Real-World Technical Hurdles of ctDNA NGS Analysis: Lessons from Clinical Implementation
Abstract
1. Liquid Biopsy in Modern Oncology: The Dawn of a New Diagnostic Era
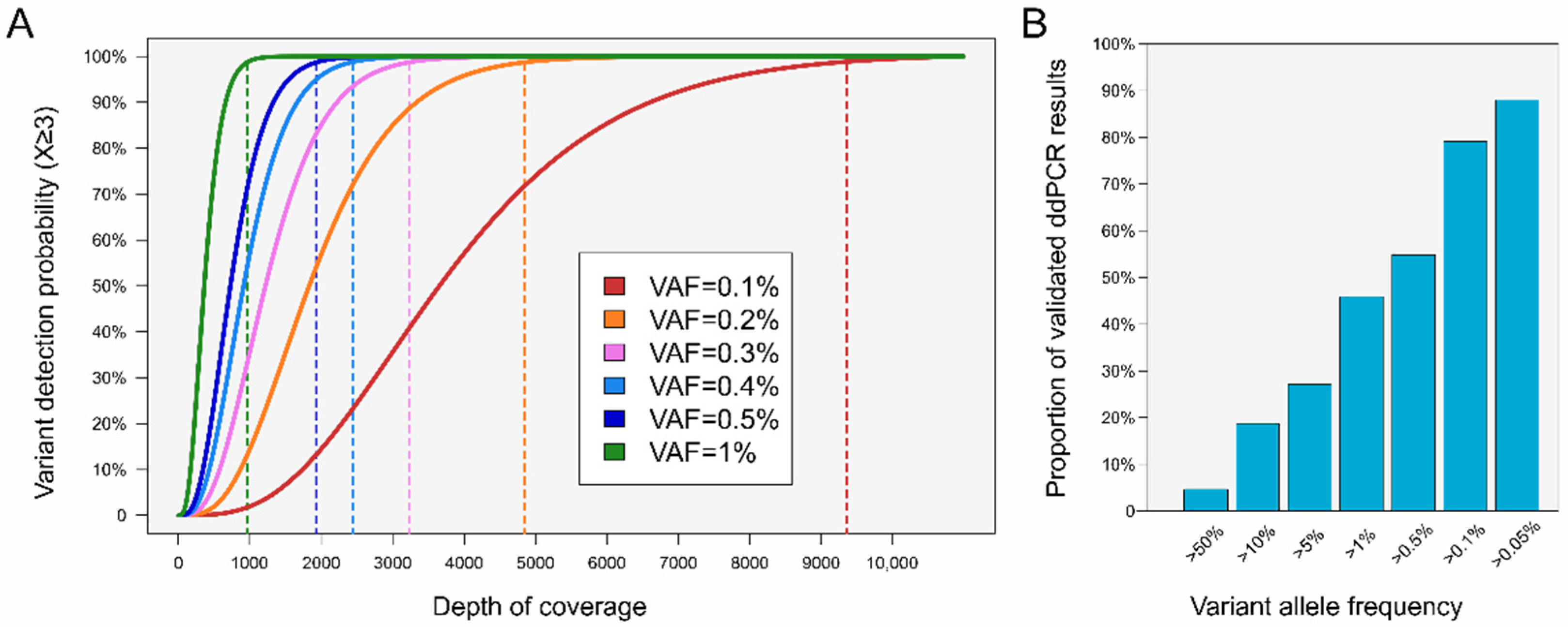
2. Variant Detection and Coverage: Navigating a Complex Relationship
3. Duplicated Reads: Avoiding Redundancy, Enhancing Sensitivity
4. DNA Quantity Drives Variant Discovery
5. Quantifying the ctDNA Fraction: An Ongoing Challenge
6. Lowering the LoD: An Essential Step to Enhance Clinical Utility
7. From Limitations to Confidence: Interpreting Negative Results in ctDNA Analysis
8. Conclusions: Unleashing the Full Potential of Cancer Care
Author Contributions
Funding
Conflicts of Interest
References
- Gibbs, S.N.; Peneva, D.; Cuyun Carter, G.; Palomares, M.R.; Thakkar, S.; Hall, D.W.; Dalglish, H.; Campos, C.; Yermilov, I. Comprehensive Review on the Clinical Impact of Next-Generation Sequencing Tests for the Management of Advanced Cancer. JCO Precis. Oncol. 2023, 7, e2200715. [Google Scholar] [CrossRef]
- Deveson, I.W.; Gong, B.; Lai, K.; LoCoco, J.S.; Richmond, T.A.; Schageman, J.; Zhang, Z.; Novoradovskaya, N.; Willey, J.C.; Jones, W.; et al. Evaluating the analytical validity of circulating tumor DNA sequencing assays for precision oncology. Nat. Biotechnol. 2021, 39, 1115–1128. [Google Scholar] [CrossRef]
- Sabnis, A.J.; Bivona, T.G. Principles of Resistance to Targeted Cancer Therapy: Lessons from Basic and Translational Cancer Biology. Trends Mol. Med. 2019, 25, 185–197. [Google Scholar] [CrossRef]
- Ko, E.Y.; Wong, J.W.H.; Shih, D.J.H.; Ho, I.; Cheung, B.M.F.; Chiu, M.K.; Leung, D.K.; Lee, A.W.; Lee, V.H.; Helali, A.E. The diagnostic accuracy of next-generation sequencing in advanced NSCLC. J. Liq. Biopsy 2025, 9, 100325. [Google Scholar] [CrossRef]
- Riely, G.J.; Wood, D.E.; Ettinger, D.S.; Aisner, D.L.; Akerley, W.; Bauman, J.R.; Bharat, A.; Bruno, D.S.; Chang, J.Y.; Chirieac, L.R.; et al. Non-Small Cell Lung Cancer, Version 4.2024, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2024, 22, 249–274. [Google Scholar] [CrossRef]
- Iams, W.T.; Mackay, M.; Ben-Shachar, R.; Drews, J.; Manghnani, K.; Hockenberry, A.J.; Cristofanilli, M.; Nimeiri, H.; Guinney, J.; Benson, A.B., 3rd. Concurrent Tissue and Circulating Tumor DNA Molecular Profiling to Detect Guideline-Based Targeted Mutations in a Multicancer Cohort. JAMA Netw. Open 2024, 7, e2351700. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.; Lingam, H.; Gao, X.; Gittleman, H.; Fiero, M.H.; Krol, D.; Biel, N.; Ricks, T.K.; Fu, W.; Hamed, S.; et al. US Food and Drug Administration Approval Summary: Elacestrant for Estrogen Receptor-Positive, Human Epidermal Growth Factor Receptor 2-Negative, ESR1-Mutated Advanced or Metastatic Breast Cancer. J. Clin. Oncol. 2024, 42, 1193–1201. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, D.; Goodman, C.; Nadauld, L.D. Clinical Utility of Genomic Testing in Cancer Care. JCO Precis. Oncol. 2022, 6, e2100349. [Google Scholar] [CrossRef]
- Vendrell, J.A.; Mau-Them, F.T.; Beganton, B.; Godreuil, S.; Coopman, P.; Solassol, J. Circulating Cell Free Tumor DNA Detection as a Routine Tool forLung Cancer Patient Management. Int. J. Mol. Sci. 2017, 18, 264. [Google Scholar] [CrossRef] [PubMed]
- Kamps, R.; Brandao, R.D.; Bosch, B.J.; Paulussen, A.D.; Xanthoulea, S.; Blok, M.J.; Romano, A. Next-Generation Sequencing in Oncology: Genetic Diagnosis, Risk Prediction and Cancer Classification. Int. J. Mol. Sci. 2017, 18, 308. [Google Scholar] [CrossRef]
- Sabari, J.K.; Offin, M.; Stephens, D.; Ni, A.; Lee, A.; Pavlakis, N.; Clarke, S.; Diakos, C.I.; Datta, S.; Tandon, N.; et al. A Prospective Study of Circulating Tumor DNA to Guide Matched Targeted Therapy in Lung Cancers. J. Natl. Cancer Inst. 2019, 111, 575–583. [Google Scholar] [CrossRef]
- Qiu, B.; Guo, W.; Zhang, F.; Lv, F.; Ji, Y.; Peng, Y.; Chen, X.; Bao, H.; Xu, Y.; Shao, Y.; et al. Dynamic recurrence risk and adjuvant chemotherapy benefit prediction by ctDNA in resected NSCLC. Nat. Commun. 2021, 12, 6770. [Google Scholar] [CrossRef]
- Rothwell, D.G.; Ayub, M.; Cook, N.; Thistlethwaite, F.; Carter, L.; Dean, E.; Smith, N.; Villa, S.; Dransfield, J.; Clipson, A.; et al. Utility of ctDNA to support patient selection for early phase clinical trials: The TARGET study. Nat. Med. 2019, 25, 738–743. [Google Scholar] [CrossRef]
- Thomas, Q.D.; Colard-Thomas, J.; Delansay, D.; Leenhardt, F.; Solassol, J.; Vendrell, J.A.; Quantin, X. Case report: Liquid biopsy, the sooner the better? Front. Oncol. 2022, 12, 1089108. [Google Scholar] [CrossRef]
- Leighl, N.B.; Page, R.D.; Raymond, V.M.; Daniel, D.B.; Divers, S.G.; Reckamp, K.L.; Villalona-Calero, M.A.; Dix, D.; Odegaard, J.I.; Lanman, R.B.; et al. Clinical Utility of Comprehensive Cell-free DNA Analysis to Identify Genomic Biomarkers in Patients with Newly Diagnosed Metastatic Non-small Cell Lung Cancer. Clin. Cancer Res. 2019, 25, 4691–4700. [Google Scholar] [CrossRef]
- Tina Shih, Y.C.; Dong, W.; Xu, Y.; Shen, Y. Assessing the Cost-Effectiveness of Updated Breast Cancer Screening Guidelines for Average-Risk Women. Value Health 2019, 22, 185–193. [Google Scholar] [CrossRef]
- Do, H.; Dobrovic, A. Sequence artifacts in DNA from formalin-fixed tissues: Causes and strategies for minimization. Clin. Chem. 2015, 61, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Larson, N.B.; Oberg, A.L.; Adjei, A.A.; Wang, L. A Clinician’s Guide to Bioinformatics for Next-Generation Sequencing. J. Thorac. Oncol. 2023, 18, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Bronkhorst, A.J.; Holdenrieder, S. The changing face of circulating tumor DNA (ctDNA) profiling: Factors that shape the landscape of methodologies, technologies, and commercialization. Med. Genet. 2023, 35, 201–235. [Google Scholar] [CrossRef] [PubMed]
- Bieler, J.; Kubik, S.; Macheret, M.; Pozzorini, C.; Willig, A.; Xu, Z. Benefits of applying molecular barcoding systems are not uniform across different genomic applications. J. Transl. Med. 2023, 21, 305. [Google Scholar] [CrossRef]
- Piovesan, A.; Pelleri, M.C.; Antonaros, F.; Strippoli, P.; Caracausi, M.; Vitale, L. On the length, weight and GC content of the human genome. BMC Res. Notes 2019, 12, 106. [Google Scholar] [CrossRef] [PubMed]
- Mattox, A.K.; Douville, C.; Wang, Y.; Popoli, M.; Ptak, J.; Silliman, N.; Dobbyn, L.; Schaefer, J.; Lu, S.; Pearlman, A.H.; et al. The Origin of Highly Elevated Cell-Free DNA in Healthy Individuals and Patients with Pancreatic, Colorectal, Lung, or Ovarian Cancer. Cancer Discov. 2023, 13, 2166–2179. [Google Scholar] [CrossRef] [PubMed]
- Adalsteinsson, V.A.; Ha, G.; Freeman, S.S.; Choudhury, A.D.; Stover, D.G.; Parsons, H.A.; Gydush, G.; Reed, S.C.; Rotem, D.; Rhoades, J.; et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat. Commun. 2017, 8, 1324. [Google Scholar] [CrossRef] [PubMed]
- Husain, H.; Pavlick, D.C.; Fendler, B.J.; Madison, R.W.; Decker, B.; Gjoerup, O.; Parachoniak, C.A.; McLaughlin-Drubin, M.; Erlich, R.L.; Schrock, A.B.; et al. Tumor Fraction Correlates With Detection of Actionable Variants Across > 23,000 Circulating Tumor DNA Samples. JCO Precis. Oncol. 2022, 6, e2200261. [Google Scholar] [CrossRef]
- Hasenleithner, S.O.; Speicher, M.R. A clinician’s handbook for using ctDNA throughout the patient journey. Mol. Cancer 2022, 21, 81. [Google Scholar] [CrossRef]
- Chen, C.; Douglas, M.P.; Ragavan, M.V.; Phillips, K.A.; Jansen, J.P. Clinical Validity and Utility of Circulating Tumor DNA (ctDNA) Testing in Advanced Non-small Cell Lung Cancer (aNSCLC): A Systematic Literature Review and Meta-analysis. Mol. Diagn. Ther. 2024, 28, 525–536. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cabello-Aguilar, S.; Vendrell, J.A.; Solassol, J. Real-World Technical Hurdles of ctDNA NGS Analysis: Lessons from Clinical Implementation. Diseases 2025, 13, 312. https://doi.org/10.3390/diseases13100312
Cabello-Aguilar S, Vendrell JA, Solassol J. Real-World Technical Hurdles of ctDNA NGS Analysis: Lessons from Clinical Implementation. Diseases. 2025; 13(10):312. https://doi.org/10.3390/diseases13100312
Chicago/Turabian StyleCabello-Aguilar, Simon, Julie A. Vendrell, and Jérôme Solassol. 2025. "Real-World Technical Hurdles of ctDNA NGS Analysis: Lessons from Clinical Implementation" Diseases 13, no. 10: 312. https://doi.org/10.3390/diseases13100312
APA StyleCabello-Aguilar, S., Vendrell, J. A., & Solassol, J. (2025). Real-World Technical Hurdles of ctDNA NGS Analysis: Lessons from Clinical Implementation. Diseases, 13(10), 312. https://doi.org/10.3390/diseases13100312






