Burdge, G.C. Epigenetic mechanisms linking early nutrition to long term health. Best Pract. Res. Clin. Endocrinol. Metab. 2012, 26, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Koletzko, B.; Chourdakis, M.; Grote, V.; Hellmuth, C.; Prell, C.; Rzehak, P.; Uhl, O.; Weber, M. Regulation of early human growth: Impact on long-term health. Ann. Nutr. Metab. 2014, 65, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Ip, S.; Chung, M.; Raman, G.; Chew, P.; Magula, N.; DeVine, D.; Trikalinos, T.; Lau, J. Breastfeeding and maternal and infant health outcomes in developed countries. Evid. Rep. Technol. Assess. (Full Rep) 2007, 153, 1–186. [Google Scholar]
- Victora, C.G.; Bahl, R.; Barros, A.J.; França, G.V.; Horton, S.; Krasevec, J.; Murch, S.; Sankar, M.J.; Walker, N.; Rollins, N.C.; Lancet Breastfeeding Series Group. Breastfeeding in the 21st century: Epidemiology, mechanisms, and lifelong effect. Lancet 2016, 387, 475–490. [Google Scholar] [CrossRef]
- Kramer, M.S. Breast is best: The evidence. Early Hum. Dev. 2010, 86, 729–732. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C.; John, S.M.; Carrera-Bastos, P.; Schmitz, G. Milk: A postnatal imprinting system stabilizing FoxP3 expression and regulatory T cell differentiation. Clin. Transl. Allergy 2016, 6, 18. [Google Scholar] [CrossRef] [PubMed]
- Alsaweed, M.; Lai, C.T.; Hartmann, P.E.; Geddes, D.T.; Kakulas, F. Human milk miRNAs primarily originate from the mammary gland resulting in unique miRNA profiles of fractionated milk. Sci. Rep. 2016, 6, 20680. [Google Scholar] [CrossRef] [PubMed]
- Bakhshandeh, B.; Kamaleddin, M.A.; Aalishah, K. A Comprehensive review on exosomes and microvesicles as epigenetic factors. Curr. Stem Cell Res. Ther. 2017, 12, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Zempleni, J.; Baier, S.R.; Howard, K.M.; Cui, J. Gene regulation by dietary microRNAs. Can. J. Physiol. Pharmacol. 2015, 93, 1097–1102. [Google Scholar] [CrossRef] [PubMed]
- Alsaweed, M.; Hartmann, P.E.; Geddes, D.T.; Kakulas, F. MicroRNAs in breastmilk and the lactating breast: Potential immunoprotectors and developmental regulators for the infant and the mother. Int. J. Environ. Res. Public Health 2015, 12, 13981–14020. [Google Scholar] [CrossRef] [PubMed]
- Floris, I.; Kraft, J.D.; Altosaar, I. Roles of microRNA across prenatal and postnatal periods. Int. J. Mol. Sci. 2016, 17, E1994. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C.; Kakulas, F.; Geddes, D.T.; Hartmann, P.E.; John, S.M.; Carrera-Bastos, P.; Cordain, L.; Schmitz, G. Milk miRNAs: Simple nutrients or systemic functional regulators? Nutr. Metab. (Lond.) 2016, 13, 42. [Google Scholar] [CrossRef] [PubMed]
- Perge, P.; Nagy, Z.; Decmann, Á.; Igaz, I.; Igaz, P. Potential relevance of microRNAs in inter-species epigenetic communication, and implications for disease pathogenesis. RNA Biol. 2016, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zempleni, J.; Aguilar-Lozano, A.; Sadri, M.; Sukreet, S.; Manca, S.; Wu, D.; Zhou, F.; Mutai, E. Biological activities of extracellular vesicles and their cargos from bovine and human milk in humans and implications for infants. J. Nutr. 2017, 147, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Chargaff, E.; West, R. The biological significance of the thromboplastic protein of blood. J. Biol. Chem. 1946, 166, 189–197. [Google Scholar] [PubMed]
- Wolf, P. The nature and significance of platelet products in human plasma. Br. J. Haematol. 1967, 13, 269–288. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.T.; Johnstone, R.M. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: Selective externalization of the receptor. Cell 1983, 33, 967–978. [Google Scholar] [CrossRef]
- Harding, C.; Heuser, J.; Stahl, P. Endocytosis and intracellular processing of transferrin and colloidal gold-transferrin in rat reticulocytes: Demonstration of a pathway for receptor shedding. Eur. J. Cell. Biol. 1984, 35, 256–263. [Google Scholar] [PubMed]
- Johnstone, R.M.; Adam, M.; Hammond, J.R.; Orr, L.; Turbide, C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 1987, 262, 9412–9420. [Google Scholar] [PubMed]
- Yáñez-Mó, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef] [PubMed]
- Van Niel, G.; Porto-Carreiro, I.; Simoes, S.; Raposo, G. Exosomes: A common pathway for a specialized function. J. Biochem. 2006, 140, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Fritz, J.V.; Heintz-Buschart, A.; Ghosal, A.; Wampach, L.; Etheridge, A.; Galas, D.; Wilmes, P. Sources and functions of extracellular small RNAs in human circulation. Annu. Rev. Nutr. 2016, 36, 301–336. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Peng, P.; Shen, K. Role of exosome shuttle RNA in cell-to-cell communication. Zhongguo Yi Xue Ke Xue Yuan Xue Bao 2016, 38, 480–483. [Google Scholar] [PubMed]
- Ratajczak, J.; Miekus, K.; Kucia, M.; Zhang, J.; Reca, R.; Dvorak, P.; Ratajczak, M.Z. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: Evidence for horizontal transfer of mRNA and protein delivery. Leukemia 2006, 20, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Turturici, G.; Tinnirello, R.; Sconzo, G.; Geraci, F. Extracellular membrane vesicles as a mechanism of cell-to-cell communication: Advantages and disadvantages. Am. J. Physiol. Cell Physiol. 2014, 306, C621–C633. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, M.; Fatima, F.; Vallabhaneni, K.C.; Penfornis, P.; Valadi, H.; Ekström, K.; Kholia, S.; Whitt, J.D.; Fernandes, J.D.; Pochampally, R.; et al. Extracellular Vesicles: Evolving factors in stem cell biology. Stem Cells Int. 2016, 2016, 1073140. [Google Scholar] [CrossRef] [PubMed]
- Admyre, C.; Johansson, S.M.; Qazi, K.R.; Filén, J.J.; Lahesmaa, R.; Norman, M.; Neve, E.P.; Scheynius, A.; Gabrielsson, S. Exosomes with immune modulatory features are present in human breast milk. J. Immunol. 2007, 179, 1969–1978. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Gao, C.; Li, H.; Huang, L.; Sun, Q.; Dong, Y.; Tian, C.; Gao, S.; Dong, H.; Guan, D.; et al. Identification and characterization of microRNAs in raw milk during different periods of lactation, commercial fluid, and powdered milk products. Cell Res. 2010, 20, 1128–1137. [Google Scholar] [CrossRef] [PubMed]
- Hata, T.; Murakami, K.; Nakatani, H.; Yamamoto, Y.; Matsuda, T.; Aoki, N. Isolation of bovine milk-derived microvesicles carrying mRNAs and microRNAs. Biochem. Biophys. Res. Commun. 2010, 396, 528–533. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Izumi, H.; Sekine, K.; Ochiya, T. microRNA as a new immune-regulatory agent on breast milk. Silence 2010, 1, 7. [Google Scholar] [CrossRef] [PubMed]
- Izumi, H.; Kosaka, N.; Shimizu, T.; Sekine, K.; Ochiya, T.; Takase, M. Bovine milk contains microRNA and messenger RNA that are stable under degradative conditions. J. Dairy Sci. 2012, 95, 4831–4841. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Li, M.; Wang, T.; Liang, Y.; Zhong, Z.; Wang, X.; Zhou, Q.; Chen, L.; Lang, Q.; He, Z.; et al. Lactation-related microRNA expression profiles of porcine breast milk exosomes. PLoS ONE 2012, 7, e43691. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, T.A.; Lippolis, J.D.; Nonnecke, B.J.; Sacco, R.E. Bovine milk exosome proteome. J. Proteom. 2012, 75, 1486–1492. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Li, M.; Wang, X.; Li, Q.; Wang, T.; Zhu, Q.; Zhou, X.; Wang, X.; Gao, X.; Li, X. Immune-related microRNAs are abundant in breast milk exosomes. Int. J. Biol. Sci. 2012, 8, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Chen, X.; Yu, J.; Zen, K.; Zhang, C.Y.; Li, L. Immune modulatory function of abundant immune-related microRNAs in microvesicles from bovine colostrum. Protein Cell 2013, 4, 197–210. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Xi, Q.Y.; Ye, R.S.; Cheng, X.; Qi, Q.E.; Wang, S.B.; Shu, G.; Wang, L.N.; Zhu, X.T.; Jiang, Q.Y.; et al. Exploration of microRNAs in porcine milk exosomes. BMC Genom. 2014, 15, 100. [Google Scholar] [CrossRef] [PubMed]
- Modepalli, V.; Kumar, A.; Hinds, L.A.; Sharp, J.A.; Nicholas, K.R.; Lefevre, C. Differential temporal expression of milk miRNA during the lactation cycle of the marsupial tammar wallaby (Macropus eugenii). BMC Genom. 2014, 15, 1012. [Google Scholar] [CrossRef] [PubMed]
- Na, R.S.; E, G.X.; Sun, W.; Sun, X.W.; Qiu, X.Y.; Chen, L.P.; Huang, Y.F. Expressional analysis of immune-related miRNAs in breast milk. Genet. Mol. Res. 2015, 14, 11371–11376. [Google Scholar] [CrossRef] [PubMed]
- Baddela, V.S.; Nayan, V.; Rani, P.; Onteru, S.K.; Singh, D. Physicochemical biomolecular insights into Buffalo milk-derived nanovesicles. Appl. Biochem. Biotechnol. 2016, 178, 544–557. [Google Scholar] [CrossRef] [PubMed]
- Munch, E.M.; Harris, R.A.; Mohammad, M.; Benham, A.L.; Pejerrey, S.M.; Showalter, L.; Hu, M.; Shope, C.D.; Maningat, P.D.; Gunaratne, P.H.; et al. Transcriptome profiling of microRNA by Next-Gen deep sequencing reveals known and novel miRNA species in the lipid fraction of human breast milk. PLoS ONE 2013, 8, e50564. [Google Scholar] [CrossRef] [PubMed]
- Alsaweed, M.; Lai, C.T.; Hartmann, P.E.; Geddes, D.T.; Kakulas, F. Human milk cells and lipids conserve numerous known and novel miRNAs, some of which are differentially expressed during lactation. PLoS ONE 2016, 11, e0152610. [Google Scholar] [CrossRef] [PubMed]
- Heid, H.W.; Keenan, T.W. Intracellular origin and secretion of milk fat globules. Eur. J. Cell Biol. 2005, 84, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Gallier, S.; Vocking, K.; Post, J.A.; Van De Heijning, B.; Acton, D.; Van Der Beek, E.M.; Van Baalen, T. A novel infant milk formula concept: Mimicking the human milk fat globule structure. Colloids Surf. B Biointerfaces 2015, 136, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, T.A.; Sacco, R.E.; Nonnecke, B.J.; Lippolis, J.D. Bovine milk proteome: Quantitative changes in normal milk exosomes, milk fat globule membranes and whey proteomes resulting from Staphylococcus aureus mastitis. J. Proteom. 2013, 82, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Hassiotou, F.; Beltran, A.; Chetwynd, E.; Stuebe, A.M.; Twigger, A.J.; Metzger, P.; Trengove, N.; Lai, C.T.; Filgueira, L.; Blancafort, P.; et al. Breastmilk is a novel source of stem cells with multilineage differentiation potential. Stem Cells 2012, 30, 2164–2174. [Google Scholar] [CrossRef] [PubMed]
- Hassiotou, F.; Geddes, D.T.; Hartmann, P.E. Cells in human milk: State of the science. J. Hum. Lact. 2013, 29, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Irmak, M.K.; Oztas, Y.; Oztas, E. Integration of maternal genome into the neonate genome through breast milk mRNA transcripts and reverse transcriptase. Theor. Biol. Med. Model. 2012, 9, 20. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Aswath, K.; Schroeder, S.G.; Lippolis, J.D.; Reinhardt, T.A.; Sonstegard, T.S. MicroRNA expression profiles of bovine milk exosomes in response to Staphylococcus aureus infection. BMC Genom. 2015, 16, 806. [Google Scholar] [CrossRef] [PubMed]
- Klopfleisch, R.; Weiss, A.T.; Gruber, A.D. Excavation of a buried treasure—DNA, mRNA, miRNA and protein analysis in formalin fixed, paraffin embedded tissues. Histol. Histopathol. 2011, 26, 797–810. [Google Scholar] [PubMed]
- Streichert, T.; Otto, B.; Lehmann, U. MicroRNA expression profiling in archival tissue specimens: Methods and data processing. Mol. Biotechnol. 2012, 50, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Record, M.; Carayon, K.; Poirot, M.; Silvente-Poirot, S. Exosomes as new vesicular lipid transporters involved in cell-cell communication and various pathophysiologies. Biochim. Biophys. Acta 2014, 1841, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Izumi, H.; Tsuda, M.; Sato, Y.; Kosaka, N.; Ochiya, T.; Iwamoto, H.; Namba, K.; Takeda, Y. Bovine milk exosomes contain microRNA and mRNA and are taken up by human macrophages. J. Dairy Sci. 2015, 98, 2920–2933. [Google Scholar] [CrossRef] [PubMed]
- Pieters, B.C.; Arntz, O.J.; Bennink, M.B.; Broeren, M.G.; van Caam, A.P.; Koenders, M.I.; van Lent, P.L.; van den Berg, W.B.; de Vries, M.; van der Kraan, P.M.; et al. Commercial cow milk contains physically stable extracellular vesicles expressing immunoregulatory TGF-β. PLoS ONE 2015, 10, e0121123. [Google Scholar] [CrossRef] [PubMed]
- Howard, K.M.; Jati Kusuma, R.; Baier, S.R.; Friemel, T.; Markham, L.; Vanamala, J.; Zempleni, J. Loss of miRNAs during processing and storage of cow’s (Bos taurus) milk. J. Agric. Food Chem. 2015, 63, 588–592. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Zhao, Z.; Sun, L.; Li, P. Fermentation results in quantitative changes in milk-derived exosomes and different effects on cell growth and survival. J. Agric. Food Chem. 2017, 65, 1220–1228. [Google Scholar] [CrossRef] [PubMed]
- Munagala, R.; Aqil, F.; Jeyabalan, J.; Gupta, R.C. Bovine milk-derived exosomes for drug delivery. Cancer Lett. 2016, 371, 48–61. [Google Scholar] [CrossRef] [PubMed]
- Benmoussa, A.; Lee, C.H.; Laffont, B.; Savard, P.; Laugier, J.; Boilard, E.; Gilbert, C.; Fliss, I.; Provost, P. Commercial dairy cow milk microRNAs resist digestion under simulated gastrointestinal tract conditions. J. Nutr. 2016, 146, 2206–2215. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Wang, Y.; Wang, H.; Zhu, Z.; Xiao, Z. Visualizing of the cellular uptake and intracellular trafficking of exosomes by live-cell microscopy. J. Cell. Biochem. 2010, 111, 488–496. [Google Scholar] [CrossRef] [PubMed]
- Vickers, K.C.; Remaley, AT. Lipid-based carriers of microRNAs and intercellular communication. Curr. Opin. Lipidol. 2012, 23, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Boon, R.A.; Vickers, K.C. Intercellular transport of microRNAs. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Zhu, Y.L.; Hu, F.H.; Wang, Y.Y.; Huang, N.P.; Xiao, Z.D. Dynamics of exosome internalization and trafficking. J. Cell. Physiol. 2013, 228, 1487–1495. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Zhu, Y.L.; Zhou, Y.Y.; Liang, G.F.; Wang, Y.Y.; Hu, F.H.; Xiao, Z.D. Exosome uptake through clathrin-mediated endocytosis and macropinocytosis and mediating miR-21 delivery. J. Biol. Chem. 2014, 289, 22258–22267. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Martin, P.; Fogarty, B.; Brown, A.; Schurman, K.; Phipps, R.; Yin, V.P.; Lockman, P.; Bai, S. Exosome delivered anticancer drugs across the blood-brain barrier for brain cancer therapy in Danio rerio. Pharm. Res. 2015, 32, 2003–2014. [Google Scholar] [CrossRef] [PubMed]
- Ha, D.; Yang, N.; Nadithe, V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: Current perspectives and future challenges. Acta Pharm. Sin. B 2016, 6, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Lässer, C.; Alikhani, V.S.; Ekström, K.; Eldh, M.; Paredes, P.T.; Bossios, A.; Sjöstrand, M.; Gabrielsson, S.; Lötvall, J.; Valadi, H. Human saliva, plasma and breast milk exosomes contain RNA: Uptake by macrophages. J. Transl. Med. 2011, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Arntz, O.J.; Pieters, B.C.; Oliveira, M.C.; Broeren, M.G.; Bennink, M.B.; de Vries, M.; van Lent, P.L.; Koenders, M.I.; van den Berg, W.B.; van der Kraan, P.M.; et al. Oral administration of bovine milk derived extracellular vesicles attenuates arthritis in two mouse models. Mol. Nutr. Food Res. 2015, 59, 1701–1712. [Google Scholar] [CrossRef] [PubMed]
- Wolf, T.; Baier, S.R.; Zempleni, J. The intestinal transport of bovine milk exosomes is mediated by endocytosis in human colon carcinoma Caco-2 cells and rat small intestinal IEC-6 cells. J. Nutr. 2015, 145, 2201–2206. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Xie, M.Y.; Sun, J.J.; Ye, R.S.; Cheng, X.; Sun, R.P.; Wei, L.M.; Li, M.; Lin, D.L.; Jiang, Q.Y.; et al. Porcine milk-derived exosomes promote proliferation of intestinal epithelial cells. Sci. Rep. 2016, 6, 33862. [Google Scholar] [CrossRef] [PubMed]
- Kusuma, R.J.; Manca, S.; Friemel, T.; Sukreet, S.; Nguyen, C.; Zempleni, J. Human vascular endothelial cells transport foreign exosomes from cow’s milk by endocytosis. Am. J. Physiol. Cell Physiol. 2016, 310, C800–C807. [Google Scholar] [CrossRef] [PubMed]
- Baier, S.R.; Nguyen, C.; Xie, F.; Wood, J.R.; Zempleni, J. MicroRNAs are absorbed in biologically meaningful amounts from nutritionally relevant doses of cow milk and affect gene expression in peripheral blood mononuclear cells, HEK-293 kidney cell cultures, and mouse livers. J. Nutr. 2014, 144, 1495–1500. [Google Scholar] [CrossRef] [PubMed]
- Shu, J.; Chiang, K.; Zempleni, J.; Cui, J. Computational characterization of exogenous microRNAs that can be transferred into human circulation. PLoS ONE 2015, 10, e0140587. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Dudemaine, P.L.; Zhao, X.; Lei, C.; Ibeagha-Awemu, E.M. Comparative analysis of the miRNome of bovine milk fat, whey and cells. PLoS ONE 2016, 11, e0154129. [Google Scholar] [CrossRef] [PubMed]
- Title, A.C.; Denzler, R.; Stoffel, M. Uptake and function studies of maternal milk-derived microRNAs. J. Biol. Chem. 2015, 290, 23680–23691. [Google Scholar] [CrossRef] [PubMed]
- Laubier, J.; Castille, J.; Le Guillou, S.; Le Provost, F. No effect of an elevated miR-30b level in mouse milk on its level in pup tissues. RNA Biol. 2015, 12, 26–29. [Google Scholar] [CrossRef] [PubMed]
- Auerbach, A.; Vyas, G.; Li, A.; Halushka, M.; Witwer, K. Uptake of dietary milk miRNAs by adult humans: A validation study. F1000Research 2016, 5, 721. [Google Scholar] [CrossRef] [PubMed]
- Alsaweed, M.; Lai, C.T.; Hartmann, P.E.; Geddes, D.T.; Kakulas, F. Human milk cells contain numerous miRNAs that may change with milk removal and regulate multiple physiological processes. Int. J. Mol. Sci. 2016, 17, E956. [Google Scholar] [CrossRef] [PubMed]
- Brenner, S.A. Extracellular ‘communicator RNA’. FEBS Lett. 1988, 233, 225–228. [Google Scholar] [CrossRef]
- Liang, H.; Huang, L.; Cao, J.; Zen, K.; Chen, X.; Zhang, C.Y. Regulation of mammalian gene expression by exogenous microRNAs. Wiley Interdiscip. Rev. RNA 2012, 3, 733–742. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liang, H.; Zhang, J.; Zen, K.; Zhang, C.Y. Secreted microRNAs: A new form of intercellular communication. Trends Cell Biol. 2012, 22, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liang, H.; Zhang, J.; Zen, K.; Zhang, C.Y. Horizontal transfer of microRNAs: Molecular mechanisms and clinical applications. Protein Cell 2012, 3, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef] [PubMed]
- Corrado, C.; Raimondo, S.; Chiesi, A.; Ciccia, F.; De Leo, G.; Alessandro, R. Exosomes as intercellular signaling organelles involved in health and disease: Basic science and clinical applications. Int. J. Mol. Sci. 2013, 14, 5338–5366. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, A.K.; Giebel, B. Exosomes: Small vesicles participating in intercellular communication. Int. J. Biochem. Cell Biol. 2012, 44, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Iraci, N.; Leonardi, T.; Gessler, F.; Vega, B.; Pluchino, S. Focus on extracellular vesicles: Physiological role and signalling properties of extracellular membrane vesicles. Int. J. Mol. Sci. 2016, 17, 171. [Google Scholar] [CrossRef] [PubMed]
- Kalra, H.; Drummen, G.P.; Mathivanan, S. Focus on extracellular vesicles: Introducing the next small big thing. Int. J. Mol. Sci. 2016, 17, 170. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, O.; Rodosthenous, R.S.; Jara, C.; Brennan, K.J.; Wright, R.O.; Baccarelli, A.A.; Wright, R.J. Detection of long non-coding RNAs in human breastmilk extracellular vesicles: Implications for early child development. Epigenetics 2016. [Google Scholar] [CrossRef] [PubMed]
- Villarroya-Beltri, C.; Baixauli, F.; Gutiérrez-Vázquez, C.; Sánchez-Madrid, F.; Mittelbrunn, M. Sorting it out: Regulation of exosome loading. Semin. Cancer Biol. 2014, 28, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Villarroya-Beltri, C.; Gutiérrez-Vázquez, C.; Sánchez-Cabo, F.; Pérez-Hernández, D.; Vázquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sánchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef] [PubMed]
- Squadrito, M.L.; Baer, C.; Burdet, F.; Maderna, C.; Gilfillan, G.D.; Lyle, R.; Ibberson, M.; De Palma, M. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 2014, 8, 1432–1446. [Google Scholar] [CrossRef] [PubMed]
- Koppers-Lalic, D.; Hackenberg, M.; Bijnsdorp, I.V.; van Eijndhoven, M.A.; Sadek, P.; Sie, D.; Zini, N.; Middeldorp, J.M.; Ylstra, B.; de Menezes, R.X.; et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 2014, 8, 1649–1658. [Google Scholar] [CrossRef] [PubMed]
- Shurtleff, M.J.; Temoche-Diaz, M.M.; Karfilis, K.V.; Ri, S.; Schekman, R. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. eLife 2016, 5, e19276. [Google Scholar] [CrossRef] [PubMed]
- Bionaz, M.; Loor, J.J. Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genom. 2008, 9, 366. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Cappello, T.; Wang, L. Emerging role of microRNAs in lipid metabolism. Acta Pharm. Sin. B 2015, 5, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Goedeke, L.; Rotllan, N.; Canfrán-Duque, A.; Aranda, J.F.; Ramírez, C.M.; Araldi, E.; Lin, C.S.; Anderson, N.N.; Wagschal, A.; de Cabo, R.; et al. MicroRNA-148a regulates LDL receptor and ABCA1 expression to control circulating lipoprotein levels. Nat. Med. 2015, 21, 1280–1289. [Google Scholar] [CrossRef] [PubMed]
- Wagschal, A.; Najafi-Shoushtari, S.H.; Wang, L.; Goedeke, L.; Sinha, S.; deLemos, A.S.; Black, J.C.; Ramírez, C.M.; Li, Y.; Tewhey, R.; et al. Genome-wide identification of microRNAs regulating cholesterol and triglyceride homeostasis. Nat. Med. 2015, 21, 1290–1297. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Liu, Q.; Zhang, X.; Ren, Y.; Lei, X.; Li, S.; Chen, Q.; Deng, K.; Wang, P.; Zhang, H.; Shi, D. Identification and analysis of the expression of microRNA from lactating and nonlactating mammary glands of the Chinese swamp buffalo. J. Dairy Sci. 2017, 100, 1971–1986. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Luo, J.; Sun, S.; Cao, D.; Shi, H.; Loor, J.J. miR-148a and miR-17-5p synergistically regulate milk TAG synthesis via PPARGC1A and PPARA in goat mammary epithelial cells. RNA Biol. 2017, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Gigli, I.; Maizon, D.O. microRNAs and the mammary gland: A new understanding of gene expression. Genet. Mol. Biol. 2013, 36, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.J.; Wang, C.M.; Wang, Y.T.; Qiao, H.; Fang, L.Q.; Wang, Z.B. Lactation-related microRNA expression in microvesicles of human umbilical cord blood. Med. Sci. Monit. 2016, 22, 4542–4554. [Google Scholar] [CrossRef] [PubMed]
- Braud, M.; Magee, D.A.; Park, S.D.; Sonstegard, T.S.; Waters, S.M.; MacHugh, D.E.; Spillane, C. Genome-wide microRNA binding site variation between extinct wild aurochs and modern cattle identifies candidate microRNA-regulated domestication genes. Front. Genet. 2017, 8, 3. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Erdman, R.A.; Swanson, K.M.; Molenaar, A.J.; Maqbool, N.J.; Wheeler, T.T.; Arias, J.A.; Quinn-Walsh, E.C.; Stelwagen, K. Epigenetic regulation of milk production in dairy cows. J. Mammary Gland Biol. Neoplasia 2010, 15, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Molenaar, A.J.; Swanson, K.M.; Gudex, B.; Arias, J.A.; Erdman, R.A.; Stelwagen, K. Epigenetics: A possible role in acute and transgenerational regulation of dairy cow milk production. Animal 2012, 6, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Luo, J.; Zhang, L.; Zhu, J. MicroRNAs synergistically regulate milk fat synthesis in mammary gland epithelial cells of dairy goats. Gene Expr. 2013, 16, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Platenburg, G.J.; Vollebregt, E.J.; Karatzas, C.N.; Kootwijk, E.P.; De Boer, H.A.; Strijker, R. Mammary gland-specific hypomethylation of Hpa II sites flanking the bovine alpha S1-casein gene. Transgenic Res. 1996, 5, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Bian, Y.; Wang, Z.; Li, D.; Wang, C.; Li, Q.; Gao, X. MicroRNA-152 regulates DNA methyltransferase 1 and is involved in the development and lactation of mammary glands in dairy cows. PLoS ONE 2014, 9, e101358. [Google Scholar] [CrossRef] [PubMed]
- Muroya, S.; Hagi, T.; Kimura, A.; Aso, H.; Matsuzaki, M.; Nomura, M. Lactogenic hormones alter cellular and extracellular microRNA expression in bovine mammary epithelial cell culture. J. Anim. Sci. Biotechnol. 2016, 7, 8. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.; Zhu, S.; Yuan, M.; Cui, H.; Wang, L.; Luo, X.; Li, J.; Zhou, H.; Tang, Y.; Shen, N. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in Lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. J. Immunol. 2010, 184, 6773–6781. [Google Scholar] [CrossRef] [PubMed]
- Long, X.R.; He, Y.; Huang, C.; Li, J. MicroRNA-148a is silenced by hypermethylation and interacts with DNA methyltransferase 1 in hepatocellular carcinogenesis. Int. J. Oncol. 2014, 44, 1915–1922. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Jiang, Y.; Yin, Y.; Li, Q.; He, J.; Jing, Y.; Qi, Y.T.; Xu, Q.; Li, W.; Lu, B.; et al. A regulatory circuit of miR-148a/152 and DNMT1 in modulating cell transformation and tumor angiogenesis through IGF-IR and IRS1. J. Mol. Cell Biol. 2013, 5, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Duursma, A.M.; Kedde, M.; Schrier, M.; le Sage, C.; Agami, R. miR-148 targets human DNMT3b protein coding region. RNA 2008, 14, 872–877. [Google Scholar] [CrossRef] [PubMed]
- Bian, Y.; Lei, Y.; Wang, C.; Wang, J.; Wang, L.; Liu, L.; Liu, L.; Gao, X.; Li, Q. Epigenetic regulation of miR-29s affects the lactation activity of dairy cow mammary epithelial cells. J. Cell. Physiol. 2015, 230, 2152–2163. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, M.; Garzon, R.; Cimmino, A.; Liu, Z.; Zanesi, N.; Callegari, E.; Liu, S.; Alder, H.; Costinean, S.; Fernandez-Cymering, C.; et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. USA 2007, 104, 15805–15810. [Google Scholar] [CrossRef] [PubMed]
- Oakes, S.R.; Naylor, M.J.; Asselin-Labat, M.L.; Blazek, K.D.; Gardiner-Garden, M.; Hilton, H.N.; Kazlauskas, M.; Pritchard, M.A.; Chodosh, L.A.; Pfeffer, P.L.; et al. The Ets transcription factor Elf5 specifies mammary alveolar cell fate. Genes Dev. 2008, 22, 581–586. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Hinshelwood, R.A.; Bouras, T.; Gallego-Ortega, D.; Valdés-Mora, F.; Blazek, K.; Visvader, J.E.; Clark, S.J.; Ormandy, C.J. Lineage specific methylation of the Elf5 promoter in mammary epithelial cells. Stem Cells 2011, 29, 1611–1619. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Shi, H.; Luo, J.; Yi, Y.; Yao, D.; Zhang, X.; Ma, G.; Loor, J.J. MiR-145 regulates lipogenesis in goat mammary cells via targeting INSIG1 and epigenetic regulation of lipid-related genes. J. Cell. Physiol. 2017, 232, 1030–1040. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Espenshade, P.J.; Wright, M.E.; Yabe, D.; Gong, Y.; Aebersold, R.; Goldstein, J.L.; Brown, M.S. Crucial step in cholesterol homeostasis: Sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell 2002, 110, 489–500. [Google Scholar] [CrossRef]
- Hayashi, A.A.; Nones, K.; Roy, N.C.; McNabb, W.C.; Mackenzie, D.S.; Pacheco, D.; McCoard, S. Initiation and elongation steps of mRNA translation are involved in the increase in milk protein yield caused by growth hormone administration during lactation. J. Dairy Sci. 2009, 92, 1889–1899. [Google Scholar] [CrossRef] [PubMed]
- Sciascia, Q.; Pacheco, D.; McCoard, S.A. Increased milk protein synthesis in response to exogenous growth hormone is associated with changes in mechanistic (mammalian) target of rapamycin (mTOR)C1-dependent and independent cell signaling. J. Dairy Sci. 2013, 96, 2327–2338. [Google Scholar] [CrossRef] [PubMed]
- Kirchner, B.; Pfaffl, M.W.; Dumpler, J.; von Mutius, E.; Ege, M.J. microRNA in native and processed cow’s milk and its implication for the farm milk effect on asthma. J. Allergy Clin. Immunol. 2016, 137, 1893–1895. [Google Scholar] [CrossRef] [PubMed]
- Simpson, M.R.; Brede, G.; Johansen, J.; Johnsen, R.; Storrø, O.; Sætrom, P.; Øien, T. Human breast milk miRNA, maternal probiotic supplementation and atopic dermatitis in offspring. PLoS ONE 2015, 10, e0143496. [Google Scholar] [CrossRef] [PubMed]
- Koh, K.P.; Rao, A. DNA methylation and methylcytosine oxidation in cell fate decisions. Curr. Opin. Cell Biol. 2013, 25, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Ko, M.; An, J.; Bandukwala, H.S.; Chavez, L.; Aijö, T.; Pastor, W.A.; Segal, M.F.; Li, H.; Koh, K.P.; Lähdesmäki, H.; et al. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature 2013, 497, 122–126. [Google Scholar] [CrossRef] [PubMed]
- Dunican, D.D.; Pennings, S.; Meeha, R.R. The CXXC-TET bridge—Mind the methylation gap! Cell Res. 2013, 23, 973–974. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C. Milk: An epigenetic amplifier of FTO-mediated transcription? Implications for Western diseases. J. Transl. Med. 2015, 13, 385. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C. Milk—A nutrient system of mammalian evolution promoting mTORC1-dependent translation. Int. J. Mol. Sci. 2015, 16, 17048–17087. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Jaffrey, S.R. The dynamic epitranscriptome: N6-Methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014, 15, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Maity, A.; Das, B. N6-methyladenosine modification in mRNA: Machinery, function and implications for health and diseases. FEBS J. 2016, 283, 1607–1630. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Jiang, D.; Wang, Y.; Wang, X. N(6)-methyladenosine (m(6)A) methylation in mRNA with a dynamic and reversible epigenetic modification. Mol. Biotechnol. 2016, 58, 450–459. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, S.; Xiao, W.; Zhao, Y.L.; Yang, Y.G. m(6)A: Signaling for mRNA splicing. RNA Biol. 2016, 13, 756–759. [Google Scholar] [CrossRef] [PubMed]
- Chhabra, R. miRNA and methylation: A multifaceted liaison. Chembiochem 2015, 16, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Y.; Toth, J.I.; Petroski, M.D.; Zhang, Z.; Zhao, J.C. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 2014, 16, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, J.C. Update: Mechanisms Underlying N6-Methyladenosine Modification of Eukaryotic mRNA. Trends Genet. 2016, 32, 763–773. [Google Scholar] [CrossRef] [PubMed]
- Boissel, S.; Reish, O.; Proulx, K.; Kawagoe-Takaki, H.; Sedgwick, B.; Yeo, G.S.; Meyre, D.; Golzio, C.; Molinari, F.; Kadhom, N.; et al. Loss-of-function mutation in the dioxygenase-encoding FTO gene causes severe growth retardation and multiple malformations. Am. J. Hum. Genet. 2009, 85, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Fischer, J.; Koch, L.; Emmerling, C.; Vierkotten, J.; Peters, T.; Brüning, J.C.; Rüther, U. Inactivation of the Fto gene protects from obesity. Nature 2009, 458, 894–898. [Google Scholar] [CrossRef] [PubMed]
- Church, C.; Moir, L.; McMurray, F.; Girard, C.; Banks, G.T.; Teboul, L.; Wells, S.; Brüning, J.C.; Nolan, P.M.; Ashcroft, F.M.; et al. Overexpression of Fto leads to increased food intake and results in obesity. Nat. Genet. 2010, 42, 1086–1092. [Google Scholar] [CrossRef] [PubMed]
- Peters, T.; Ausmeier, K.; Rüther, U. Cloning of Fatso (Fto), a novel gene deleted by the Fused toes (Ft) mouse mutation. Mamm. Genome 1999, 10, 983–986. [Google Scholar] [PubMed]
- Gao, X.; Shin, Y.H.; Li, M.; Wang, F.; Tong, Q.; Zhang, P. The fat mass and obesity associated gene FTO functions in the brain to regulate postnatal growth in mice. PLoS ONE 2010, 5, e14005. [Google Scholar] [CrossRef] [PubMed]
- Speakman, J.R. The ‘Fat Mass and Obesity related’ (FTO) gene: Mechanisms of impact on obesity and energy balance. Curr. Obes. Rep. 2015, 4, 73–91. [Google Scholar] [CrossRef] [PubMed]
- Berulava, T.; Horsthemke, B. The obesity-associated SNPs in intron 1 of the FTO gene affect primary transcript levels. Eur. J. Hum. Genet. 2010, 18, 1054–1056. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yang, Y.; Sun, B.F.; Shi, Y.; Yang, X.; Xiao, W.; Hao, Y.J.; Ping, X.L.; Chen, Y.S.; Wang, W.J.; et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014, 24, 1403–1419. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Lee, H.S.; Kim, Y.K.; Park, S.; Kim, J.M.; Yun, J.H.; Yu, H.Y.; Kim, B.J. Association of metabolites with obesity and type 2 diabetes based on FTO genotype. PLoS ONE 2016, 11, e0156612. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.W.; Zhang, J.T.; Cai, Q.Y.; Zhang, H.X.; Wang, Y.H.; Yan, H.T.; Wu, H.M.; Yang, X.J. Birth weight is associated with placental fat mass- and obesity-associated gene expression and promoter methylation in a Chinese population. J. Matern. Fetal Neonatal Med. 2016, 29, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Dayeh, T.; Volkov, P.; Salö, S.; Hall, E.; Nilsson, E.; Olsson, A.H.; Kirkpatrick, C.L.; Wollheim, C.B.; Eliasson, L.; Rönn, T.; et al. Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. PLoS Genet. 2014, 10, e1004160. [Google Scholar] [CrossRef] [PubMed]
- Toperoff, G.; Kark, J.D.; Aran, D.; Nassar, H.; Ahmad, W.A.; Sinnreich, R.; Azaiza, D.; Glaser, B.; Hellman, A. Premature aging of leukocyte DNA methylation is associated with type 2 diabetes prevalence. Clin. Epigenet. 2015, 7, 35. [Google Scholar] [CrossRef] [PubMed]
- Rönn, T.; Ling, C. DNA methylation as a diagnostic and therapeutic target in the battle against type 2 diabetes. Epigenomics 2015, 7, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Howell, J.J.; Ricoult, S.J.; Ben-Sahra, I.; Manning, B.D. A growing role for mTOR in promoting anabolic metabolism. Biochem. Soc. Trans. 2013, 41, 906–912. [Google Scholar] [CrossRef] [PubMed]
- Gulati, P.; Cheung, M.K.; Antrobus, R.; Church, C.D.; Harding, H.P.; Tung, Y.C.; Rimmington, D.; Ma, M.; Ron, D.; Lehner, P.J.; et al. Role for the obesity-related FTO gene in the cellular sensing of amino acids. Proc. Natl. Acad. Sci. USA 2013, 110, 2557–2562. [Google Scholar] [CrossRef] [PubMed]
- Gulati, P.; Yeo, G.S. The biology of FTO: From nucleic acid demethylase to amino acid sensor. Diabetologia 2013, 56, 2113–2121. [Google Scholar] [CrossRef] [PubMed]
- Manifava, M.; Smith, M.; Rotondo, S.; Walker, S.; Niewczas, I.; Zoncu, R.; Clark, J.; Ktistakis, N.T. Dynamics of mTORC1 activation in response to amino acids. eLife 2016, 5, e19960. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Wang, L.; Chen, B.; Zheng, P.; He, Y.; Ding, Y.; Deng, Y.; Lu, X.; Guo, X.; Zhang, Y.; et al. DNA demethylation upregulated Nrf2 expression in Alzheimer’s disease cellular model. Front. Aging Neurosci. 2016, 7, 244. [Google Scholar] [CrossRef] [PubMed]
- Bendavit, G.; Aboulkassim, T.; Hilmi, K.; Shah, S.; Batist, G. Nrf2 transcription factor can directly regulate mTOR: Linking cytoprotective gene expression to a major metabolic regulator that generates redoox activity. J. Biol. Chem. 2016, 291, 25476–25488. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Zhang, W.; Zhou, Y.; Li, F.; Wie, H.; Peng, J. Recent advances in understanding amino acid sensing mechanisms that regulate mTORC1. Int. J. Mol. Sci. 2016, 17, E1636. [Google Scholar] [CrossRef] [PubMed]
- Shibata, T.; Saito, S.; Kokubu, A.; Suzuki, T.; Yamamoto, M.; Hirohashi, S. Global downstream pathway analysis reveals a dependence of oncogenic NF-E2-related factor 2 mutation on the mTOR growth signaling pathway. Cancer Res. 2010, 70, 9095–9105. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, H.; Shitara, M.; Yokota, K.; Hikosaka, Y.; Moriyama, S.; Yano, M.; Fujii, Y. RagD gene expression and NRF2 mutations in lung squamous cell carcinomas. Oncol. Lett. 2012, 4, 1167–1170. [Google Scholar] [CrossRef] [PubMed]
- Kurinna, S.; Schäfer, M.; Ostano, P.; Karouzakis, E.; Chiorino, G.; Bloch, W.; Bachmann, A.; Gay, S.; Garrod, D.; Lefort, K.; et al. A novel Nrf2-miR-29-desmocollin-2 axis regulates desmosome function in keratinocytes. Nat. Commun. 2014, 5, 5099. [Google Scholar] [CrossRef] [PubMed]
- Kurinna, S.; Werner, S. NRF2 and microRNAs: New but awaited relations. Biochem. Soc. Trans. 2015, 43, 595–601. [Google Scholar] [CrossRef] [PubMed]
- Dibble, C.C.; Cantley, L.C. Regulation of mTORC1 by PI3K signaling. Trends Cell Biol. 2015, 25, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, A.; Rauch, T.A.; Todorov, I.; Ku, H.T.; Al-Abdullah, I.H.; Kandeel, F.; Mullen, Y.; Pfeifer, G.P.; Ferreri, K. Insulin gene expression is regulated by DNA methylation. PLoS ONE 2009, 4, e6953. [Google Scholar] [CrossRef]
- Dooley, J.; Garcia-Perez, J.E.; Sreenivasan, J.; Schlenner, S.M.; Vangoitsenhoven, R.; Papadopoulou, A.S.; Tian, L.; Schonefeldt, S.; Serneels, L.; Deroose, C.; et al. The microRNA-29 family dictates the balance between homeostatic and pathological glucose handling in diabetes and obesity. Diabetes 2016, 65, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.Q.; He, K.; Xu, J.Y. Milk consumption and circulating insulin-like growth factor-I level: A systematic literature review. Int. J. Food Sci. Nutr. 2009, 60 (Suppl. 7), 330–340. [Google Scholar] [CrossRef] [PubMed]
- Hoppe, C.; Mølgaard, C.; Michaelsen, K.F. Cow’s milk and linear growth in industrialized and developing countries. Annu. Rev. Nutr. 2006, 26, 131–173. [Google Scholar] [CrossRef] [PubMed]
- Ouni, M.; Gunes, Y.; Belot, M.P.; Castell, A.L.; Fradin, D.; Bougnères, P. The IGF1 P2 promoter is an epigenetic QTL for circulating IGF1 and human growth. Clin. Epigenet. 2015, 7, 22. [Google Scholar] [CrossRef] [PubMed]
- Ouni, M.; Belot, M.P.; Castell, A.L.; Fradin, D.; Bougnères, P. The P2 promoter of the IGF1 gene is a major epigenetic locus for GH responsiveness. Pharmacogen. J. 2016, 16, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Tahir, S.A.; Yang, G.; Goltsov, A.; Song, K.D.; Ren, C.; Wang, J.; Chang, W.; Thompson, T.C. Caveolin-1-LRP6 signaling module stimulates aerobic glycolysis in prostate cancer. Cancer Res. 2013, 73, 1900–1911. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Feng, X.; Zhang, S.; Ren, Z.; Liu, Y.; Yang, B.; lv, B.; Cai, Y.; Xia, J.; Ge, N. Caveolin-1 confers resistance of hepatoma cells to anoikis by activating IGF-1 pathway. Cell. Physiol. Biochem. 2015, 36, 1223–1236. [Google Scholar] [CrossRef] [PubMed]
- Palacios-Ortega, S.; Varela-Guruceaga, M.; Martínez, J.A.; de Miguel, C.; Milagro, F.I. Effects of high glucose on caveolin-1 and insulin signaling in 3T3-L1 adipocytes. Adipocyte 2015, 5, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Palacios-Ortega, S.; Varela-Guruceaga, M.; Milagro, F.I.; Martínez, J.A.; de Miguel, C. Expression of caveolin 1 is enhanced by DNA demethylation during adipocyte differentiation. Status of insulin signaling. PLoS ONE 2014, 9, e95100. [Google Scholar] [CrossRef] [PubMed]
- Palomares, O.; Yaman, G.; Azkur, A.K.; Akkoc, T.; Akdis, M.; Akdis, C.A. Role of Treg in immune regulation of allergic diseases. Eur. J. Immunol. 2010, 40, 1232–1240. [Google Scholar] [CrossRef] [PubMed]
- Pellerin, L.; Jenks, J.A.; Bégin, P.; Bacchetta, R.; Nadeau, K.C. Regulatory T cells and their roles in immune dysregulation and allergy. Immunol. Res. 2014, 58, 358–368. [Google Scholar] [CrossRef] [PubMed]
- Alroqi, F.J.; Chatila, T.A. T regulatory cell biology in health and disease. Curr. Allergy Asthma Rep. 2016, 16, 27. [Google Scholar] [CrossRef] [PubMed]
- Huehn, J.; Beyer, M. Epigenetic and transcriptional control of Foxp3+ regulatory T cells. Semin. Immunol. 2015, 27, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Polansky, J.K.; Kretschmer, K.; Freyer, J.; Floess, S.; Garbe, A.; Baron, U.; Olek, S.; Hamann, A.; von Boehmer, H.; Huehn, J. DNA methylation controls Foxp3 gene expression. Eur. J. Immunol. 2008, 38, 1654–1663. [Google Scholar] [CrossRef] [PubMed]
- Polansky, J.K.; Schreiber, L.; Thelemann, C.; Ludwig, L.; Krüger, M.; Baumgrass, R.; Cording, S.; Floess, S.; Hamann, A.; Huehn, J. Methylation matters: Binding of Ets-1 to the demethylated Foxp3 gene contributes to the stabilization of Foxp3 expression in regulatory T cells. J. Mol. Med. 2010, 88, 1029–1040. [Google Scholar] [CrossRef] [PubMed]
- Paparo, L.; Nocerino, R.; Cosenza, L.; Aitoro, R.; D’Argenio, V.; Del Monaco, V.; Di Scala, C.; Amoroso, A.; Di Costanzo, M.; Salvatore, F.; et al. Epigenetic features of FoxP3 in children with cow’s milk allergy. Clin. Epigenet. 2016, 8, 86. [Google Scholar] [CrossRef] [PubMed]
- Janson, P.C.; Winerdal, M.E.; Marits, P.; Thörn, M.; Ohlsson, R.; Winqvist, O. FOXP3 promoter demethylation reveals the committed Treg population in humans. PLoS ONE 2008, 3, e1612. [Google Scholar] [CrossRef] [PubMed]
- Bacchetta, R.; Gambineri, E.; Roncarolo, M.G. Role of regulatory T cells and FOXP3 in human diseases. J. Allergy Clin. Immunol. 2007, 120, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Nadeau, K.; McDonald-Hyman, C.; Noth, E.M.; Pratt, B.; Hammond, S.K.; Balmes, J.; Tager, I. Ambient air pollution impairs regulatory T-cell function in asthma. J. Allergy Clin. Immunol. 2010, 126, 845–852. [Google Scholar] [CrossRef] [PubMed]
- Hinz, D.; Bauer, M.; Röder, S.; Olek, S.; Huehn, J.; Sack, U.; Borte, M.; Simon, J.C.; Lehmann, I.; Herberth, G.; LINA study group. Cord blood Tregs with stable FOXP3 expression are influenced by prenatal environment and associated with atopic dermatitis at the age of one year. Allergy 2012, 67, 380–389. [Google Scholar] [CrossRef] [PubMed]
- Lal, G.; Bromberg, J.S. Epigenetic mechanisms of regulation of Foxp3 expression. Blood 2009, 114, 3727–3735. [Google Scholar] [CrossRef] [PubMed]
- Lal, G.; Zhang, N.; van der Touw, W.; Ding, Y.; Ju, W.; Bottinger, E.P.; Reid, S.P.; Levy, D.E.; Bromberg, J.S. Epigenetic regulation of Foxp3 expression in regulatory T cells by DNA methylation. J. Immunol. 2009, 182, 259–273. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C.; John, S.M.; Schmitz, G. Milk: An exosomal microRNA transmitter promoting thymic regulatory T cell maturation preventing the development of atopy? J. Transl. Med. 2014, 12, 43. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C.; John, S.M.; Schmitz, G. Milk: An epigenetic inducer of FoxP3 expression. J. Allergy Clin. Immunol. 2016, 138, 937–938. [Google Scholar] [CrossRef] [PubMed]
- Parigi, S.M.; Eldh, M.; Larssen, P.; Gabrielsson, S.; Villablanca, E.J. Breast milk and solid food shaping intestinal immunity. Front. Immunol. 2015, 6, 415. [Google Scholar] [CrossRef] [PubMed]
- Petrus, N.C.; Henneman, P.; Venema, A.; Mul, A.; van Sinderen, F.; Haagmans, M.; Mook, O.; Hennekam, R.C.; Sprikkelman, A.B.; Mannens, M. Cow’s milk allergy in Dutch children: An epigenetic pilot survey. Clin. Transl. Allergy 2016, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Tooley, K.L.; El-Merhibi, A.; Cummins, A.G.; Grose, R.H.; Lymn, K.A.; DeNichilo, M.; Penttila, I.A. Maternal milk, but not formula, regulates the immune response to beta-lactoglobulin in allergy-prone rat pups. J. Nutr. 2009, 139, 2145–2151. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Pastrana, J.; Shao, Y.; Chernaya, V.; Wang, H.; Yang, X.F. Epigenetic enzymes are the therapeutic targets for CD4(+)CD25(+/high)Foxp3(+) regulatory T cells. Transl. Res. 2015, 165, 221–240. [Google Scholar] [CrossRef] [PubMed]
- Bluestone, J.A. FOXP3, the transcription factor at the heart of the rebirth of immune tolerance. J. Immunol. 2017, 198, 979–980. [Google Scholar] [CrossRef] [PubMed]
- Ranhotra, H.S. The NR4A orphan nuclear receptors: Mediators in metabolism and diseases. J. Recept. Signal Transduct. Res. 2015, 35, 184–188. [Google Scholar] [CrossRef] [PubMed]
- Sekiya, T.; Kashiwagi, I.; Yoshida, R.; Fukaya, T.; Morita, R.; Kimura, A.; Ichinose, H.; Metzger, D.; Chambon, P.; Yoshimura, A. Nr4a receptors are essential for thymic regulatory T cell development and immune homeostasis. Nat. Immunol. 2013, 14, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Won, H.Y.; Hwang, E.S. Transcriptional modulation of regulatory T cell development by novel regulators NR4As. Arch. Pharm. Res. 2016, 39, 1530–1536. [Google Scholar] [CrossRef] [PubMed]
- Sekiya, T.; Kondo, T.; Shichita, T.; Morita, R.; Ichinose, H.; Yoshimura, A. Suppression of Th2 and Tfh immune reactions by Nr4a receptors in mature T reg cells. J. Exp. Med. 2015, 212, 1623–1640. [Google Scholar] [CrossRef] [PubMed]
- Sekiya, T.; Nakatsukasa, H.; Lu, Q.; Yoshimura, A. Roles of transcription factors and epigenetic modifications in differentiation and maintenance of regulatory T cells. Microbes Infect. 2016, 18, 378–386. [Google Scholar] [CrossRef] [PubMed]
- Bandukwala, H.S.; Rao, A. ‘Nurr’ishing Treg cells: Nr4a transcription factors control Foxp3 expression. Nat. Immunol. 2013, 14, 201–203. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Fatima, N.; Dufau, M.L. Coordinated changes in DNA methylation and histone modifications regulate silencing/derepression of luteinizing hormone receptor gene transcription. Mol. Cell. Biol. 2005, 25, 7929–7939. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.M.; Chang, L.Y.; Lin, S.H.; Chou, J.L.; Hsieh, H.Y.; Zeng, L.H.; Chuang, S.Y.; Wang, H.W.; Dittner, C.; Lin, C.Y.; et al. Epigenetic silencing of the NR4A3 tumor suppressor, by aberrant JAK/STAT signaling, predicts prognosis in gastric cancer. Sci. Rep. 2016, 6, 31690. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Nomiyama, T.; Findeisen, H.M.; Qing, H.; Aono, J.; Jones, K.L.; Heywood, E.B.; Bruemmer, D. Epigenetic regulation of the NR4A orphan nuclear receptor NOR1 by histone acetylation. FEBS Lett. 2014, 588, 4825–4830. [Google Scholar] [CrossRef] [PubMed]
- El-Osta, A.; Wolffe, A.P. DNA methylation and histone deacetylation in the control of gene expression: Basic biochemistry to human development and disease. Gene Expr. 2000, 9, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Dobosy, J.R.; Selker, E.U. Emerging connections between DNA methylation and histone acetylation. Cell. Mol. Life Sci. 2001, 58, 721–727. [Google Scholar] [CrossRef] [PubMed]
- Fuks, F.; Hurd, P.J.; Wolf, D.; Nan, X.; Bird, A.P.; Kouzarides, T. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. 2003, 278, 4035–4040. [Google Scholar] [CrossRef] [PubMed]
- Drewell, R.A.; Goddard, C.J.; Thomas, J.O.; Surani, M.A. Methylation-dependent silencing at the H19 imprinting control region by MeCP2. Nucleic Acids Res. 2002, 30, 1139–1144. [Google Scholar] [CrossRef] [PubMed]
- Sisk, P.M.; Lovelady, C.A.; Dillard, R.G.; Gruber, K.J.; O’Shea, T.M. Early human milk feeding is associated with a lower risk of necrotizing enterocolitis in very low birth weight infants. J. Perinatol. 2007, 27, 428–433. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xue, L.; Ma, T.; Li, Y.; Li, Z. Non-intervention observation: Dynamic evolution laws of inflammatory response in necrotizing enterocolitis. Exp. Ther. Med. 2016, 12, 1770–1774. [Google Scholar] [CrossRef] [PubMed]
- Ruan, W.Y.; Bi, M.Y.; Feng, W.W.; Wang, Y.J.; Bu, W.Q.; Lu, L. Effect of human breast milk on the expression of proinflammatory cytokines in Caco-2 cells after hypoxia/re-oxygenation. Rev. Investig. Clin. 2016, 68, 105–111. [Google Scholar]
- Ferreiro, D.U.; Komives, E.A. Molecular mechanisms of system control of NF-kappaB signaling by IkappaBalpha. Biochemistry 2010, 49, 1560–1567. [Google Scholar] [CrossRef] [PubMed]
- Verma, I.M.; Stevenson, J.K.; Schwarz, E.M.; Van Antwerp, D.; Miyamoto, S. Rel/NF-kappa B/I kappa B family: Intimate tales of association and dissociation. Genes Dev. 1995, 9, 2723–2735. [Google Scholar] [CrossRef] [PubMed]
- O’Gorman, A.; Colleran, A.; Ryan, A.; Mann, J.; Egan, L.J. Regulation of NF-kappaB responses by epigenetic suppression of IkappaBalpha expression in HCT116 intestinal epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 299, G96–G105. [Google Scholar] [CrossRef] [PubMed]
- Scheinman, R.I.; Cogswell, P.C.; Lofquist, A.K.; Baldwin, A.S., Jr. Role of transcriptional activation of I kappa B alpha in mediation of immunosuppression by glucocorticoids. Science 1995, 270, 283–286. [Google Scholar] [CrossRef] [PubMed]
- Auphan, N.; DiDonato, J.A.; Rosette, C.; Helmberg, A.; Karin, M. Immunosuppression by glucocorticoids: Inhibition of NF-kappa B activity through induction of I kappa B synthesis. Science 1995, 270, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Labrie, V.; Buske, O.J.; Oh, E.; Jeremian, R.; Ptak, C.; Gasiūnas, G.; Maleckas, A.; Petereit, R.; Žvirbliene, A.; Adamonis, K.; et al. Lactase nonpersistence is directed by DNA-variation-dependent epigenetic aging. Nat. Struct. Mol. Biol. 2016, 23, 566–573. [Google Scholar] [CrossRef] [PubMed]
- Swallow, D.M.; Troelsen, J.T. Escape from epigenetic silencing of lactase expression is triggered by a single-nucleotide change. Nat. Struct. Mol. Biol. 2016, 23, 505–507. [Google Scholar] [CrossRef] [PubMed]
- Olds, L.C.; Sibley, E. Lactase persistence DNA variant enhances lactase promoter activity in vitro: Functional role as a cis regulatory element. Hum. Mol. Genet. 2003, 12, 2333–2340. [Google Scholar] [CrossRef] [PubMed]
- Troelsen, J.T.; Olsen, J.; Møller, J.; Sjöström, H. An upstream polymorphism associated with lactase persistence has increased enhancer activity. Gastroenterology 2003, 125, 1686–1694. [Google Scholar] [CrossRef] [PubMed]
- Lewinsky, R.H.; Jensen, T.G.; Møller, J.; Stensballe, A.; Olsen, J.; Troelsen, J.T. T-13910 DNA variant associated with lactase persistence interacts with Oct-1 and stimulates lactase promoter activity in vitro. Hum. Mol. Genet. 2005, 14, 3945–3953. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Maravelias, C.; Sibley, E. Lactase gene promoter fragments mediate differential spatial and temporal expression patterns in transgenic mice. DNA Cell Biol. 2006, 25, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Merkestein, M.; McTaggart, J.S.; Lee, S.; Kramer, H.B.; McMurray, F.; Lafond, M.; Boutens, L.; Cox, R.; Ashcroft, F.M. Changes in gene expression associated with FTO overexpression in mice. PLoS ONE 2014, 9, e97162. [Google Scholar]
- Karra, E.; O’Daly, O.G.; Choudhury, A.I.; Yousseif, A.; Millership, S.; Neary, M.T.; Scott, W.R.; Chandarana, K.; Manning, S.; Hess, M.E.; et al. A link between FTO, ghrelin, and impaired brain food-cue responsivity. J. Clin. Investig. 2013, 123, 3539–3551. [Google Scholar] [CrossRef] [PubMed]
- Burger, K.S.; Berner, L.A. A functional neuroimaging review of obesity, appetitive hormones and ingestive behavior. Physiol. Behav. 2014, 136, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Hewson, A.K.; Dickson, S.L. Systemic administration of ghrelin induces Fos and Egr-1 proteins in the hypothalamic arcuate nucleus of fasted and fed rats. J. Neuroendocrinol. 2000, 12, 1047–1049. [Google Scholar] [CrossRef] [PubMed]
- Perello, M.; Dickson, S.L. Ghrelin signalling on food reward: A salient link between the gut and the mesolimbic system. J. Neuroendocrinol. 2015, 27, 424–434. [Google Scholar] [CrossRef] [PubMed]
- Skibicka, K.P.; Shirazi, R.H.; Hansson, C.; Dickson, S.L. Ghrelin interacts with neuropeptide Y Y1 and opioid receptors to increase food reward. Endocrinology 2012, 153, 1194–1205. [Google Scholar] [CrossRef] [PubMed]
- Caruso, V.; Chen, H.; Morris, M.J. Early hypothalamic FTO overexpression in response to maternal obesity—Potential contribution to postweaning hyperphagia. PLoS ONE 2011, 6, e25261. [Google Scholar] [CrossRef] [PubMed]
- Wiley, A.S. Dairy and milk consumption and child growth: Is BMI involved? An analysis of NHANES 1999–2004. Am. J. Hum. Biol. 2010, 22, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Parmar, M.S.; Jaumotte, J.D.; Zigmond, M.J.; Cavanaugh, J.E. ERK1, 2, and 5 expression and activation in dopaminergic brain regions during postnatal development. Int. J. Dev. Neurosci. 2015, 46, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Mastwal, S.; Cao, V.Y.; Ren, M.; Liu, Q.; Zhang, W.; Elkahloun, A.G.; Wang, K.H. Dopamine is required for activity-dependent amplification of Arc mRNA in developing postnatal frontal cortex. Cereb. Cortex 2016. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.X.; Huang, E.J. Dopaminergic neurons and brain reward pathways: From neurogenesis to circuit assembly. Am. J. Pathol. 2016, 186, 478–488. [Google Scholar] [CrossRef] [PubMed]
- Heni, M.; Kullmann, S.; Ahlqvist, E.; Wagner, R.; Machicao, F.; Staiger, H.; Häring, H.U.; Almgren, P.; Groop, L.C.; Small, D.M.; et al. Interaction between the obesity-risk gene FTO and the dopamine D2 receptor gene ANKK1/TaqIA on insulin sensitivity. Diabetologia 2016, 59, 2622–2631. [Google Scholar] [CrossRef] [PubMed]
- Hess, M.E.; Hess, S.; Meyer, K.D.; Verhagen, L.A.; Koch, L.; Brönneke, H.S.; Dietrich, M.O.; Jordan, S.D.; Saletore, Y.; Elemento, O.; et al. The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat. Neurosci. 2013, 16, 1042–1048. [Google Scholar] [CrossRef] [PubMed]
- Kodama, N.; Iwao, T.; Kabeya, T.; Horikawa, T.; Niwa, T.; Kondo, Y.; Nakamura, K.; Matsunaga, T. Inhibition of mitogen-activated protein kinase kinase, DNA methyltransferase, and transforming growth factor-β promotes differentiation of human induced pluripotent stem cells into enterocytes. Drug Metab. Pharmacokinet. 2016, 31, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Spalding, K.L.; Arner, E.; Westermark, P.O.; Bernard, S.; Buchholz, B.A.; Bergmann, O.; Blomqvist, L.; Hoffstedt, J.; Näslund, E.; Britton, T.; et al. Dynamics of fat cell turnover in humans. Nature 2008, 453, 783–787. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.M.; Jones, P.A. Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell 1979, 17, 771–779. [Google Scholar] [CrossRef]
- Noer, A.; Sørensen, A.L.; Boquest, A.C.; Collas, P. Stable CpG hypomethylation of adipogenic promoters in freshly isolated, cultured, and differentiated mesenchymal stem cells from adipose tissue. Mol. Biol. Cell 2006, 17, 3543–3556. [Google Scholar] [CrossRef] [PubMed]
- Londoño Gentile, T.; Lu, C.; Lodato, P.M.; Tse, S.; Olejniczak, S.H.; Witze, E.S.; Thompson, C.B.; Wellen, K.E. DNMT1 is regulated by ATP-citrate lyase and maintains methylation patterns during adipocyte differentiation. Mol. Cell. Biol. 2013, 33, 3864–3878. [Google Scholar]
- An, X.; Ma, K.; Zhang, Z.; Zhao, T.; Zhang, X.; Tang, B.; Li, Z. miR-17, miR-21, and miR-143 enhance adipogenic differentiation from porcine bone marrow-derived mesenchymal stem cells. DNA Cell Biol. 2016, 35, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.C.; Chia, S.Y.; Jin, S.; Han, W.; Ding, C.; Sun, L. Dynamic DNA methylation landscape defines brown and white cell specificity during adipogenesis. Mol. Metab. 2016, 5, 1033–1041. [Google Scholar] [CrossRef] [PubMed]
- Merkestein, M.; Laber, S.; McMurray, F.; Andrew, D.; Sachse, G.; Sanderson, J.; Li, M.; Usher, S.; Sellayah, D.; Ashcroft, F.M.; et al. FTO influences adipogenesis by regulating mitotic clonal expansion. Nat. Commun. 2015, 6, 6792. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhou, B.; Luo, Y.; Huang, Z.; Jia, G.; Liu, G.; Zhao, H. Tissue distribution of porcine FTO and its effect on porcine intramuscular preadipocytes proliferation and differentiation. PLoS ONE 2016, 11, e0151056. [Google Scholar] [CrossRef] [PubMed]
- Grunnet, L.G.; Nilsson, E.; Ling, C.; Hansen, T.; Pedersen, O.; Groop, L.; Vaag, A.; Poulsen, P. Regulation and function of FTO mRNA expression in human skeletal muscle and subcutaneous adipose tissue. Diabetes 2009, 58, 2402–2408. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhou, X.; Wu, W.; Wang, X.; Wang, Y. FTO-dependent function of N6-methyladenosine is involved in the hepatoprotective effects of betaine on adolescent mice. J. Physiol. Biochem. 2015, 71, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Goodman, C.A. The role of mTORC1 in regulating protein synthesis and skeletal muscle mass in response to various mechanical stimuli. Rev. Physiol. Biochem. Pharmacol. 2014, 166, 43–95. [Google Scholar] [PubMed]
- Bond, P. Regulation of mTORC1 by growth factors, energy status, amino acids and mechanical stimuli at a glance. J. Int. Soc. Sports Nutr. 2016, 13, 8. [Google Scholar] [CrossRef] [PubMed]
- Petrie, M.A.; Kimball, A.L.; McHenry, C.L.; Suneja, M.; Yen, C.L.; Sharma, A.; Shields, R.K. Distinct skeletal muscle gene regulation from active contraction, passive vibration, and whole body heat stress in humans. PLoS ONE 2016, 11, e0160594. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Sieira, S.; López, M.; Nogueiras, R.; Tovar, S. Regulation of NR4A by nutritional status, gender, postnatal development and hormonal deficiency. Sci. Rep. 2014, 4, 4264. [Google Scholar] [CrossRef] [PubMed]
- Szyf, M.; Rouleau, J.; Theberge, J.; Bozovic, V. Induction of myogenic differentiation by an expression vector encoding the DNA methyltransferase cDNA sequence in the antisense orientation. J. Biol. Chem. 1992, 267, 12831–12836. [Google Scholar] [PubMed]
- Lucarelli, M.; Fuso, A.; Strom, R.; Scarpa, S. The dynamics of myogenin site-specific demethylation is strongly correlated with its expression and with muscle differentiation. J. Biol. Chem. 2001, 276, 7500–7506. [Google Scholar] [CrossRef] [PubMed]
- Palacios, D.; Puri, P.L. The epigenetic network regulating muscle development and regeneration. J. Cell. Physiol. 2006, 207, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Park, I.H.; Chen, J. Mammalian target of rapamycin (mTOR) signaling is required for a late-stage fusion process during skeletal myotube maturation. J. Biol. Chem. 2005, 280, 32009–32017. [Google Scholar] [CrossRef] [PubMed]
- Hatfield, I.; Harvey, I.; Yates, E.R.; Redd, J.R.; Reiter, L.T.; Bridges, D. The role of TORC1 in muscle development in Drosophila. Sci. Rep. 2015, 5, 9676. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ying, Z.Z.; Tang, Z.L.; Long, L.Q.; Li, K. MicroRNA-148a promotes myogenic differentiation by targeting the ROCK1 gene. J. Biol. Chem. 2012, 287, 21093–21101. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Sun, H.; Guttridge, D.C. microRNAs: Novel components in a muscle gene regulatory network. Cell Cycle 2009, 8, 1833–1837. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Wang, L.; Lu, L.; Jiang, P.; Sun, H.; Wang, H. A novel target of microRNA-29, Ring1 and YY1-binding protein (Rybp), negatively regulates skeletal myogenesis. J. Biol. Chem. 2012, 287, 25255–25265. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Singhal, V.; Biswal, S.; Thimmulappa, R.K.; DiGirolamo, D.J. Nrf2 is required for normal postnatal bone acquisition in mice. Bone Res. 2014, 2, 14033. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Hassan, M.Q.; Jafferji, M.; Aqeilan, R.I.; Garzon, R.; Croce, C.M.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; Lian, J.B. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J. Biol. Chem. 2009, 284, 15676–15684. [Google Scholar] [CrossRef] [PubMed]
- Jeon, E.J.; Lee, K.Y.; Choi, N.S.; Lee, M.H.; Kim, H.N.; Jin, Y.H.; Ryoo, H.M.; Choi, J.Y.; Yoshida, M.; Nishino, N.; et al. Bone morphogenetic protein-2 stimulates Runx2 acetylation. J. Biol. Chem. 2006, 281, 16502–16511. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.S.; Alliston, T.; Delston, R.; Derynck, R. Repression of Runx2 function by TGF-beta through recruitment of class II histone deacetylases by Smad3. EMBO J. 2005, 24, 2543–2555. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Wang, Z. Adrenaline inhibits osteogenesis via repressing miR-21 expression. Cell Biol. Int. 2017, 41, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Bian, C.; Li, J.; Du, Z.; Zhou, H.; Yang, Z.; Zhao, R.C. miR-21 modulates the ERK-MAPK signaling pathway by regulating SPRY2 expression during human mesenchymal stem cell differentiation. J. Cell. Biochem. 2013, 114, 1374–1384. [Google Scholar] [CrossRef] [PubMed]
- Trohatou, O.; Zagoura, D.; Bitsika, V.; Pappa, K.I.; Antsaklis, A.; Anagnou, N.P.; Roubelakis, M.G. Sox2 suppression by miR-21 governs human mesenchymal stem cell properties. Stem Cells Transl. Med. 2014, 3, 54–68. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.F.; Yang, G.H.; Pan, X.H.; Zhang, S.J.; Zhao, C.; Qiu, B.S.; Gu, H.F.; Hong, J.F.; Cao, L.; Chen, Y.; et al. Altered microRNA expression profile in exosomes during osteogenic differentiation of human bone marrow-derived mesenchymal stem cells. PLoS ONE 2014, 9, e114627. [Google Scholar] [CrossRef] [PubMed]
- Ishitsuka, Y.; Huebner, A.J.; Rice, R.H.; Koch, P.J.; Speransky, V.V.; Steven, A.C.; Roop, D.R. Lce1 family members are Nrf2-target genes that are induced to compensate for the loss of loricrin. J. Investig. Dermatol. 2016, 136, 1656–1663. [Google Scholar] [CrossRef] [PubMed]
- Huebner, A.J.; Dai, D.; Morasso, M.; Schmidt, E.E.; Schäfer, M.; Werner, S.; Roop, D.R. Amniotic fluid activates the nrf2/keap1 pathway to repair an epidermal barrier defect in utero. Dev. Cell 2012, 23, 1238–1246. [Google Scholar] [CrossRef] [PubMed]
- Pastore, S.; Mascia, F.; Mariani, V.; Girolomoni, G. The epidermal growth factor receptor system in skin repair and inflammation. J. Investig. Dermatol. 2008, 128, 1365–1374. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Pickin, K.A.; Bose, R.; Jura, N.; Cole, P.A.; Kuriyan, J. Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface. Nature 2007, 450, 741–744. [Google Scholar] [CrossRef] [PubMed]
- Ying, H.; Zheng, H.; Scott, K.; Wiedemeyer, R.; Yan, H.; Lim, C.; Huang, J.; Dhakal, S.; Ivanova, E.; Xiao, Y.; et al. Mig-6 controls EGFR trafficking and suppresses gliomagenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 6912–6917. [Google Scholar] [CrossRef] [PubMed]
- Fiorentino, L.; Pertica, C.; Fiorini, M.; Talora, C.; Crescenzi, M.; Castellani, L.; Alemà, S.; Benedetti, P.; Segatto, O. Inhibition of ErbB-2 mitogenic and transforming activity by RALT, a mitogen-induced signal transducer which binds to the ErbB-2 kinase domain. Mol. Cell. Biol. 2000, 20, 7735–7750. [Google Scholar] [CrossRef] [PubMed]
- Ferby, I.; Reschke, M.; Kudlacek, O.; Knyazev, P.; Pantè, G.; Amann, K.; Sommergruber, W.; Kraut, N.; Ullrich, A.; Fässler, R.; et al. Mig6 is a negative regulator of EGF receptor-mediated skin morphogenesis and tumor formation. Nat. Med. 2006, 12, 568–573. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Zhang, Y.; Skalski, M.; Hayes, J.; Kefas, B.; Schiff, D.; Purow, B.; Parsons, S.; Lawler, S.; Abounader, R. microRNA-148a is a prognostic oncomiR that targets MIG6 and BIM to regulate EGFR and apoptosis in glioblastoma. Cancer Res. 2014, 74, 1541–1553. [Google Scholar] [CrossRef] [PubMed]
- Vu, H.L.; Rosenbaum, S.; Capparelli, C.; Purwin, T.J.; Davies, M.A.; Berger, A.C.; Aplin, A.E. MIG6 is MEK regulated and affects EGF-induced migration in mutant NRAS melanoma. J. Investig. Dermatol. 2016, 136, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C.; John, S.M.; Schmitz, G. Milk consumption during pregnancy increases birth weight, a risk factor for the development of diseases of civilization. J. Transl. Med. 2015, 13, 13. [Google Scholar] [CrossRef] [PubMed]
- Van Deutekom, A.W.; Chinapaw, M.J.; Vrijkotte, T.G.; Gemke, R.J. The association of birth weight and postnatal growth with energy intake and eating behavior at 5 years of age—A birth cohort study. Int. J. Behav. Nutr. Phys. Act. 2016, 13, 15. [Google Scholar] [CrossRef] [PubMed]
- Frayling, T.M. Genome-wide association studies provide new insights into type 2 diabetes aetiology. Nat. Rev. Genet. 2007, 8, 657–662. [Google Scholar] [CrossRef]
- Lindgren, C.M.; McCarthy, M.I. Mechanisms of disease: Genetic insights into the etiology of type 2 diabetes and obesity. Nat. Clin. Pract. Endocrinol. Metab. 2008, 4, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Basile, K.J.; Johnson, M.E.; Xia, Q.; Grant, S.F. Genetic susceptibility to type 2 diabetes and obesity: Follow-up of findings from genome-wide association studies. Int. J. Endocrinol. 2014, 2014, 769671. [Google Scholar] [CrossRef] [PubMed]
- Zdrojowy-Wełna, A.; Tupikowska, M.; Kolackov, K.; Bednarek-Tupikowska, G. The role of fat mass and obesity-associated gene (FTO) in obesity—An overview. Endokrynol. Pol. 2014, 65, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yang, Y.; Sun, B.F.; Zhao, Y.L.; Yang, Y.G. FTO and obesity: Mechanisms of association. Curr. Diabetes Rep. 2014, 14, 486. [Google Scholar] [CrossRef] [PubMed]
- Sebert, S.; Salonurmi, T.; Keinänen-Kiukaanniemi, S.; Savolainen, M.; Herzig, K.H.; Symonds, M.E.; Järvelin, M.R. Programming effects of FTO in the development of obesity. Acta Physiol. 2014, 210, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Wiley, A.S. Milk intake and total dairy consumption: Associations with early menarche in NHANES 1999–2004. PLoS ONE 2011, 6, e14685. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, E.L.; Grove, K.L. Metabolic imprinting in obesity. Forum Nutr. 2010, 63, 186–194. [Google Scholar] [PubMed]
- Koletzko, B. Childhood obesity: Current situation and future opportunities. J. Pediatr. Gastroenterol. Nutr. 2016, 63 (Suppl. S1), S18–S21. [Google Scholar] [PubMed]
- Smego, A.; Woo, J.G.; Klein, J.; Suh, C.; Bansal, D.; Bliss, S.; Daniels, S.R.; Bolling, C.; Crimmins, N.A. High body mass index in infancy may predict severe obesity in early childhood. J. Pediatr. 2016. [Google Scholar] [CrossRef] [PubMed]
- Gruszfeld, D.; Socha, P. Early nutrition and health: Short- and long-term outcomes. World Rev. Nutr. Diet. 2013, 108, 32–39. [Google Scholar] [PubMed]
- Reynolds, C.M.; Gray, C.; Li, M.; Segovia, S.A.; Vickers, M.H. Early life nutrition and energy balance disorders in offspring in later life. Nutrients 2015, 7, 8090–8111. [Google Scholar] [CrossRef] [PubMed]
- Egan, K.B.; Ettinger, A.S.; Bracken, M.B. Childhood body mass index and subsequent physician-diagnosed asthma: A systematic review and meta-analysis of prospective cohort studies. BMC Pediatr. 2013, 13, 121. [Google Scholar] [CrossRef] [PubMed]
- Brüske, I.; Flexeder, C.; Heinrich, J. Body mass index and the incidence of asthma in children. Curr. Opin. Allergy Clin. Immunol. 2014, 14, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C. The potential mechanistic link between allergy and obesity development and infant formula feeding. Allergy Asthma Clin. Immunol. 2014, 10, 37. [Google Scholar] [CrossRef] [PubMed]
- Günther, A.L.; Schulze, M.B.; Kroke, A.; Diethelm, K.; Joslowski, G.; Krupp, D.; Wudy, S.; Buyken, A.E. Early diet and later cancer risk: Prospective associations of dietary patterns during critical periods of childhood with the GH-IGF axis, insulin resistance and body fatness in younger adulthood. Nutr. Cancer 2015, 67, 877–892. [Google Scholar] [CrossRef] [PubMed]
- Von Bonsdorff, M.B.; Törmäkangas, T.; Rantanen, T.; Salonen, M.K.; Osmond, C.; Kajantie, E.; Eriksson, J.G. Early life body mass trajectories and mortality in older age: Findings from the Helsinki Birth Cohort Study. Ann. Med. 2015, 47, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Burdge, G.C.; Lillycrop, K.A.; Jackson, A.A. Nutrition in early life, and risk of cancer and metabolic disease: Alternative endings in an epigenetic tale? Br. J. Nutr. 2009, 101, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Bag, S.; Ramaiah, S.; Anbarasu, A. fabp4 is central to eight obesity associated genes: A functional gene Network-based polymorphic study. J. Theor. Biol. 2015, 364, 344–354. [Google Scholar] [CrossRef] [PubMed]
- Khalyfa, A.; Bhushan, B.; Hegazi, M.; Kim, J.; Kheirandish-Gozal, L.; Bhattacharjee, R.; Capdevila, O.S.; Gozal, D. Fatty-acid binding protein 4 gene variants and childhood obesity: Potential implications for insulin sensitivity and CRP levels. Lipids Health Dis. 2010, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- Erbay, E.; Babaev, V.R.; Mayers, J.R.; Makowski, L.; Charles, K.N.; Snitow, M.E.; Fazio, S.; Wiest, M.M.; Watkins, S.M.; Linton, M.F.; et al. Reducing endoplasmic reticulum stress through a macrophage lipid chaperone alleviates atherosclerosis. Nat. Med. 2009, 15, 1383–1391. [Google Scholar] [CrossRef] [PubMed]
- Zamaninour, N.; Mirzaei, K.; Keshavarz, S.A.; Ansar, H.; Hossein-Nezhad, A. New insight into determining indicators of metabolic status in women: Expression of PPARγ and FABP4 in PBMCs. Women Health 2016, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Cabré, A.; Babio, N.; Lázaro, I.; Bulló, M.; Garcia-Arellano, A.; Masana, L.; Salas-Salvadó, J. FABP4 predicts atherogenic dyslipidemia development. The PREDIMED study. Atherosclerosis 2012, 222, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Seeßle, J.; Liebisch, G.; Schmitz, G.; Stremmel, W.; Chamulitrat, W. Palmitate activation by fatty acid transport protein 4 as a model system for hepatocellular apoptosis and steatosis. Biochim. Biophys. Acta 2015, 1851, 549–565. [Google Scholar] [CrossRef] [PubMed]
- Terra, X.; Quintero, Y.; Auguet, T.; Porras, J.A.; Hernández, M.; Sabench, F.; Aguilar, C.; Luna, A.M.; Del Castillo, D.; Richart, C. FABP 4 is associated with inflammatory markers and metabolic syndrome in morbidly obese women. Eur. J. Endocrinol. 2011, 164, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Zhang, H.; Sun, Y.; Wang, Y.; Yang, X.; Yang, X.; Zhang, H.; Guo, W.; Zhu, G.; Tian, J.; et al. Modulation of FABP4 hypomethylation by DNMT1 and its inverse interaction with miR-148a/152 in the placenta of preeclamptic rats and HTR-8 cells. Placenta 2016, 46, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Michaëlsson, K.; Wolk, A.; Langenskiöld, S.; Basu, S.; Warensjö Lemming, E.; Melhus, H.; Byberg, L. Milk intake and risk of mortality and fractures in women and men: Cohort studies. BMJ 2014, 349, g6015. [Google Scholar] [CrossRef] [PubMed]
- Sekar, D.; Venugopal, B.; Sekar, P.; Ramalingam, K. Role of microRNA 21 in diabetes and associated/related diseases. Gene 2016, 582, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Bai, C.; Li, X.; Gao, Y.; Wang, K.; Fan, Y.; Zhang, S.; Ma, Y.; Guan, W. Role of microRNA-21 in the formation of insulin-producing cells from pancreatic progenitor cells. Biochim. Biophys. Acta 2016, 1859, 280–293. [Google Scholar] [CrossRef] [PubMed]
- Calo, N.; Ramadori, P.; Sobolewski, C.; Romero, Y.; Maeder, C.; Fournier, M.; Rantakari, P.; Zhang, F.P.; Poutanen, M.; Dufour, J.F.; et al. Stress-activated miR-21/miR-21* in hepatocytes promotes lipid and glucose metabolic disorders associated with high-fat diet consumption. Gut 2016. [Google Scholar] [CrossRef] [PubMed]
- Seeger, T.; Fischer, A.; Muhly-Reinholz, M.; Zeiher, A.M.; Dimmeler, S. Long-term inhibition of miR-21 leads to reduction of obesity in db/db mice. Obesity (Silver Spring) 2014, 22, 2352–2360. [Google Scholar] [CrossRef] [PubMed]
- Janghorbani, M.; Mansourian, M.; Hosseini, E. Systematic review and meta-analysis of age at menarche and risk of type 2 diabetes. Acta Diabetol. 2014, 51, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Sluijs, I.; Forouhi, N.G.; Beulens, J.W.; van der Schouw, Y.T.; Agnoli, C.; Arriola, L.; Balkau, B.; Barricarte, A.; Boeing, H.; Bueno-de-Mesquita, H.B.; et al. The amount and type of dairy product intake and incident type 2 diabetes: Results from the EPIC-InterAct Study. Am. J. Clin. Nutr. 2012, 96, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Hoppe, C.; Mølgaard, C.; Vaag, A.; Barkholt, V.; Michaelsen, K.F. High intakes of milk, but not meat, increase s-insulin and insulin resistance in 8-year-old boys. Eur. J. Clin. Nutr. 2005, 59, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Chavarro, J.E.; Cao, Y.; Qiu, W.; Mucci, L.; Sesso, H.D.; Stampfer, M.J.; Giovannucci, E.; Pollak, M.; Liu, S.; et al. Whole milk intake is associated with prostate cancer-specific mortality among U.S. male physicians. J. Nutr. 2013, 143, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C. The pathogenic role of persistent milk signaling in mTORC1- and milk-microRNA-driven type 2 diabetes mellitus. Curr. Diabetes Rev. 2015, 11, 46–62. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Zhong, M.; Zhao, W.; Wang, C.; Zhang, J.; Liu, X.; Li, Y.; Paudel, S.D.; Wang, Q.; Lou, T. Urinary miR-29 correlates with albuminuria and carotid intima-media thickness in type 2 diabetes patients. PLoS ONE 2013, 8, e82607. [Google Scholar] [CrossRef] [PubMed]
- Arnold, N.; Koppula, P.R.; Gul, R.; Luck, C.; Pulakat, L. Regulation of cardiac expression of the diabetic marker microRNA miR-29. PLoS ONE. 2014, 9, e103284. [Google Scholar] [CrossRef] [PubMed]
- Deiuliis, J.A. MicroRNAs as regulators of metabolic disease: Pathophysiologic significance and emerging role as biomarkers and therapeutics. Int. J. Obes. 2016, 40, 88–101. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, C.L.; Peck, B.C.; Fannin, E.E.; Beysen, C.; Miao, J.; Landstreet, S.R.; Ding, S.; Turaga, V.; Lund, P.K.; Turner, S.; et al. MicroRNA-29 fine-tunes the expression of key FOXA2-activated lipid metabolism genes and is dysregulated in animal models of insulin resistance and diabetes. Diabetes 2014, 63, 3141–3148. [Google Scholar] [CrossRef] [PubMed]
- Nicholas, L.M.; Rattanatray, L.; MacLaughlin, S.M.; Ozanne, S.E.; Kleemann, D.O.; Walker, S.K.; Morrison, J.L.; Zhang, S.; Muhlhäusler, B.S.; Martin-Gronert, M.S.; et al. Differential effects of maternal obesity and weight loss in the periconceptional period on the epigenetic regulation of hepatic insulin-signaling pathways in the offspring. FASEB J. 2013, 27, 3786–3796. [Google Scholar] [CrossRef] [PubMed]
- Shen, F.; Huang, W.; Huang, J.T.; Xiong, J.; Yang, Y.; Wu, K.; Jia, G.F.; Chen, J.; Feng, Y.Q.; Yuan, B.F.; et al. Decreased N(6)-methyladenosine in peripheral blood RNA from diabetic patients is associated with FTO expression rather than ALKBH5. J. Clin. Endocrinol. Metab. 2015, 100, E148–E154. [Google Scholar] [CrossRef] [PubMed]
- Su, D.; Zhang, C.L.; Gao, Y.C.; Liu, X.Y.; Li, C.P.; Huangfu, J.; Xiao, R. Gene expression and correlation of Pten and Fabp4 in liver, muscle, and adipose tissues of type 2 diabetes rats. Med. Sci. Monit. 2015, 21, 3616–3621. [Google Scholar] [CrossRef] [PubMed]
- Bagheri, R.; Qasim, A.N.; Mehta, N.N.; Terembula, K.; Kapoor, S.; Braunstein, S.; Schutta, M.; Iqbal, N.; Lehrke, M.; Reilly, M.P. Relation of plasma fatty acid binding proteins 4 and 5 with the metabolic syndrome, inflammation and coronary calcium in patients with type-2 diabetes mellitus. Am. J. Cardiol. 2010, 106, 1118–1123. [Google Scholar] [CrossRef] [PubMed]
- Furuhashi, M.; Saitoh, S.; Shimamoto, K.; Miura, T. Fatty acid-binding protein 4 (FABP4): Pathophysiological insights and potent clinical biomarker of metabolic and cardiovascular diseases. Clin. Med. Insights Cardiol. 2015, 8 (Suppl. 3), 23–33. [Google Scholar] [CrossRef] [PubMed]
- Zielke, L.G.; Bortfeldt, R.H.; Reissmann, M.; Tetens, J.; Thaller, G.; Brockmann, G.A. Impact of variation at the FTO locus on milk fat yield in Holstein dairy cattle. PLoS ONE 2013, 8, e63406. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Cheng, L.; Azimu, W.; Hodge, S.; Edwards, G.R.; Hickford, J.G. Variation in the bovine FABP4 gene affects milk yield and milk protein content in dairy cows. Sci. Rep. 2015, 5, 10023. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Sundquist, J.; Sundquist, K. Lactose intolerance and risk of lung, breast and ovarian cancers: Aetiological clues from a population-based study in Sweden. Br. J. Cancer 2015, 112, 149–152. [Google Scholar] [CrossRef] [PubMed]
- Gaard, M.; Tretli, S.; Løken, E.B. Dietary fat and the risk of breast cancer: A prospective study of 25,892 Norwegian women. Int. J. Cancer 1995, 63, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.; Dang, Y.; Chen, G.; Mo, Z. Overexpression of the fat mass and obesity associated gene (FTO) in breast cancer and its clinical implications. Int. J. Clin. Exp. Pathol. 2015, 8, 13405–13410. [Google Scholar] [PubMed]
- Guaita-Esteruelas, S.; Bosquet, A.; Saavedra, P.; Gumà, J.; Girona, J.; Lam, E.W.; Amillano, K.; Borràs, J.; Masana, L. Exogenous FABP4 increases breast cancer cell proliferation and activates the expression of fatty acid transport proteins. Mol. Carcinog. 2017, 56, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Chen, H.; Niu, Y.; Wu, H.; Xia, D.; Wu, Y. Dairy products intake and cancer mortality risk: A meta-analysis of 11 population-based cohort studies. Nutr. J. 2016, 15, 91. [Google Scholar] [CrossRef] [PubMed]
- Murata, T.; Takayama, K.; Katayama, S.; Urano, T.; Horie-Inoue, K.; Ikeda, K.; Takahashi, S.; Kawazu, C.; Hasegawa, A.; Ouchi, Y.; et al. miR-148a is an androgen-responsive microRNA that promotes LNCaP prostate cell growth by repressing its target CAND1 expression. Prostate Cancer Prostatic Dis. 2010, 13, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Tate, P.L.; Bibb, R.; Larcom, L.L. Milk stimulates growth of prostate cancer cells in culture. Nutr. Cancer 2011, 63, 1361–1366. [Google Scholar] [CrossRef] [PubMed]
- Day, K.C.; Hiles, G.L.; Kozminsky, M.; Dawsey, S.J.; Paul, A.; Broses, L.J.; Shah, R.; Kunja, L.P.; Hall, C.; Palanisamy, N.; et al. HER2 and EGFR overexpression support metastatic progression of prostate cancer to bone. Cancer Res. 2017, 77, 74–85. [Google Scholar] [CrossRef] [PubMed]
- Siu, A.; Virtanen, C.; Jongstra, J. PIM kinase isoform specific regulation of MIG6 expression and EGFR signaling in prostate cancer cells. Oncotarget 2011, 2, 1134–1144. [Google Scholar] [CrossRef] [PubMed]
- Ralston, R.A.; Truby, H.; Palermo, C.E.; Walker, K.Z. Colorectal cancer and nonfermented milk, solid cheese, and fermented milk consumption: A systematic review and meta-analysis of prospective studies. Crit. Rev. Food Sci. Nutr. 2014, 54, 1167–1179. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Cuatrecasas, M.; Balaguer, F.; Hur, K.; Toiyama, Y.; Castells, A.; Boland, C.R.; Goel, A. The clinical significance of MiR-148a as a predictive biomarker in patients with advanced colorectal cancer. PLoS ONE 2012, 7, e46684. [Google Scholar] [CrossRef] [PubMed]
- Hibino, Y.; Sakamoto, N.; Naito, Y.; Goto, K.; Oo, H.Z.; Sentani, K.; Hinoi, T.; Ohdan, H.; Oue, N.; Yasui, W. Significance of miR-148a in colorectal neoplasia: Downregulation of miR-148a contributes to the carcinogenesis and cell invasion of colorectal cancer. Pathobiology 2015, 82, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Deng, X.; Zeng, X.; Peng, X. The role of mir-148a in cancer. J. Cancer 2016, 7, 1233–1241. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.L.; Yang, L.F.; Zhu, Y.; Yao, X.D.; Zhang, S.L.; Dai, B.; Zhu, Y.P.; Shen, Y.J.; Shi, G.H.; Ye, D.W. Serum miRNA-21: Elevated levels in patients with metastatic hormone-refractory prostate cancer and potential predictive factor for the efficacy of docetaxel-based chemotherapy. Prostate 2011, 71, 326–331. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Hruby, G.W.; McKiernan, J.M.; Gurvich, I.; Lipsky, M.J.; Benson, M.C.; Santella, R.M. Dysregulation of circulating microRNAs and prediction of aggressive prostate cancer. Prostate 2012, 72, 1469–1477. [Google Scholar] [CrossRef] [PubMed]
- Filella, X.; Foj, L. Prostate cancer detection and prognosis: From prostate specific antigen (PSA) to exosomal biomarkers. Int. J. Mol. Sci. 2016, 17, E1784. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Guo, Y.; Wang, Z.; Dai, Z.; Xu, Y.; Zhang, W.; Liu, Z.; Li, S. Analysis of circulating miRNAs 21 and 375 as potential biomarkers for early diagnosis of prostate cancer. Neoplasma 2016, 63, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.H.; Tsao, C.J. Emerging role of microRNA-21 in cancer. Biomed. Rep. 2016, 5, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C. MiR-21: An environmental driver of malignant melanoma? J. Transl. Med. 2015, 13, 202. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.R.; Yang, C.H.; Pfeffer, L.M. The role of miR-21 in cancer. Drug Dev. Res. 2015, 76, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Shi, J. Considering exosomal miR-21 as a biomarker for cancer. J. Clin. Med. 2016, 5, E42. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Liu, H.; Zou, H.; Chen, R.; Dou, Y.; Sheng, S.; Dai, S.; Ai, J.; Melson, J.; Kittles, R.A.; et al. Evaluation of plasma miR-21 and miR-152 as diagnostic biomarkers for common types of human cancers. J. Cancer 2016, 7, 490–499. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.T.; Liu, S.M.; Ma, H.; Yang, Y.; Zhang, X.; Sun, H.; Zhang, X.; Xu, J.; Wang, J. Systematic review and meta-analysis: Circulating miRNAs for diagnosis of hepatocellular carcinoma. J. Cell. Physiol. 2016, 231, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Shen, Q.; Li, C.; Li, D.; Chen, J.; He, M. Identification of circulating microRNAs as novel potential biomarkers for hepatocellular carcinoma detection: A systematic review and meta-analysis. Clin. Transl. Oncol. 2015, 17, 684–693. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Hou, L.; Li, A.; Duan, Y.; Gao, H.; Song, X. Expression of serum exosomal microRNA-21 in human hepatocellular carcinoma. Biomed. Res. Int. 2014, 2014, 864894. [Google Scholar] [CrossRef] [PubMed]
- Duarte-Salles, T.; Fedirko, V.; Stepien, M.; Trichopoulou, A.; Bamia, C.; Lagiou, P.; Lukanova, A.; Trepo, E.; Overvad, K.; Tjønneland, A.; et al. Dairy products and risk of hepatocellular carcinoma: The European Prospective Investigation into Cancer and Nutrition. Int. J. Cancer 2014, 135, 1662–1672. [Google Scholar] [CrossRef] [PubMed]
- Chouliaras, L.; Rutten, B.P.; Kenis, G.; Peerbooms, O.; Visser, P.J.; Verhey, F.; van Os, J.; Steinbusch, H.W.; van den Hove, D.L. Epigenetic regulation in the pathophysiology of Alzheimer’s disease. Prog. Neurobiol. 2010, 90, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Otaegui-Arrazola, A.; Amiano, P.; Elbusto, A.; Urdaneta, E.; Martínez-Lage, P. Diet, cognition, and Alzheimer’s disease: Food for thought. Eur. J. Nutr. 2014, 53, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Davinelli, S.; Calabrese, V.; Zella, D.; Scapagnini, G. Epigenetic nutraceutical diets in Alzheimer’s disease. J. Nutr. Health Aging 2014, 18, 800–805. [Google Scholar] [CrossRef] [PubMed]
- Lardenoije, R.; Iatrou, A.; Kenis, G.; Kompotis, K.; Steinbusch, H.W.; Mastroeni, D.; Coleman, P.; Lemere, C.A.; Hof, P.R.; van den Hove, D.L.; et al. The epigenetics of aging and neurodegeneration. Prog. Neurobiol. 2015, 131, 21–64. [Google Scholar] [CrossRef] [PubMed]
- Wüllner, U.; Kaut, O.; deBoni, L.; Piston, D.; Schmitt, I. DNA methylation in Parkinson‘s disease. J. Neurochem. 2016, 139 (Suppl. 1), 108–120. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Bereczki, E.; Zhang, H.; Wang, S.; Li, C.; Ji, X.; Branca, R.M.; Lehtiö, J.; Guan, Z.; Filipcik, P.; et al. Mammalian target of rapamycin (mTor) mediates tau protein dyshomeostasis: Implication for Alzheimer disease. J. Biol. Chem. 2013, 288, 15556–15570. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Ioja, E.; Bereczki, E.; Hultenby, K.; Li, C.; Guan, Z.; Winblad, B.; Pei, J.J. mTor mediates tau localization and secretion: Implication for Alzheimer’s disease. Biochim. Biophys Acta 2015, 1853, 1646–1657. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.X.; Ji, X.; Mao, X.; Xie, L.; Jia, J.; Galvan, V.; Greenberg, D.A.; Jin, K. Differential activation of mTOR complex 1 signaling in human brain with mild to severe Alzheimer’s disease. J. Alzheimers Dis. 2014, 38, 437–444. [Google Scholar] [PubMed]
- Leszek, J.; Trypka, E.; Tarasov, V.V.; Md Ashraf, G. Type 3 diabetes mellitus: A novel implication of Alzheimer Disease. Curr. Top. Med. Chem. 2017. [Google Scholar] [CrossRef]
- Ho, A.J.; Stein, J.L.; Hua, X.; Lee, S.; Hibar, D.P.; Leow, A.D.; Dinov, I.D.; Toga, A.W.; Saykin, A.J.; Shen, L.; et al. Alzheimer’s Disease Neuroimaging Initiative. A commonly carried allele of the obesity-related FTO gene is associated with reduced brain volume in the healthy elderly. Proc. Natl. Acad. Sci. USA 2010, 107, 8404–8409. [Google Scholar] [CrossRef] [PubMed]
- Benedict, C.; Jacobsson, J.A.; Rönnemaa, E.; Sällman-Almén, M.; Brooks, S.; Schultes, B.; Fredriksson, R.; Lannfelt, L.; Kilander, L.; Schiöth, H.B. The fat mass and obesity gene is linked to reduced verbal fluency in overweight and obese elderly men. Neurobiol. Aging 2011, 32, 1159.e1–1159.e5. [Google Scholar] [CrossRef] [PubMed]
- De Groot, C.; Felius, A.; Trompet, S.; de Craen, A.J.; Blauw, G.J.; van Buchem, M.A.; Delemarre-van de Waal, H.A.; van der Grond, J. Association of the fat mass and obesity-associated gene risk allele, rs9939609A, and reward-related brain structures. Obesity (Silver Spring) 2015, 23, 2118–2122. [Google Scholar] [CrossRef] [PubMed]
- Keller, L.; Xu, W.; Wang, H.X.; Winblad, B.; Fratiglioni, L.; Graff, C. The obesity related gene, FTO, interacts with APOE, and is associated with Alzheimer’s disease risk: A prospective cohort study. J. Alzheimers Dis. 2011, 23, 461–469. [Google Scholar] [PubMed]
- Yamagata, K.; Urakami, K.; Ikeda, K.; Ji, Y.; Adachi, Y.; Arai, H.; Sasaki, H.; Sato, K.; Nakashima, K. High expression of apolipoprotein E mRNA in the brains with sporadic Alzheimer’s disease. Dement. Geriatr. Cogn. Disord. 2001, 12, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Caesar, I.; Gandy, S. Evidence that an APOE ε4 ‘double whammy’ increases risk for Alzheimer’s disease. BMC Med. 2012, 10, 36. [Google Scholar] [CrossRef] [PubMed]
- Castellano, J.M.; Kim, J.; Stewart, F.R.; Jiang, H.; DeMattos, R.B.; Patterson, B.W.; Fagan, A.M.; Morris, J.C.; Mawuenyega, K.G.; Cruchaga, C.; et al. Human apoE isoforms differentially regulate brain amyloid-β peptide clearance. Sci. Transl. Med. 2011, 3, 89ra57. [Google Scholar] [CrossRef] [PubMed]
- Pankiewicz, J.E.; Guridi, M.; Kim, J.; Asuni, A.A.; Sanchez, S.; Sullivan, P.M.; Holtzman, D.M.; Sadowski, M.J. Blocking the apoE/Aβ interaction ameliorates Aβ-related pathology in APOE ε2 and ε4 targeted replacement Alzheimer model mice. Acta Neuropathol. Commun. 2014, 2, 75. [Google Scholar] [CrossRef] [PubMed]
- Foraker, J.; Millard, S.P.; Leong, L.; Thomson, Z.; Chen, S.; Keene, C.D.; Bekris, L.M.; Yu, C.E. The APOE gene is differentially methylated in Alzheimer’s Disease. J. Alzheimers Dis. 2015, 48, 745–755. [Google Scholar] [CrossRef] [PubMed]
- Reitz, C.; Tosto, G.; Mayeux, R.; Luchsinger, J.A.; NIA-LOAD/NCRAD Family Study Group; Alzheimer’s Disease Neuroimaging Initiative. Genetic variants in the Fat and Obesity Associated (FTO) gene and risk of Alzheimer’s disease. PLoS ONE 2012, 7, e50354. [Google Scholar] [CrossRef] [PubMed]
- Adebakin, A.; Bradley, J.; Gümüsgöz, S.; Waters, E.J.; Lawrence, C.B. Impaired satiation and increased feeding behaviour in the triple-transgenic Alzheimer’s disease mouse model. PLoS ONE 2012, 7, e45179. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, S.M.; Hernán, M.A.; Willett, W.C.; Ascherio, A. Diet and Parkinson’s disease: A potential role of dairy products in men. Ann. Neurol. 2002, 52, 793–801. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Ross, G.W.; Petrovitch, H.; White, L.R.; Masaki, K.H.; Nelson, J.S.; Tanner, C.M.; Curb, J.D.; Blanchette, P.L.; Abbott, R.D. Consumption of milk and calcium in midlife and the future risk of Parkinson disease. Neurology 2005, 64, 1047–1051. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; O’Reilly, E.; McCullough, M.L.; Rodriguez, C.; Schwarzschild, M.A.; Calle, E.E.; Thun, M.J.; Ascherio, A. Consumption of dairy products and risk of Parkinson’s disease. Am. J. Epidemiol. 2007, 165, 998–1006. [Google Scholar] [CrossRef] [PubMed]
- Kyrozis, A.; Ghika, A.; Stathopoulos, P.; Vassilopoulos, D.; Trichopoulou, D.; Trichopoulou, A. Dietary and lifestyle variables in relation to incidence of Parkinson’s disease in Greece. Eur. J. Epidemiol. 2013, 28, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Sääksjärvi, K.; Knekt, P.; Lundqvist, A.; Männistö, S.; Heliövaara, M.; Rissanen, H.; Järvinen, R. A cohort study on diet and the risk of Parkinson’s disease: The role of food groups and diet quality. Br. J. Nutr. 2013, 109, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Ju, C.; Jiang, H.; Zhang, D. Dairy foods intake and risk of Parkinson’s disease: A dose-response meta-analysis of prospective cohort studies. Eur. J. Epidemiol. 2014, 29, 613–619. [Google Scholar] [CrossRef] [PubMed]
- Abbott, R.D.; Ross, G.W.; Petrovitch, H.; Masaki, K.H.; Launer, L.J.; Nelson, J.S.; White, L.R.; Tanner, C.M. Midlife milk consumption and substantia nigra neuron density at death. Neurology 2016, 86, 512–519. [Google Scholar] [CrossRef] [PubMed]
- Xu, X. DNA methylation and cognitive aging. Oncotarget 2015, 6, 13922–13932. [Google Scholar] [CrossRef] [PubMed]
- Jowaed, A.; Schmitt, I.; Kaut, O.; Wüllner, U. Methylation regulates alpha-synuclein expression and is decreased in Parkinson’s disease patients’ brains. J. Neurosci. 2010, 30, 6355–6359. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, L.; Takuma, H.; Tamaoka, A.; Kurisaki, H.; Date, H.; Tsuji, S.; Iwata, A. CpG demethylation enhances alpha-synuclein expression and affects the pathogenesis of Parkinson’s disease. PLoS ONE 2010, 5, e15522. [Google Scholar] [CrossRef] [PubMed]
- Swinnen, E.; Büttner, S.; Outeiro, T.F.; Galas, M.C.; Madeo, F.; Winderickx, J.; Franssens, V. Aggresome formation and segregation of inclusions influence toxicity of α-synuclein and synphilin-1 in yeast. Biochem. Soc. Trans. 2011, 39, 1476–1481. [Google Scholar] [CrossRef] [PubMed]
- Engelender, S.; Kaminsky, Z.; Guo, X.; Sharp, A.H.; Amaravi, R.K.; Kleiderlein, J.J.; Margolis, R.L.; Troncoso, J.C.; Lanahan, A.A.; Worley, P.F.; et al. Synphilin-1 associates with alpha-synuclein and promotes the formation of cytosolic inclusions. Nat. Genet. 1999, 22, 110–114. [Google Scholar] [PubMed]
- Outeiro, T.F.; Lindquist, S. Yeast cells provide insight into alpha-synuclein biology and pathobiology. Science 2003, 302, 1772–1775. [Google Scholar] [CrossRef] [PubMed]
- Goris, A.; Williams-Gray, C.H.; Clark, G.R.; Foltynie, T.; Lewis, S.J.; Brown, J.; Ban, M.; Spillantini, M.G.; Compston, A.; Burn, D.J.; et al. Tau and alpha-synuclein in susceptibility to, and dementia in, Parkinson’s disease. Ann. Neurol. 2007, 62, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Collins, L.M.; Williams-Gray, C.H. The genetic basis of cognitive impairment and dementia in Parkinson’s disease. Front. Psychiatry 2016, 7, 89. [Google Scholar] [CrossRef] [PubMed]
- Desplats, P.; Spencer, B.; Coffee, E.; Patel, P.; Michael, S.; Patrick, C.; Adame, A.; Rockenstein, E.; Masliah, E. Alpha-synuclein sequesters Dnmt1 from the nucleus: A novel mechanism for epigenetic alterations in Lewy body diseases. J. Biol. Chem. 2011, 286, 9031–9037. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.W.; Tseng, C.W.; Chien, C.W.; Huang, H.C.; Ku, W.C.; Lee, S.J.; Chen, Y.J.; Juan, H.F. Quantitative proteomics reveals diverse roles of miR-148a from gastric cancer progression to neurological development. J. Proteome Res. 2013, 12, 3993–4004. [Google Scholar] [CrossRef] [PubMed]
- Zoncu, R.; Efeyan, A.; Sabatini, D.M. mTOR: From growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 2011, 12, 21–35. [Google Scholar] [CrossRef] [PubMed]
- Dazert, E.; Hall, M.N. mTOR signaling in disease. Curr. Opin. Cell Biol. 2011, 23, 744–755. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C.; John, S.M.; Carrera-Bastos, P.; Cordain, L. The impact of cow’s milk-mediated mTORC1-signaling in the initiation and progression of prostate cancer. Nutr. Metab. (Lond.) 2012, 9, 74. [Google Scholar] [CrossRef] [PubMed]
- Oddo, S. The role of mTOR signaling in Alzheimer disease. Front. Biosci. (Schol. Ed.) 2012, 4, 941–952. [Google Scholar] [CrossRef] [PubMed]
- Cornu, M.; Albert, V.; Hall, M.N. mTOR in aging, metabolism, and cancer. Curr. Opin. Genet. Dev. 2013, 23, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Perl, A. mTOR activation is a biomarker and a central pathway to autoimmune disorders, cancer, obesity, and aging. Ann. N. Y. Acad. Sci. 2015, 1346, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Steelman, L.S.; Martelli, A.M.; Cocco, L.; Libra, M.; Nicoletti, F.; Abrams, S.L.; McCubrey, J.A. The therapeutic potential of mTOR inhibitors in breast cancer. Br. J. Clin. Pharmacol. 2016, 82, 1189–1212. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C.; Schmitz, G. Metformin: An inhibitor of mTORC1 signaling. J. Endocrinol. Diabetes Obes. 2014, 2, 1029. [Google Scholar]
- Zhong, T.; Men, Y.; Lu, L.; Geng, T.; Zhou, J.; Mitsuhashi, A.; Shozu, M.; Maihle, N.J.; Carmichael, G.G.; Taylor, H.S.; et al. Metformin alters DNA methylation genome-wide via the H19/SAHH axis. Oncogene 2016. [Google Scholar] [CrossRef] [PubMed]
- Tehlivets, O.; Malanovic, N.; Visram, M.; Pavkov-Keller, T.; Keller, W. S-adenosyl-l-homocysteine hydrolase and methylation disorders: Yeast as a model system. Biochim. Biophys. Acta 2013, 1832, 204–215. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yang, L.; Zhong, T.; Mueller, M.; Men, Y.; Zhang, N.; Xie, J.; Giang, K.; Chung, H.; Sun, X.; et al. H19 lncRNA alters DNA methylation genome wide by regulating S-adenosylhomocysteine hydrolase. Nat. Commun. 2015, 6, 10221. [Google Scholar] [CrossRef] [PubMed]
- Saha, A.; Blando, J.; Tremmel, L.; DiGiovanni, J. Effect of metformin, rapamycin, and their combination on growth and progression of prostate tumors in HiMyc mice. Cancer Prev. Res. (Phila.) 2015, 8, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Revuelta, B.I.; Hettich, M.M.; Ciociaro, A.; Rotermund, C.; Kahle, P.J.; Krauss, S.; Di Monte, D.A. Metformin lowers Ser-129 phosphorylated α-synuclein levels via mTOR-dependent protein phosphatase 2A activation. Cell Death Dis. 2014, 5, e1209. [Google Scholar] [CrossRef] [PubMed]
- Kickstein, E.; Krauss, S.; Thornhill, P.; Rutschow, D.; Zeller, R.; Sharkey, J.; Williamson, R.; Fuchs, M.; Köhler, A.; Glossmann, H.; et al. Biguanide metformin acts on tau phosphorylation via mTOR/protein phosphatase 2A (PP2A) signaling. Proc. Natl. Acad. Sci. USA 2010, 107, 21830–21835. [Google Scholar] [CrossRef] [PubMed]
- Rotllan, N.; Price, N.; Pati, P.; Goedeke, L.; Fernández-Hernando, C. microRNAs in lipoprotein metabolism and cardiometabolic disorders. Atherosclerosis 2016, 246, 352–360. [Google Scholar] [CrossRef] [PubMed]
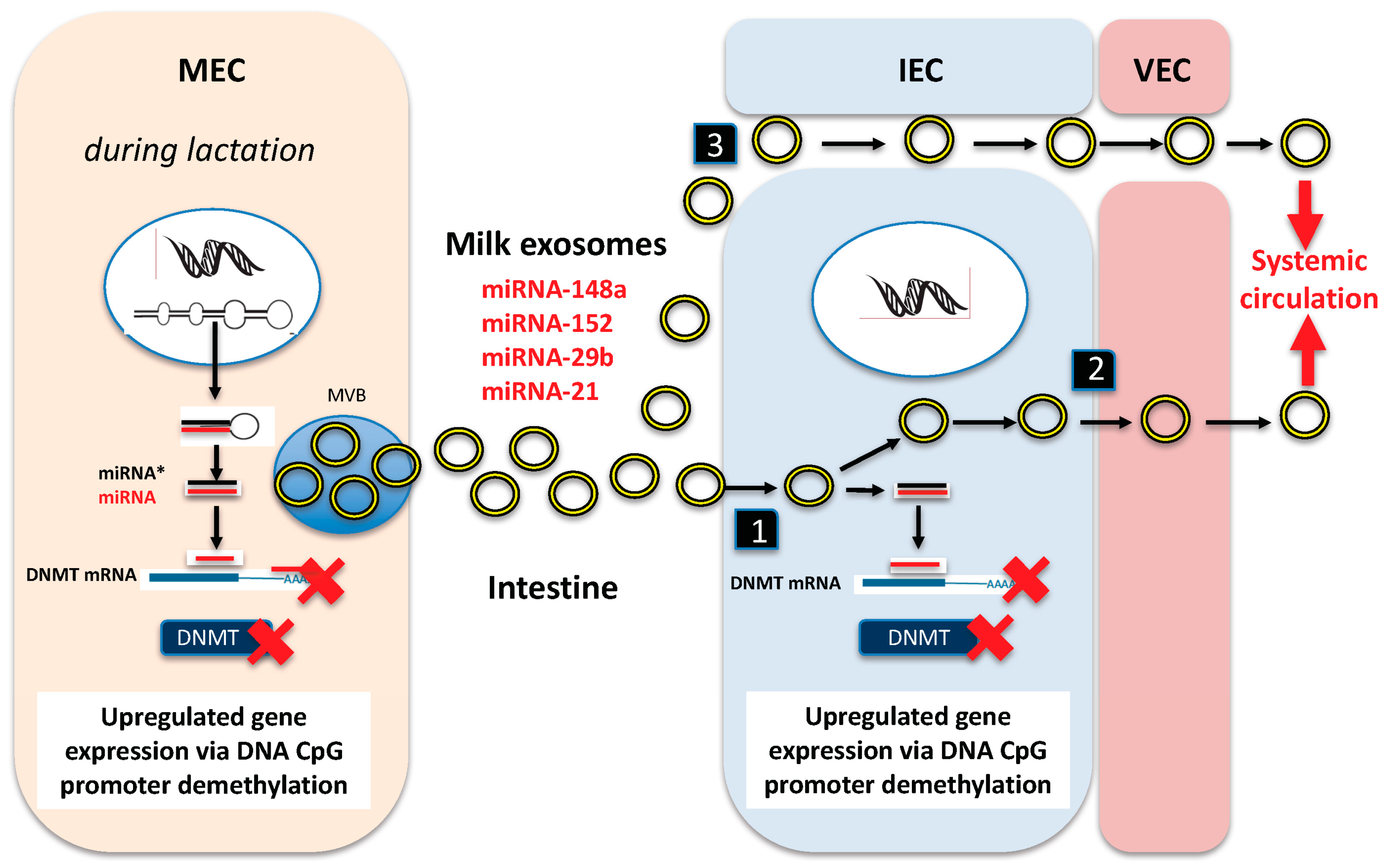
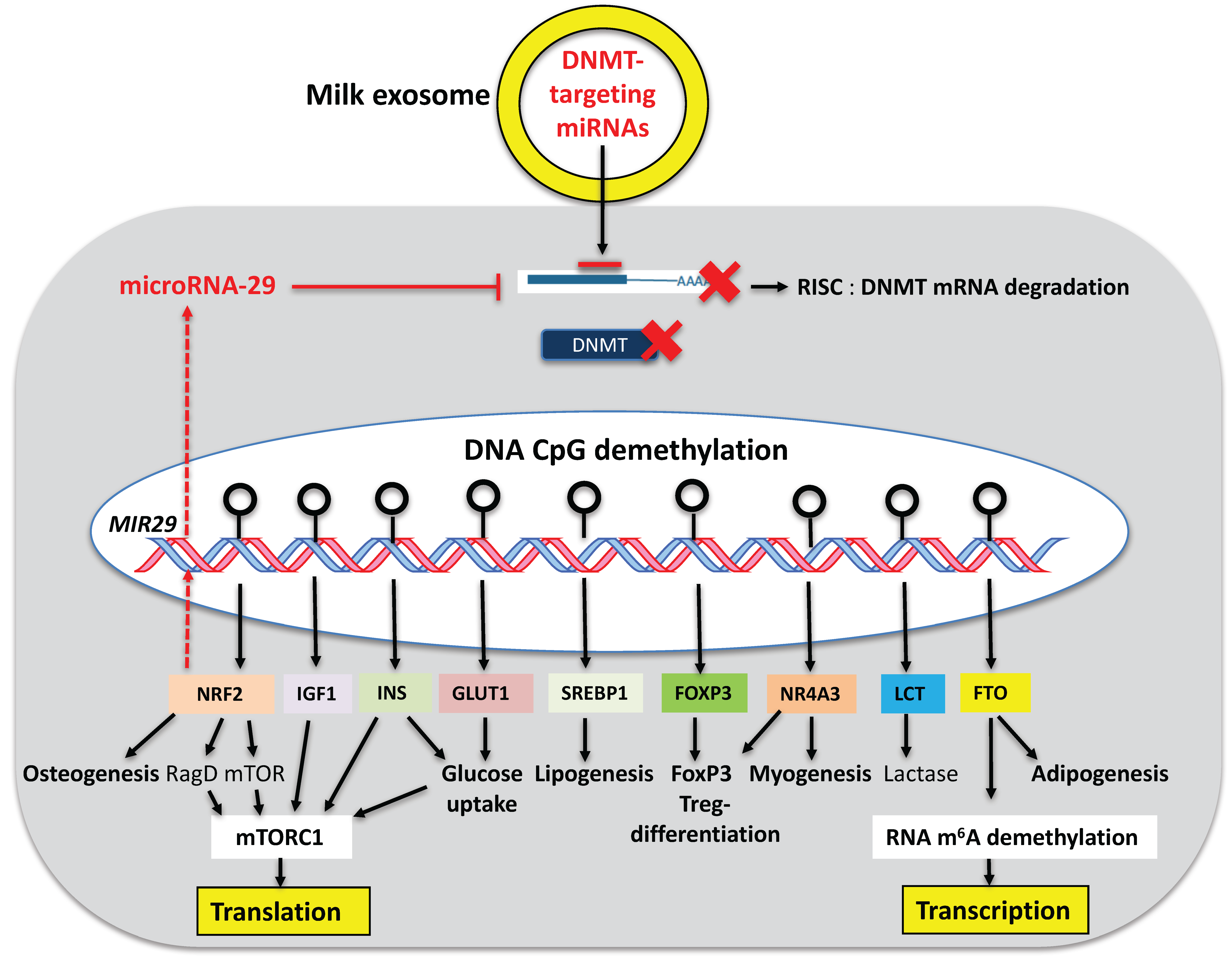
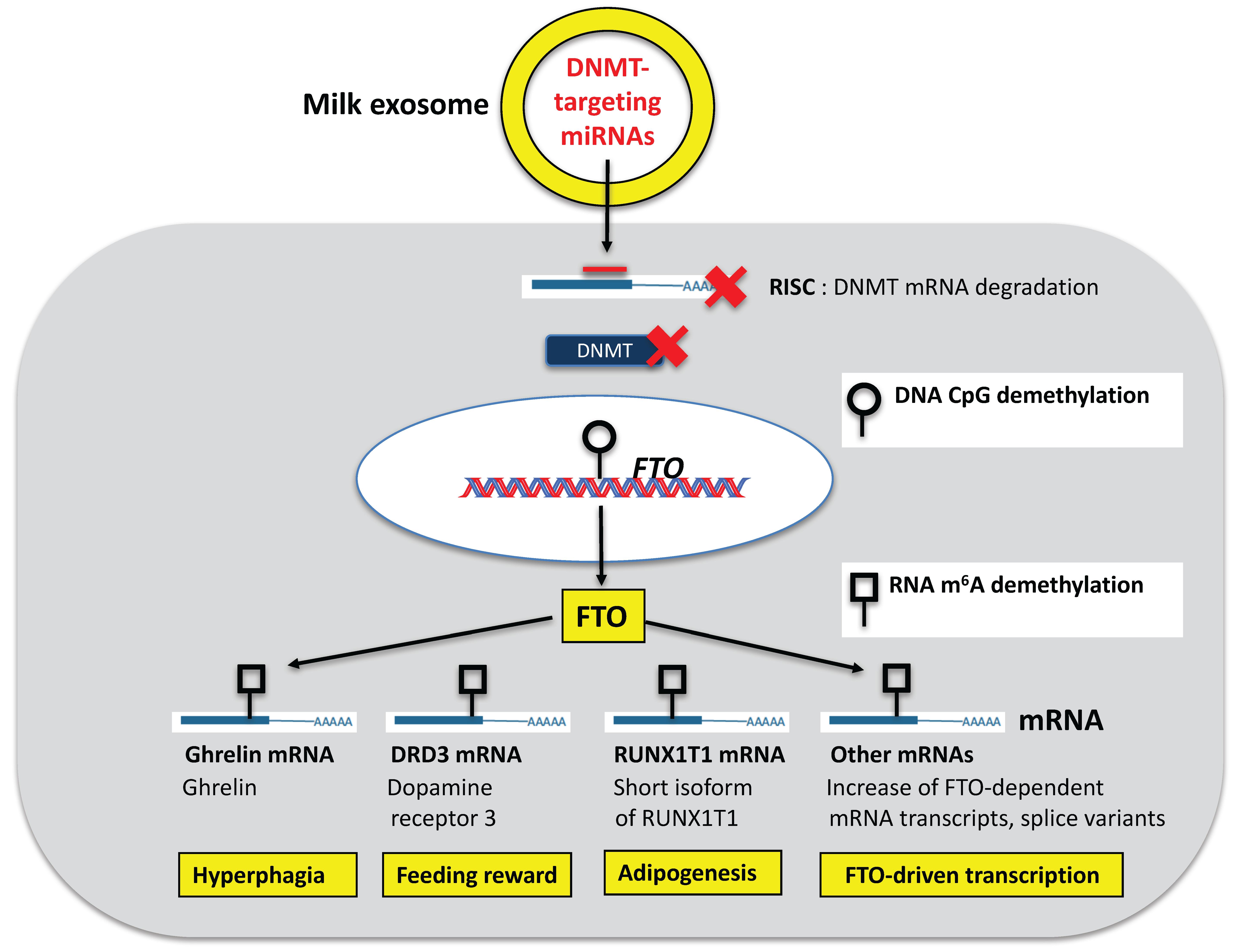
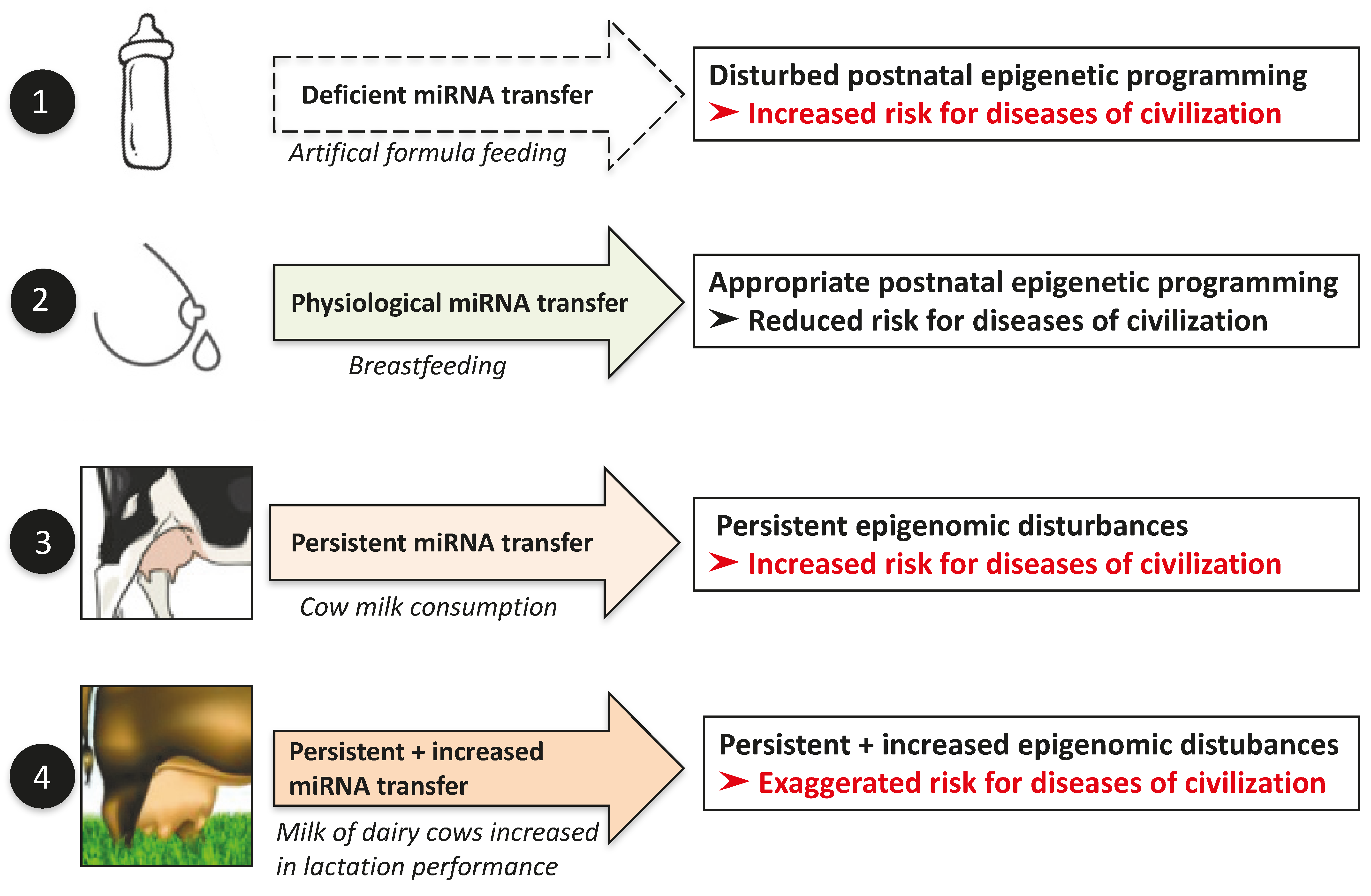
| The majority of milk exosomes is secreted by mammary epithelial cells | [14] |
| Milk exsomes resist intestinal degradation | [38,40,43,61,62,66] |
| Milk exosomes are taken up by intestinal epithelial cells | [75,76,77] |
| Milk exosomes are taken up by vascular endothelial cells | [78] |
| Increased serum levels of milk-derived miRNAs during lactation | [41,46] |
| Dose-dependent increase of miRNA-29b and miRNA-200c in the serum of cow’s milk consumers | [79] |
| Increase of miRNA-29b and miRNA-200c in peripheral blood mononuclear cells of human volunteers 6 h after commercial milk intake | [79] |
| Increased expression of RUNX2, a regulatory target of miRNA-29b, in PBMCs of healthy humans after cow’s milk consumption | [79] |
| Detection of bovine milk exosomes in murine splenocytes | [75] |
| Predicted role of milk miRNAs in organismal development and organ maturation | [17,80,81] |
| Human miRNAs Targeting DNMTs | Bovine miRNAs Targeting DNMTs |
|---|---|
| hsa-miRNA-148a-5p 6-aaaguucugagacacuccgacu-27 hsa-miRNA-148a-3p 44-ucagugcacuacagaacuuugu-65 | bta-miRNA-148a-3p 44-ucagugcacuacagaacuuugu-65 |
| hsa-miRNA-21-5p 8-uagcuuaucagacugauguuga-29 hsa-miRNA-21-3p 46-caacaccagucgaugggcugu-66 | bta-miRNA-21-5p 8-uagcuuaucagacugauguugacu-31 bta-miRNA-21-3p 47-aacagcagucgaugggcugucu-68 |
| hsa-miRNA-29b-1-5p 10-gcugguuucauauggugguuuaga-33 hsa-miRNA-29b-1-3p 51-uagcaccauuugaaaucaguguu-73 | bta-miRNA-29-1-3p 51-uagcaccauuugaaaucaguguu-73 |
| mRNA | Function | References |
|---|---|---|
| RUNX1T1 | Promotion of adipogenesis, mitotic clonal expansion increasing adipocyte numbers | [152,247] |
| PPARγ | Promotion of adipogenesis | [248] |
| CEBPα | Promotion of adipogenesis | [248] |
| PGC1α | Promotion of adipogenesis | [249] |
| Ghrelin | Increased ghrelin mRNA and protein expression, increased orexigenic signaling | [228] |
| Dopamine receptor 2 and 3 | Increased dopaminergic signaling potentially involved in feeding reward | [239] |
| Target | Reported Biological Effects of miRNA-148a | References |
|---|---|---|
| DNMT1 | Reduced maintenance DNA methylation during cell division | [118,244] |
| DNMT3B | Reduced de novo DNA methylation | [121] |
| ABCA1 | Reduced reverse cholesterol transport, risk of dyslipidemia | [106] |
| LDLR | Reduced hepatic uptake of LDL, risk of dyslipidemia | [106] |
| CPT1A | Reduced mitochondrial fatty acid β-oxidation, risk of dyslipidemia | [106] |
| MIG6 | Reduced inhibition of EGFR, increased cell proliferation | [278] |
| ROCK1 | Reduced suppression of myogenesis, enhanced myogenesis | [261] |
| Gene | Functions | References |
|---|---|---|
| FTO | Increased RNA m6A demethylation, resulting in increased transcription, generation of adipogenic splice variant (short form) of RUNX1T1 | [152,154,155,156] |
| INS | Increased insulin expression, activation of mTORC1, increased glucose uptake, anabolism | [170] |
| IGF1 | Increased IGF-1 expression, activation of mTORC1, promotion of growth and GH signaling | [174,175] |
| CAV1 | Stimulation of insulin- and IGF-1 receptor signal transduction, promotion of adipocyte differentiation | [179] |
| FABP4 | Adipogenic differentiation | [243] |
| LPL | Adipogenic differentiation | [243] |
| NRF2 | Increased expression of mTOR, RagD, promotion of mTORC1 signaling, promotion of osteogenesis | [162,163,164,165,166] |
| NR4A3 | Promotion of myogenesis and FoxP3 expression | [201,202,207,253] |
| FOXP3 | Increased FoxP3 expression, differentiation and stable expression of regulatory T cells, induction of immune tolerance, prevention of allergy | [183,184,185,186,187] |
| APOE | Decreased isotype-specific APOE methylation in brains of patients with Alzheimer’s disease | [372] |
| SNCA | Decreased methylation at SNCA intron 1 in patients with Parkinson’s disease | [383,384] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Melnik, B.C.; Schmitz, G. Milk’s Role as an Epigenetic Regulator in Health and Disease. Diseases 2017, 5, 12. https://doi.org/10.3390/diseases5010012
Melnik BC, Schmitz G. Milk’s Role as an Epigenetic Regulator in Health and Disease. Diseases. 2017; 5(1):12. https://doi.org/10.3390/diseases5010012
Chicago/Turabian StyleMelnik, Bodo C., and Gerd Schmitz. 2017. "Milk’s Role as an Epigenetic Regulator in Health and Disease" Diseases 5, no. 1: 12. https://doi.org/10.3390/diseases5010012
APA StyleMelnik, B. C., & Schmitz, G. (2017). Milk’s Role as an Epigenetic Regulator in Health and Disease. Diseases, 5(1), 12. https://doi.org/10.3390/diseases5010012





