An Integrated Outlook on the Metagenome and Metabolome of Intestinal Diseases
Abstract
:1. The Gut Microbiota
2. What Is Metabolomics?
3. Using Metabolomics to Understand the Gut Microbiota
4. Relationships among the Gut Ecosystem, Colorectal Cancer and Inflammatory Bowel Disease
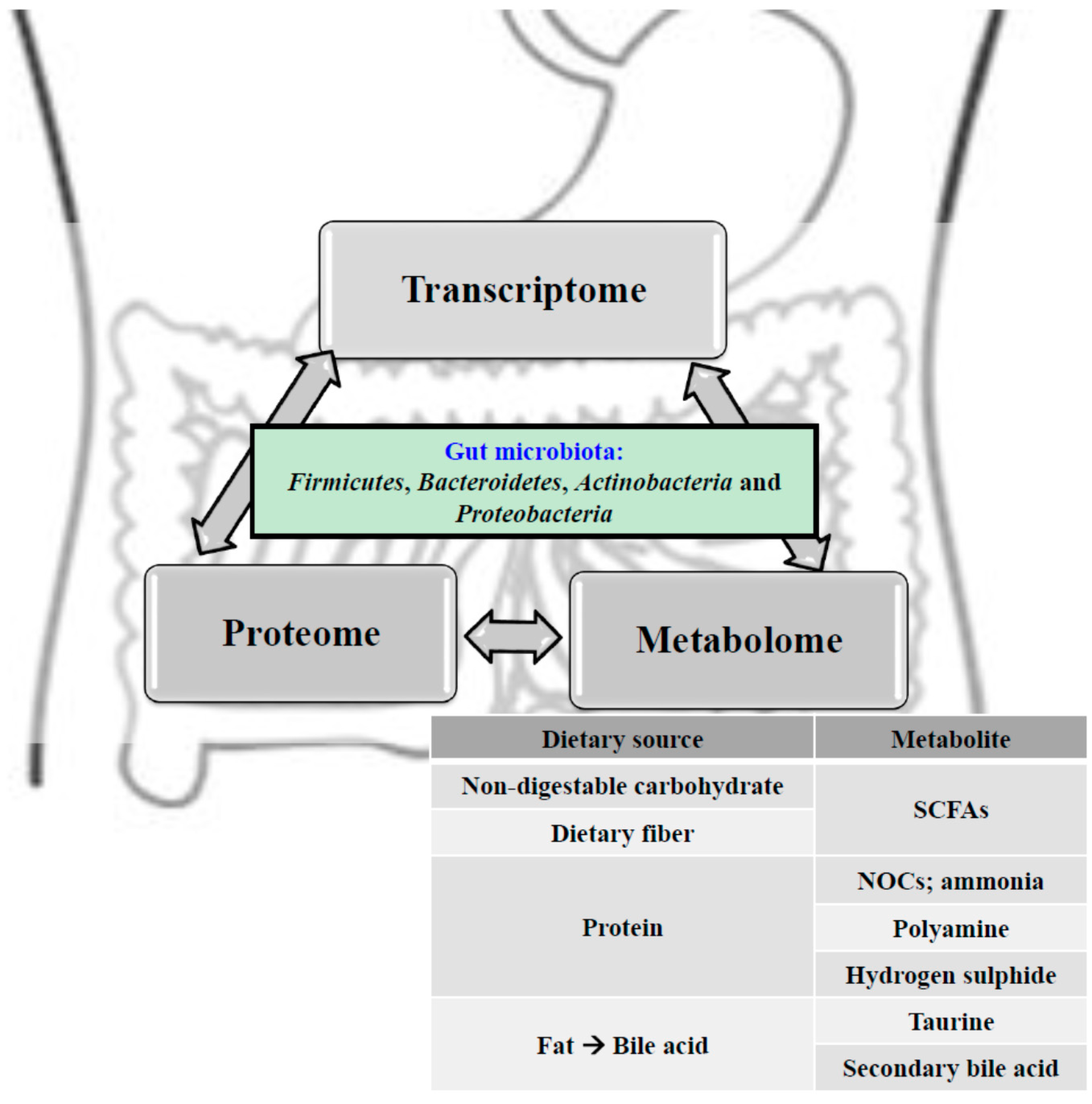
5. Colorectal Cancer
6. Inflammatory Bowel Disease
7. Microbial Metabolism in the Gut
8. Protective and Detrimental Metabolites
9. Impact of SCFAs on Host Cells
10. Detrimental Metabolites: Products of Protein Fermentation
11. Detrimental Metabolites: Bile Acid Metabolism
12. Detrimental Metabolites: Hydrogen Sulphide
13. Therapeutic Interventions of Intestinal Diseases
13.1. Probiotic Interventions
13.2. Prebiotic Interventions
13.3. Fecal Microbiota Transplantations

14. Conclusions
Conflicts of Interest
References
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Bianconi, E.; Piovesan, A.; Facchin, F.; Beraudi, A.; Casadei, R.; Frabetti, F.; Vitale, L.; Pelleri, M.C.; Tassani, S.; Piva, F.; et al. An estimation of the number of cells in the human body. Ann. Hum. Biol. 2013, 40, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, S.; Ohno, H. Gut microbiome and metabolic diseases. Semin. Immunopathol. 2014, 36, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Mahowald, M.; Ley, R.; Lozupone, C.; Hamady, M.; Martens, E.; Henrissat, B.; Coutinho, P.; Minx, P.; Latreille, P.; et al. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 2007, 5, 1574–1586. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Cheng, J.; Duncan, A.E.; Kau, A.L.; Griffin, N.W.; Lombard, V.; Henrissat, B.; Bain, J.R.; et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 2013, 341, 1241214. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Hamady, M.; Yatsunenko, T.; Cantarel, B.L.; Duncan, A.; Ley, R.E.; Sogin, M.L.; Jones, W.J.; Roe, B.A.; Affourtit, J.P.; et al. A core gut microbiome in obese and lean twins. Nature 2009, 457, 480–484. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.; Ley, R.E.; Volchkov, P.Y.; Stranges, P.B.; Avanesyan, L.; Stonebraker, A.C.; Hu, C.; Wong, F.S.; Szot, G.L.; Bluestone, J.A.; et al. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature 2008, 455, 1109–1113. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zhang, P.; Jiang, H.; Cheng, S. Gut bacterial translocation contributes to microinflammation in experimental uremia. Dig. Dis. Sci. 2012, 57, 2856–2862. [Google Scholar] [CrossRef] [PubMed]
- Molodecky, N.A.; Soon, I.S.; Rabi, D.M.; Ghali, W.A.; Ferris, M.; Chernoff, G.; Benchimol, E.I.; Panaccione, R.; Ghosh, S.; Barkema, H.W.; et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology 2012, 142, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Scanlan, P.D.; Shanahan, F.; Clune, Y.; Collins, J.K.; O’Sullivan, G.C.; O'Riordan, M.; Holmes, E.; Wang, Y.; Marchesi, J.R. Culture-independent analysis of the gut microbiota in colorectal cancer and polyposis. Environ. Microbiol. 2008, 10, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Kirjavainen, P.; Arvola, T.; Salminen, S.; Isolauri, E. Aberrant composition of gut microbiota of allergic infants: A target of bifidobacterial therapy at weaning? Gut 2002, 51, 51–55. [Google Scholar] [CrossRef] [PubMed]
- Berer, K.; Mues, M.; Koutrolos, M.; Rasbi, Z.A.; Boziki, M.; Johner, C.; Wekerle, H.; Krishnamoorthy, G. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature 2011, 479, 538–541. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.H.; Wang, Z.; Levison, B.S.; Koeth, R.A.; Britt, E.B.; Fu, X.; Wu, Y.; Hazen, S.L. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N. Engl. J. Med. 2013, 368, 1575–1584. [Google Scholar] [CrossRef] [PubMed]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L.; et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef] [PubMed]
- Finegold, S.M. Therapy and epidemiology of autism—Clostridial spores as key elements. Med. Hypotheses 2008, 70, 508–511. [Google Scholar] [CrossRef] [PubMed]
- Ellis, D.I.; Dunn, W.B.; Griffin, J.L.; Allwood, J.W.; Goodacre, R. Metabolic fingerprinting as a diagnostic tool. Pharmacogenomics 2007, 8, 1243–1266. [Google Scholar] [CrossRef] [PubMed]
- Tringe, S.; Hugenholtz, P. A renaissance for the pionnering 16s rrna gene. Curr. Opin. Microbiol. 2008, 11, 442–446. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-gut microbiota metabolic interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef] [PubMed]
- Dettmer, K.; Aronov, P.A.; Hammock, B.D. Mass spectrometry based metabolomics. Mass. Spectrom. Rev. 2007, 26, 51–78. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.P.; Wang, Y.; Sprenger, N.; Yap, I.K.; Lundstedt, T.; Lek, P.; Rezzi, S.; Ramadan, Z.; van Bladeren, P.; Fay, L.B.; et al. Probiotic modulation of symbiotic gut microbial-host metabolic interactions in a humanized microbiome mouse model. Mol. Syst. Biol. 2008, 4, 157. [Google Scholar] [CrossRef] [PubMed]
- Wikoff, W.R.; Anfora, A.T.; Liu, J.; Schultz, P.G.; Lesley, S.A.; Peters, E.C.; Siuzdak, G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA 2009, 106, 3698–3703. [Google Scholar] [CrossRef] [PubMed]
- Aw, W.; Fukuda, S. Toward the comprehensive understanding of the gut ecosystem via metabolomics-based integrated omics approach. Semin. Immunopathol. 2015, 37, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A.J.; Kinzler, K.W. Cancer genome landscapes. Science 2013, 339, 1546–1558. [Google Scholar] [CrossRef] [PubMed]
- Sears, C.L.; Garrett, W.S. Microbes, microbiota, and colon cancer. Cell Host Microbe 2014, 15, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Jemal, A.; Bray, F.; Center, M.M.; Ferlay, J.; Ward, E.; Forman, D. Global cancer statistics. CA Cancer J. Clin. 2011, 61, 69–90. [Google Scholar] [CrossRef] [PubMed]
- Belcheva, A.; Irrazabal, T.; Robertson, S.J.; Streutker, C.; Maughan, H.; Rubino, S.; Moriyama, E.H.; Copeland, J.K.; Kumar, S.; Green, B.; et al. Gut microbial metabolism drives transformation of msh2-deficient colon epithelial cells. Cell 2014, 158, 288–299. [Google Scholar] [CrossRef] [PubMed]
- Jess, T.; Gamborg, M.; Matzen, P.; Munkholm, P.; Sorensen, T.I. Increased risk of intestinal cancer in crohn’s disease: A meta-analysis of population-based cohort studies. Am. J. Gastroenterol. 2005, 100, 2724–2729. [Google Scholar] [CrossRef] [PubMed]
- Danese, S.; Malesci, A.; Vetrano, S. Colitis-associated cancer: The dark side of inflammatory bowel disease. Gut 2011, 60, 1609–1610. [Google Scholar] [CrossRef] [PubMed]
- Arthur, J.C.; Perez-Chanona, E.; Muhlbauer, M.; Tomkovich, S.; Uronis, J.M.; Fan, T.J.; Campbell, B.J.; Abujamel, T.; Dogan, B.; Rogers, A.B.; et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 2012, 338, 120–123. [Google Scholar] [CrossRef] [PubMed]
- Schwabe, R.F.; Jobin, C. The microbiome and cancer. Nat. Rev. Cancer 2013, 13, 800–812. [Google Scholar] [CrossRef] [PubMed]
- Baena, R.; Salinas, P. Diet and colorectal cancer. Maturitas 2015, 80, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Baumgart, D.C.; Sandborn, W.J. Inflammatory bowel disease: Clinical aspects and established and evolving therapies. Lancet 2007, 369, 1641–1657. [Google Scholar] [CrossRef]
- Loftus, E.V. Clinical epidemiology of inflammatory bowel disease: Incidence, prevalence, and environmental influences. Gastroenterology 2004, 126, 1504–1517. [Google Scholar] [CrossRef] [PubMed]
- M'Koma, A.E. Inflammatory bowel disease: An expanding global health problem. Clin. Med. Insights Gastroenterol. 2013, 6, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.C.; Hansoul, S.; Nicolae, D.L.; Cho, J.H.; Duerr, R.H.; Rioux, J.D.; Brant, S.R.; Silverberg, M.S.; Taylor, K.D.; Barmada, M.M.; et al. Genome-wide association defines more than 30 distinct susceptibility loci for crohn’s disease. Nat. Genet. 2008, 40, 955–962. [Google Scholar] [CrossRef] [PubMed]
- Frank, D.N.; St Amand, A.L.; Feldman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 2007, 104, 13780–13785. [Google Scholar] [CrossRef] [PubMed]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermudez-Humaran, L.G.; Gratadoux, J.J.; Blugeon, S.; Bridonneau, C.; Furet, J.P.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of crohn disease patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef] [PubMed]
- Norman, J.M.; Handley, S.A.; Baldridge, M.T.; Droit, L.; Liu, C.Y.; Keller, B.C.; Kambal, A.; Monaco, C.L.; Zhao, G.; Fleshner, P.; et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 2015, 160, 447–460. [Google Scholar] [CrossRef] [PubMed]
- Marcobal, A.; Kashyap, P.C.; Nelson, T.A.; Aronov, P.A.; Donia, M.S.; Spormann, A.; Fischbach, M.A.; Sonnenburg, J.L. A metabolomic view of how the human gut microbiota impacts the host metabolome using humanized and gnotobiotic mice. ISME J. 2013, 7, 1933–1943. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Hold, G.L.; Flint, H.J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014, 12, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Flint, H.J.; Scott, K.P.; Duncan, S.H.; Louis, P.; Forano, E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 2012, 3, 289–306. [Google Scholar] [CrossRef] [PubMed]
- Sieber, J.R.; McInerney, M.J.; Gunsalus, R.P. Genomic insights into syntrophy: The paradigm for anaerobic metabolic cooperation. Ann. Rev. Microbiol. 2012, 66, 429–452. [Google Scholar] [CrossRef] [PubMed]
- Baughn, A.D.; Malamy, M.H. The strict anaerobe bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature 2004, 427, 441–444. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.T.; Duncan, S.H.; Stams, A.J.; van Dijl, J.M.; Flint, H.J.; Harmsen, H.J. The gut anaerobe faecalibacterium prausnitzii uses an extracellular electron shuttle to grow at oxic-anoxic interphases. ISME J. 2012, 6, 1578–1585. [Google Scholar] [CrossRef] [PubMed]
- Nava, G.M.; Carbonero, F.; Croix, J.A.; Greenberg, E.; Gaskins, H.R. Abundance and diversity of mucosa-associated hydrogenotrophic microbes in the healthy human colon. ISME J. 2012, 6, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Carbonero, F.; Benefiel, A.C.; Gaskins, H.R. Contributions of the microbial hydrogen economy to colonic homeostasis. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 504–518. [Google Scholar] [CrossRef] [PubMed]
- Sleeth, M.L.; Thompson, E.L.; Ford, H.E.; Zac-Varghese, S.E.; Frost, G. Free fatty acid receptor 2 and nutrient sensing: A proposed role for fibre, fermentable carbohydrates and short-chain fatty acids in appetite regulation. Nutr. Res. Rev. 2010, 23, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Fung, K.Y.; Cosgrove, L.; Lockett, T.; Head, R.; Topping, D.L. A review of the potential mechanisms for the lowering of colorectal oncogenesis by butyrate. Br. J. Nutr. 2012, 108, 820–831. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.J.; Chueh, A.C.; Togel, L.; Corner, G.A.; Ahmed, N.; Goel, S.; Byun, D.S.; Nasser, S.; Houston, M.A.; Jhawer, M.; et al. Apoptotic sensitivity of colon cancer cells to histone deacetylase inhibitors is mediated by an sp1/sp3-activated transcriptional program involving immediate-early gene induction. Cancer Res. 2010, 70, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Hamer, H.M.; Jonkers, D.; Venema, K.; Vanhoutvin, S.; Troost, F.J.; Brummer, R.J. Review article: The role of butyrate on colonic function. Aliment. Pharmacol. Ther. 2008, 27, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.V.; Hao, L.; Offermanns, S.; Medzhitov, R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. USA 2014, 111, 2247–2252. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly, Y.M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic treg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory t cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of colonic regulatory t cells by indigenous clostridium species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Geuking, M.B.; Cahenzli, J.; Lawson, M.A.; Ng, D.C.; Slack, E.; Hapfelmeier, S.; McCoy, K.D.; Macpherson, A.J. Intestinal bacterial colonization induces mutualistic regulatory t cell responses. Immunity 2011, 34, 794–806. [Google Scholar] [CrossRef] [PubMed]
- Round, J.L.; Mazmanian, S.K. Inducible foxp3+ regulatory t-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. USA 2010, 107, 12204–12209. [Google Scholar] [CrossRef] [PubMed]
- O'Mahony, C.; Scully, P.; O'Mahony, D.; Murphy, S.; O'Brien, F.; Lyons, A.; Sherlock, G.; MacSharry, J.; Kiely, B.; Shanahan, F.; et al. Commensal-induced regulatory t cells mediate protection against pathogen-stimulated nf-kappab activation. PLoS Pathog. 2008, 4, e1000112. [Google Scholar] [CrossRef] [PubMed]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; deRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory t-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef] [PubMed]
- Latham, T.; Mackay, L.; Sproul, D.; Karim, M.; Culley, J.; Harrison, D.J.; Hayward, L.; Langridge-Smith, P.; Gilbert, N.; Ramsahoye, B.H. Lactate, a product of glycolytic metabolism, inhibits histone deacetylase activity and promotes changes in gene expression. Nucleic Acids Res. 2012, 40, 4794–4803. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.J.; Goldsworthy, S.M.; Barnes, A.A.; Eilert, M.M.; Tcheang, L.; Daniels, D.; Muir, A.I.; Wigglesworth, M.J.; Kinghorn, I.; Fraser, N.J.; et al. The orphan g protein-coupled receptors gpr41 and gpr43 are activated by propionate and other short chain carboxylic acids. J. Biol. Chem. 2003, 278, 11312–11319. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, P.D.; Manicassamy, S.; Munn, D.H.; et al. Activation of gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Thangaraju, M.; Cresci, G.A.; Liu, K.; Ananth, S.; Gnanaprakasam, J.P.; Browning, D.D.; Mellinger, J.D.; Smith, S.B.; Digby, G.J.; Lambert, N.A.; et al. Gpr109a is a g-protein-coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer Res. 2009, 69, 2826–2832. [Google Scholar] [CrossRef] [PubMed]
- Maslowski, K.M.; Vieira, A.T.; Ng, A.; Kranich, J.; Sierro, F.; Yu, D.; Schilter, H.C.; Rolph, M.S.; Mackay, F.; Artis, D.; et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor gpr43. Nature 2009, 461, 1282–1286. [Google Scholar] [PubMed]
- Ganapathy, V.; Thangaraju, M.; Prasad, P.D.; Martin, P.M.; Singh, N. Transporters and receptors for short-chain fatty acids as the molecular link between colonic bacteria and the host. Curr. Opin. Pharmacol. 2013, 13, 869–874. [Google Scholar] [CrossRef] [PubMed]
- Buda, A.; Qualtrough, D.; Jepson, M.A.; Martines, D.; Paraskeva, C.; Pignatelli, M. Butyrate downregulates alpha2beta1 integrin: A possible role in the induction of apoptosis in colorectal cancer cell lines. Gut 2003, 52, 729–734. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.M.; Topping, D.L.; Bird, A.R.; Young, G.P.; Cobiac, L. Effects of high-amylose maize starch and butyrylated high-amylose maize starch on azoxymethane-induced intestinal cancer in rats. Carcinogenesis 2008, 29, 2190–2194. [Google Scholar] [CrossRef] [PubMed]
- Blachier, F.; Nepelska, M.; Cultrone, A.; Béguet-Crespel, F.; Le Roux, K.; Doré, J.; Arulampalam, V.; Blottière, H.M. Butyrate produced by commensal bacteria potentiates phorbol esters induced ap-1 response in human intestinal epithelial cells. PLoS ONE 2012, 7, e52869. [Google Scholar] [CrossRef]
- Ou, J.; Carbonero, F.; Zoetendal, E.G.; DeLany, J.P.; Wang, M.; Newton, K.; Gaskins, H.R.; O'Keefe, S.J. Diet, microbiota, and microbial metabolites in colon cancer risk in rural africans and african americans. Am. J. Clin. Nutr. 2013, 98, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Russell, W.R.; Gratz, S.W.; Duncan, S.H.; Holtrop, G.; Ince, J.; Scobbie, L.; Duncan, G.; Johnstone, A.M.; Lobley, G.E.; Wallace, R.J.; et al. High-protein, reduced-carbohydrate weight-loss diets promote metabolite profiles likely to be detrimental to colonic health. Am. J. Clin. Nutr. 2011, 93, 1062–1072. [Google Scholar] [CrossRef] [PubMed]
- Windey, K.; de Preter, V.; Verbeke, K. Relevance of protein fermentation to gut health. Mol. Nutr. Food Res. 2012, 56, 184–196. [Google Scholar] [CrossRef] [PubMed]
- Russell, W.R.; Duncan, S.H.; Scobbie, L.; Duncan, G.; Cantlay, L.; Calder, A.G.; Anderson, S.E.; Flint, H.J. Major phenylpropanoid-derived metabolites in the human gut can arise from microbial fermentation of protein. Mol. Nutr. Food Res. 2013, 57, 523–535. [Google Scholar] [CrossRef] [PubMed]
- Loh, Y.H.; Jakszyn, P.; Luben, R.N.; Mulligan, A.A.; Mitrou, P.N.; Khaw, K.T. N-nitroso compounds and cancer incidence: The european prospective investigation into cancer and nutrition (epic)-norfolk study. Am. J. Clin. Nutr. 2011, 93, 1053–1061. [Google Scholar] [CrossRef] [PubMed]
- Hughes, R. Metabolic activities of the gut microflora in relation to cancer. Microb. Ecol. Health Dis. 2000, 12, 179–185. [Google Scholar] [CrossRef]
- Di Martino, M.L.; Campilongo, R.; Casalino, M.; Micheli, G.; Colonna, B.; Prosseda, G. Polyamines: Emerging players in bacteria-host interactions. Int. J. Med. Microbiol. IJMM 2013, 303, 484–491. [Google Scholar] [CrossRef] [PubMed]
- Pegg, A.E. Toxicity of polyamines and their metabolic products. Chem. Res. Toxicol. 2013, 26, 1782–1800. [Google Scholar] [CrossRef] [PubMed]
- Hanfrey, C.C.; Pearson, B.M.; Hazeldine, S.; Lee, J.; Gaskin, D.J.; Woster, P.M.; Phillips, M.A.; Michael, A.J. Alternative spermidine biosynthetic route is critical for growth of campylobacter jejuni and is the dominant polyamine pathway in human gut microbiota. J. Biol. Chem. 2011, 286, 43301–43312. [Google Scholar] [CrossRef] [PubMed]
- Kibe, R.; Kurihara, S.; Sakai, Y.; Suzuki, H.; Ooga, T.; Sawaki, E.; Muramatsu, K.; Nakamura, A.; Yamashita, A.; Kitada, Y.; et al. Upregulation of colonic luminal polyamines produced by intestinal microbiota delays senescence in mice. Sci. Rep. 2014, 4, 4548. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, M.; Kurihara, S.; Kibe, R.; Ashida, H.; Benno, Y. Longevity in mice is promoted by probiotic-induced suppression of colonic senescence dependent on upregulation of gut bacterial polyamine production. PLoS ONE 2011, 6, e23652. [Google Scholar] [CrossRef] [PubMed]
- Swann, J.R.; Want, E.J.; Geier, F.M.; Spagou, K.; Wilson, I.D.; Sidaway, J.E.; Nicholson, J.K.; Holmes, E. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4523–4530. [Google Scholar] [CrossRef] [PubMed]
- Ridlon, J.M.; Kang, D.J.; Hylemon, P.B. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 2006, 47, 241–259. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Houten, S.M.; Mataki, C.; Christoffolete, M.A.; Kim, B.W.; Sato, H.; Messaddeq, N.; Harney, J.W.; Ezaki, O.; Kodama, T.; et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature 2006, 439, 484–489. [Google Scholar] [CrossRef] [PubMed]
- Barrasa, J.I.; Olmo, N.; Lizarbe, M.A.; Turnay, J. Bile acids in the colon, from healthy to cytotoxic molecules. Toxicol. Vitro Int. J. Publ. Assoc. BIBRA 2013, 27, 964–977. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, H. Bile acids as endogenous etiologic agents in gastrointestinal cancer. World J. Gastroenterol. 2009, 15, 3329. [Google Scholar] [CrossRef] [PubMed]
- Ou, J.; DeLany, J.P.; Zhang, M.; Sharma, S.; O’Keefe, S.J. Association between low colonic short-chain fatty acids and high bile acids in high colon cancer risk populations. Nutr. Cancer 2012, 64, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Islam, K.B.; Fukiya, S.; Hagio, M.; Fujii, N.; Ishizuka, S.; Ooka, T.; Ogura, Y.; Hayashi, T.; Yokota, A. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology 2011, 141, 1773–1781. [Google Scholar] [CrossRef] [PubMed]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Magee, E.A.; Richardson, C.J.; Hughes, R.; Cummings, J.H. Contribution of dietary protein to sulfide production in the large intestine: An in vitro and a controlled feeding study in humans. Am. J. Clin. Nutr. 2000, 72, 1488–1494. [Google Scholar] [PubMed]
- Attene-Ramos, M.S.; Nava, G.M.; Muellner, M.G.; Wagner, E.D.; Plewa, M.J.; Gaskins, H.R. DNA damage and toxicogenomic analyses of hydrogen sulfide in human intestinal epithelial fhs 74 int cells. Environ. Mol. Mutagen. 2010, 51, 304–314. [Google Scholar] [CrossRef] [PubMed]
- Attene-Ramos, M.S.; Wagner, E.D.; Gaskins, H.R.; Plewa, M.J. Hydrogen sulfide induces direct radical-associated DNA damage. Mol. Cancer Res. MCR 2007, 5, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Health and nutritional properties and guidelines for evaluation. Available online: ftp://ftp.fao.org/docrep/fao/009/a0512e/a0512e00.pdf (accessed on 5 November 2015).
- Kostic, A.D.; Xavier, R.J.; Gevers, D. The microbiome in inflammatory bowel disease: Current status and the future ahead. Gastroenterology 2014, 146, 1489–1499. [Google Scholar] [CrossRef] [PubMed]
- Rembacken, B.J.; Snelling, A.M.; Hawkey, P.M.; Chalmers, D.M.; Axon, A.T.R. Non-pathogenic escherichia coli versus mesalazine for the treatment of ulcerative colitis: A randomised trial. Lancet 1999, 354, 635–639. [Google Scholar] [CrossRef]
- Matthes, H.; Krummenerl, T.; Giensch, M.; Wolff, C.; Schulze, J. Clinical trial: Probiotic treatment of acute distal ulcerative colitis with rectally administered escherichia coli nissle 1917 (ecn). BMC Complement. Altern. Med. 2010, 10. [Google Scholar] [CrossRef] [PubMed]
- Zocco, M.A.; dal Verme, L.Z.; Cremonini, F.; Piscaglia, A.C.; Nista, E.C.; Candelli, M.; Novi, M.; Rigante, D.; Cazzato, I.A.; Ojetti, V.; et al. Efficacy of lactobacillus gg in maintaining remission of ulcerative colitis. Aliment. Pharmacol. Ther. 2006, 23, 1567–1574. [Google Scholar] [CrossRef] [PubMed]
- Veiga, P.; Gallini, C.A.; Beal, C.; Michaud, M.; Delaney, M.L.; DuBois, A.; Khlebnikov, A.; van Hylckama Vlieg, J.E.; Punit, S.; Glickman, J.N.; et al. Bifidobacterium animalis subsp. Lactis fermented milk product reduces inflammation by altering a niche for colitogenic microbes. Proc. Natl. Acad. Sci. USA 2010, 107, 18132–18137. [Google Scholar] [CrossRef] [PubMed]
- Guarner, F. Prebiotics, probiotics and helminths: The “natural” solution? Dig. Dis. 2009, 27, 412–417. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, S.; Furuya, H.; Suzuki, Y.; Asanuma, N.; Hino, T. A new strain of butyrivibrio fibrisolvens that has high ability to isomerize linoleic acid to conjugated linoleic acid. J. Gen. Appl. Microbiol. 2005, 51, 105–113. [Google Scholar] [CrossRef] [PubMed]
- West, D.; Delany, J.; Camet, P.; Blohm, F.; Truett, A.; Scimeca, J. Effects of conjugated linoleic acid on body fat and energy metabolism in the mouse. Am. J. Physiol. 1998, 275, R667–R672. [Google Scholar] [PubMed]
- Fukuda, S.; Suzuki, Y.; Murai, M.; Asanuma, N.; Hino, T. Isolation of a novel strain of butyrivibrio fibrisolvens that isomerizes linoleic acid to conjugated linoleic acid without hydrogenation, and its utilization as a probiotic for animals. J. Appl. Microbiol. 2006, 100, 787–794. [Google Scholar] [CrossRef] [PubMed]
- Sugahara, H.; Odamaki, T.; Fukuda, S.; Kato, T.; Xiao, J.Z.; Abe, F.; Kikuchi, J.; Ohno, H. Probiotic bifidobacterium longum alters gut luminal metabolism through modification of the gut microbial community. Sci. Rep. 2015, 5, 13548. [Google Scholar] [CrossRef]
- Kanauchi, O.; Agata, K. Protein, and dietary fiber-rich new foodstuff from brewer’s spent grain increased excretion of feces and jejunum mucosal protein content in rats. Biosci. Biotechnol. Biochem. 1997, 61, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Kanauchi, O.; Iwanaga, T.; Andoh, A.; Araki, Y.; Nakamura, T.; Mitsuyama, K.; Suzuki, A.; Hibi, T.; Bamba, T. Dietary fiber fraction of germinated barley foodstuff attenuated mucosal damage and diarrhea, and accelerated the repair of the colonic mucosa in an experimental colitis. J. Gastroenterol. Hepatol. 2001, 16, 160–168. [Google Scholar] [CrossRef]
- Mitsuyama, K.; Saiki, T.; Kanauchi, O.; Iwanaga, T.; Tomiyasu, N.; Nishiyama, T.; Tateishi, H.; Shirachi, A.; Ide, M.; Suzuki, A.; et al. Treatment of ulcerative colitis with germinated barley foodstuff feeding: A pilot study. Aliment. Pharmacol. Ther. 1998, 12, 1225–1230. [Google Scholar] [CrossRef] [PubMed]
- Faghfoori, Z.; Navai, L.; Shakerhosseini, R.; Somi, M.H.; Nikniaz, Z.; Norouzi, M.F. Effects of an oral supplementation of germinated barley foodstuff on serum tumour necrosis factor-alpha, interleukin-6 and -8 in patients with ulcerative colitis. Ann. Clin. Biochem. 2011, 48, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Joossens, M.; de Preter, V.; Ballet, V.; Verbeke, K.; Rutgeerts, P.; Vermeire, S. Effect of oligofructose-enriched inulin (of-in) on bacterial composition and disease activity of patients with crohn's disease: Results from a double-blinded randomised controlled trial. Gut 2012, 61, 958. [Google Scholar] [CrossRef] [PubMed]
- Verghese, M.; Rao, D.R.; Chawan, C.B.; Williams, L.L.; Shackelford, L. Dietary inulin suppresses azoxymethane-induced aberrant crypt foci and colon tumors at the promotion stage in young fisher 344 rat. J. Nutr. 2002, 132, 2809–2813. [Google Scholar] [PubMed]
- Hsu, C.K.; Liao, J.W.; Chung, Y.C.; Hsieh, C.P.; Chan, Y.C. Xylooligosaccharides and fructooligosaccharides affect the intestinal microbiota and precancerous colonic lesion development in rats. J. Nutr. 2004, 134, 1523–1528. [Google Scholar] [PubMed]
- Smits, L.; Bouter, K.; de Vos, W.; Borody, T.; Nieuwdorp, M. Therapeutic potential of fecal microbiota transplantation. Gastroenterology 2013, 145, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Borody, T. “Flora power”—Fecal bacteria cure chronic c. Difficile diarrhea. Am. J. Gastroenterol. 2000, 95, 3028–3029. [Google Scholar] [PubMed]
- Khoruts, A.; Dicksved, J.; Jansson, J.; Sadowsky, M. Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent clostridium difficile-associated diarrhea. J. Clin. Gastroenterol. 2010, 44, 354–360. [Google Scholar] [CrossRef] [PubMed]
- Van Nood, E.; Vrieze, A.; Nieuwdorp, M.; Fuentes, S.; Zoetendal, E.G.; de Vos, W.M.; Visser, C.E.; Kuijper, E.J.; Bartelsman, J.F.; Tijssen, J.G.; et al. Duodenal infusion of donor feces for recurrent clostridium difficile. N. Engl. J. Med. 2013, 368, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Zhu, W.; Gong, J.; Guo, D.; Gu, L.; Li, N.; Li, J. Fecal microbiota transplantation improves the quality of life in patients with inflammatory bowel disease. Gastroenterol. Res. Pract. 2015, 2015, 517597. [Google Scholar] [CrossRef] [PubMed]
- Kellermayer, R.; Nagy-Szakal, D.; Harris, R.A.; Luna, R.A.; Pitashny, M.; Schady, D.; Mir, S.A.; Lopez, M.E.; Gilger, M.A.; Belmont, J.; et al. Serial fecal microbiota transplantation alters mucosal gene expression in pediatric ulcerative colitis. Am. J. Gastroenterol. 2015, 110, 604–606. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aw, W.; Fukuda, S. An Integrated Outlook on the Metagenome and Metabolome of Intestinal Diseases. Diseases 2015, 3, 341-359. https://doi.org/10.3390/diseases3040341
Aw W, Fukuda S. An Integrated Outlook on the Metagenome and Metabolome of Intestinal Diseases. Diseases. 2015; 3(4):341-359. https://doi.org/10.3390/diseases3040341
Chicago/Turabian StyleAw, Wanping, and Shinji Fukuda. 2015. "An Integrated Outlook on the Metagenome and Metabolome of Intestinal Diseases" Diseases 3, no. 4: 341-359. https://doi.org/10.3390/diseases3040341
APA StyleAw, W., & Fukuda, S. (2015). An Integrated Outlook on the Metagenome and Metabolome of Intestinal Diseases. Diseases, 3(4), 341-359. https://doi.org/10.3390/diseases3040341






