Abstract
Tuberculosis (TB) remains an impactful infectious disease, leading to millions of deaths every year. Mycobacterium tuberculosis causes the formation of granulomas, which will determine, through the host–pathogen relationship, if the infection will remain latent or evolve into active disease. Early TB diagnosis is life-saving, especially among immunocompromised individuals, and leads to proper treatment, preventing transmission. This review addresses different approaches to diagnosing TB, from traditional methods such as sputum smear microscopy to more advanced molecular techniques. Integrating these techniques, such as polymerase chain reaction (PCR) and loop-mediated isothermal amplification (LAMP), has significantly improved the sensitivity and specificity of M. tuberculosis identification. Additionally, exploring novel biomarkers and applying artificial intelligence in radiological imaging contribute to more accurate and rapid diagnosis. Furthermore, we discuss the challenges of existing diagnostic methods, including limitations in resource-limited settings and the emergence of drug-resistant strains. While the primary focus of this review is on TB diagnosis, we also briefly explore the challenges and strategies for diagnosing non-tuberculous mycobacteria (NTM). In conclusion, this review provides an overview of the current landscape of TB diagnostics, emphasizing the need for ongoing research and innovation. As the field evolves, it is crucial to ensure that these advancements are accessible and applicable in diverse healthcare settings to effectively combat tuberculosis worldwide.
1. Introduction
Tuberculosis (TB) is a bacterial disease caused by Mycobacterium tuberculosis (sensu stricto) and other members of the Mycobacterium tuberculosis complex (MTBC) [1,2]. The disease caused 1.3 million deaths in 2022 [3], with over 31 million TB-related deaths estimated for the coming years [4].
The lungs are the most commonly affected organs (pulmonary tuberculosis, PTB) [5]. However, the disease can affect other sites, such as pleura, lymph nodes, abdomen, genitourinary tract, skin, joints, bones, and meninges, a condition known as extrapulmonary TB [6], which afflicts between 15 and 20% of TB patients, especially HIV-positive individuals [7].
Typical symptoms of PTB include persistent cough, hemoptysis, chest discomfort, fatigue, weight loss, night sweats, and fever, although mild or no symptoms may occur in the initial stages [8,9]. Moreover, individuals with latent tuberculosis infection (LTBI) exhibit no active TB signs [10] and constitute a potential source for future active TB cases [11].
Transmission, mainly through cough-generated aerosols [12], is influenced by environmental conditions, microbial viability, and host immune response [13]. Risk factors include alcohol abuse [14], coinfection with HIV [15], anemia [16], diabetes [17], silicosis [18], smoking and exposure to air pollution [19], homelessness [20], and incarceration [21].
Upon infection, M. tuberculosis triggers the formation of granulomas [2,22], after which the pathogen can remain in a latent state or lead to active disease depending on the host’s immune response [2,23]. Since recent studies suggest a potential association between PTB and lung cancer, effective TB control is needed to reduce this risk [24,25,26].
The diagnosis of TB encompasses difficulties such as underdiagnosis and limited access to testing [27], a scenario that was worsened by the COVID-19 pandemic, although the last Global Tuberculosis Report signals some progress in reversing its impact on TB control [3]. Additionally, the growing incidence of non-tuberculous mycobacterial (NTM) infections, partially due to the aging of the population, as well as an increase in immunosuppressed individuals [28], poses challenges for the diagnosis of TB. In fact, non-tuberculous mycobacteria share some features with M. tuberculosis, and non-tuberculous pulmonary disease usually presents with nonspecific signs and radiological findings similar to those caused by the TB causative agent. Therefore, diagnosing non-tuberculous mycobacterial pulmonary disease in regions with high TB prevalence is a complex endeavor. Recent reviews have outlined methods for diagnosing NTM, and the reader is referred to these reviews for a greater understanding of the methods that are used or are being developed for the diagnosis of NTM [29,30]. We briefly summarized the main techniques, and they are shown in Supplementary Table S1 [31,32,33,34,35,36,37,38,39,40,41,42,43,44,45].
Early TB diagnosis is critical for global health since it prevents TB transmission [46,47,48] and reduces mortality, especially for people living with HIV [49]. Rapid diagnostic techniques enable prompt treatment initiation [50,51], aligning with the End TB Strategy, which emphasizes effective diagnosis and treatment for people with TB [52].
Therefore, this non-systematic literature review discusses the diagnosis of tuberculosis, particularly PTB, and identifies future research directions regarding TB diagnostics.
2. Traditional Diagnostic Methods
2.1. Sputum Smear Microscopy
Sputum smear microscopy (SSM) is one of the techniques which has been widely used to diagnose active PTB [53,54,55], identifying acid-fast bacilli through acid-fast stainings such as Ziehl–Neelsen (hot stain) or Kinyoun (cold stain), whose principle involves the binding of carbol fuchsin to the mycobacterial mycolic acids [6].
Ziehl–Neelsen staining is cost-effective (mean cost USD 13.31) [56] but may underestimate bacterial burden [57,58], as well as presenting variable sensitivity (32% to 89%) [56]. Furthermore, this technique cannot differentiate between dead and live bacteria, drug-susceptible from drug-resistant strains, nor distinguish M. tuberculosis from other mycobacteria [46], exhibiting compromised sensitivity whenever the bacterial load is less than 10,000 bacilli/mL sputum sample [59]. A study performed by Chopra et al. [60] showed that Kinyoun’s method presented higher sensitivity (98.37%) compared to Ziehl–Neelsen staining (89.25%), indicating it is more effective at detecting tubercle bacilli.
Fluorescence or light-emitting diode (LED) microscopy [61,62,63], automated microscopy, and artificial intelligence (AI) [64,65,66,67] techniques have been employed to improve the performance of SSM.
Notably, SSM should be combined with other diagnostic tools to enhance the efficacy and reliability of TB diagnosis [68]. Despite the diffused employment of molecular approaches, researchers underscore that SSM remains fundamental as the primary diagnostic technique for TB globally [69,70,71], with high-burden countries conducting millions of smears annually [72,73].
2.2. Chest Radiography
Chest radiography/chest X-ray (CXR) is very useful for TB diagnosis, particularly in resource-limited settings, showing high sensitivity in detecting PTB but limited specificity [74,75]. According to the World Health Organization (WHO) [76], this technique exhibits a sensitivity ranging from 87% to 98% and is considered a valuable tool for screening PTB [77]. Moreover, CXR provides diagnostic and prognostic information in children with TB [78] despite its limitations in detecting lymphadenopathy, which requires additional methods [79]. Key radiographic findings frequently observed on CXRs in patients diagnosed with PTB encompass cavitation, consolidation, masses, pleural effusion, calcification, and nodules [80], with predominant features often comprising cavitation, bronchiectasis, and fibrosis [81]. Due to the overlap of radiographic findings, another limitation of CXR is its inability to differentiate PTB from pulmonary diseases caused by NTM [82,83].
CXR as a screening test shortens the period required to diagnose PTB [84]. Furthermore, the rise of AI, particularly deep learning algorithms, has revolutionized TB detection, showing superior performance in both screening and diagnosis [85,86]. AI approaches have opened new possibilities for improved accuracy and efficiency in TB diagnosis, aligning with the WHO’s recommendation to use Computer-Aided Detections (CAD) in conjunction with human interpretation for TB screening in individuals aged 15 years or older [87].
A notable breakthrough is the deep convolutional neural network (DCNN)-based AI algorithm developed by Nijiati et al. [88], which demonstrated an impressive accuracy of 96.73% in diagnosing TB from CXR, surpassing the performance of other models. Notably, incorporating demographic variables such as age, weight, height, and gender has enhanced the performance of deep learning models [89]. Additionally, Rahman et al. [90] underscored the importance of segmenting lung regions in CXR images before applying deep learning classification. Their study revealed that the DenseNet201 model achieved a higher accuracy of 98.6% when using segmented lung images compared to 96.47% when using whole X-ray images.
Despite the advancements in CAD for TB detection, concerns persist regarding potential biases, limitations in implementation, and challenges in its broader applicability. Ongoing evaluation, consideration of version updates, and a tailored approach to implementation that considers local data and patient characteristics are crucial to ensure the effectiveness and safety of CAD software in clinical practice [91]. Challenges include limited disease detection, suboptimal accuracy for specific populations, and the need for validation for pediatric use [92].
Currently, 16 CADs for digital chest radiography are planned for WHO policy review [93], signaling the collaboration between AI and conventional diagnostic methods.
3. Culture-Based Diagnostic
M. tuberculosis is classified as a slow-growing mycobacteria, requiring seven days to six weeks to produce visible colonies on solid media. The procedure for culturing M. tuberculosis must be performed only in Biosafety Level 3 or 4 laboratories due to the high transmissibility of the mycobacteria, which is a limiting diagnostic capacity in many low and middle-income countries. Also, adequate training of laboratory analysts is essential for reducing pre-analytical risks associated with the entire diagnostic process [94].
The use of selective media enhances accuracy compared to non-selective media, as the most frequent sample is sputum, which may carry non-tuberculous bacteria from normal microbiota [3,95]. The Petroff method has been used for the digestion-decontamination of contaminated clinical samples before culturing on solid media. Clinical specimens are incubated with an equal volume of a 2 to 4% NaOH solution for approximately 15 min, followed by centrifugation, and then inoculated onto solid media [96]. Kudoh and Kudoh [97] proposed a simplified method for the decontamination step and subsequent culturing. The clinical samples are picked up using a sterile cotton swab, which is subsequently immersed into a 4% NaOH solution. After 2 min, the swab is inoculated onto slightly acidified solid media (such as Kudoh–Ogawa medium). This method has been evaluated and has proven to be as effective and sensitive as the Petroff decontamination method for sputum samples [98,99,100]. It has been used in developing countries [99,101,102], as well as in rural areas without adequate infrastructure for culturing mycobacteria [100]. Additionally, due to its speed, this method is favored in laboratories that handle a high volume of clinical samples [98].
Solid media that have been used to cultivate M. tuberculosis involve Löwenstein–Jensen (LJ), described by Ernest Löwenstein in 1931 and modified by Kai Adolf Jensen in 1932 [103], and Kudoh–Ogawa (KO), an alternative medium used to grow this bacterium [97]. Both LJ and KO are selective for Mycobacterium species due to the presence of malachite green (a triarylmethane dye) [104]. According to Madeira et al. [102], the growth of M. tuberculosis in these media presents high and similar sensibility and specificity. These researchers observed a lower contamination rate with KO medium (4.1%) compared to LJ medium (9.0%), suggesting a potentially higher risk of false-positive results with LJ due to contamination by NTM or other microorganisms. However, both media are time-consuming, which might lead to delays in the diagnosis of TB [102].
Regarding liquid media, Middlebrook is a medium that may vary in composition, resulting in different formulations, such as Middlebrook 7H9, 7H10, and 7H11, which primarily differ in the composition and concentration of organic compounds [105]. Traditionally, Middlebrook medium has been enriched with various compounds to enhance sensitivity [106]. Middlebrook 7H10, in particular, can be supplemented with oleic acid–albumin–dextrose–catalase. This medium supports faster mycobacteria growth and allows further biofilm studies [107,108].
In addressing the limitations of liquid media, Becton Dickinson (BD) introduced the Mycobacteria Growth Indicator Tube (MGIT), combining Middlebrook 7H9 broth with fluorescent compounds metabolized by M. tuberculosis. This method, revealed under ultraviolet (UV) light, shortened incubation time and enhanced detection rates during routine laboratory procedures [109,110]. Some studies have reported that MGIT 960 outperforms the use of LJ to detect M. tuberculosis [111,112,113].
Despite the time-consuming nature of culture, it remains the gold standard for diagnosing TB (including extrapulmonary infections) and monitoring TB treatment, offering advantages in identifying the pathogen and the antimicrobial susceptibility profile determination [114]. Furthermore, culturing mycobacteria is also the gold standard for differentiating between MTBC and NTM [69,115].
4. Molecular Diagnostic Techniques
Advances in molecular biology have driven research and development in methods for the rapid and accurate detection of MTBC and NTM and their drug resistance, providing important tools for TB control [116]. In fact, the introduction of molecular tests has significantly improved TB diagnosis, particularly for cases with negative microscopy [117]. However, the WHO emphasizes that molecular tests are not substitutes for microbiological cultures and phenotypic antimicrobial susceptibility testing [53].
4.1. Molecular WHO-Recommended Rapid Diagnostic Tests
WHO recommends the use of molecular rapid diagnostic tests to detect M. tuberculosis and resistance to rifampicin (RIF) and fluoroquinolones. These tests are used to diagnose and guide TB treatment, and can also be used to monitor the response to treatment [118].
4.1.1. Nucleic-Acid Amplification Tests
Most WHO-recommended rapid diagnostic tests for TB rely on nucleic-acid amplification technology (NAAT) (Table 1) [119]. Critical examples for initial TB diagnosis without antimicrobial resistance assessment include the Loopamp™ MTBC Detection Kit and FluoroType® MTB [118].

Table 1.
Nucleic acid amplification tests (NAATs) recommended by the World Health Organization for tuberculosis diagnosis.
The Loopamp™ MTBC Detection Kit (TB-LAMP; Eiken Chemical, Tokyo, Japan) is a manual assay that provides results in less than one hour. The loop-mediated isothermal amplification (LAMP) technology [138] utilizes four primer sets to amplify six distinct regions within the DNA of MTBC. This process forms stem-loop structures detectable by dyes like SYBR green and calcein [120,139,140]. Nagai et al. [119] reported an approximate sensitivity of 80.9% and specificity of 96.5%. TB-LAMP holds potential as a substitute for SSM in diagnosing PTB in adults with symptoms and as a follow-up test to SSM, especially for further examination of smear-negative samples [120].
FluoroType® MTB VER 1.0 and VER 2.0 (Bruker/Hain Lifescience, Nehren, Germany) utilize FluoroType® technology for rapid molecular genetic testing, allowing direct detection of MTBC from patient samples. These automated systems deliver results within three hours, using fluorescence-based technology for enhanced reliability and efficiency in TB diagnostics [121]. The assay utilizes high-resolution melt analysis to identify and automatically record fluorescence signals linked to probes that target the MTBC insertion element IS6110 [141]. It is important to highlight that a remarkable characteristic of FluoroType® MTB VER 1.0 relies on its ability to detect clinically relevant NTM species [142].
Moreover, WHO has endorsed several mWRD tests for the initial TB diagnosis and the detection of resistance to RIF or the combination RIF/INH [118]. Among these, the Xpert® MTB/RIF Assay (Cepheid, Sunnyvale, CA, USA) is an automated diagnostic test using nested real-time PCR (qPCR) to detect the MTBC and RIF resistance qualitatively. It amplifies a specific segment of the rpoB gene and distinguishes between wild-type and mutation-associated RIF resistance. The assay is compatible with GeneXpert® systems and automates sample processing and nucleic acid amplification [125]. Furthermore, the Xpert® MTB/RIF Ultra assay, also developed by Cepheid, is an improved version of the Xpert® MTB/RIF test, offering higher sensitivity and reliability for detecting MTBC and RIF resistance. This is achieved through two multicopy amplification targets (IS6110 and IS1081), a larger PCR chamber, and melting curve analysis. Xpert Ultra exhibits a lower LoD (16 CFU/mL) compared to 131 CFU/mL for Xpert® MTB/RIF) [69].
Chip-based qPCR technology is employed by Truenat® MTB and Truenat® MTB Plus (Molbio Diagnostics, Goa, India) to detect M. tuberculosis in specimens from pulmonary and extrapulmonary TB [69]. Truenat® MTB targets the nrdB gene (codes for R2-like ligand binding oxidase) with an LoD of 100 CFU/mL [128]. At the same time, Truenat® MTB Plus utilizes the nrdZ gene and the IS6110 sequence, offering a significantly lower LoD of 29 CFU/mL [129]. Following a positive Truenat™ MTB Plus test, an aliquot of the extracted DNA undergoes further testing with the Truenat MTB-RIF-Dx assay to detect mutations related to RIF resistance [69]. An important drawback of Truenat® MTB is its inability to detect mycobacterial species other than M. tuberculosis [128].
The RealTime MTB assay (Abbott Molecular, Des Plaines, IL, USA) uses PCR to detect MTBC DNA, exhibiting a sensitivity of 93% in culture-positive samples, 81% in smear-negative culture-positive samples, and a specificity of 97%. With an LoD of 17 CFU/mL, the target regions include IS6110 and pab gene (codes for protein antigen B, Pab) [130]. The RealTime MTB RIF/INH (Abbott Molecular, Des Plaines, IL, USA) assay detects resistance to RIF (rpoB gene) and different levels of INH resistance (katG gene and inhA promoter region) [132]. It allows for the identification of MTBC with or without rifampicin-resistant TB (RR-TB), isoniazid-resistant rifampicin-susceptible TB (Hr-TB), or multidrug-resistant TB (MDR-TB) within 10.5 h using raw or processed sputum specimens or DNA eluates from positive samples [69].
The BD MAX™ MDR-TB assay (Becton Dickinson, Sparks, MD, USA) integrates MTBC detection and assesses resistance to RIF and INH. The automated process involves specimen treatment with BD MAX STR reagent, transfer to the BD MAX MDR-TB Sample Tube, and utilization of the BD MAX System for DNA extraction, amplification, and detection using qPCR. Targeting IS6110 and IS1081 for MTBC detection (LoD = 0.5 CFU/mL; 92.6% overall sensitivity, and 98.6% overall specificity), the system also identifies resistance, with the rpoB gene for RIF resistance, and the inhA promoter region and katG gene for INH resistance [133].
The FluoroType® MTBDR VER 2.0 (Bruker/Hain Lifescience, Nehren, Germany) is a multiplex qPCR assay for the rapid detection of MTBC and resistance to RIF and INH, based on the LiquidArray® technology. DNA extraction can be manual (FluoroLyse) or automated (GenoXtract® X2 cartridge). The targeted sequences include rpoB, katG, and inhA, allowing the identification of mutations associated with RIF and INH resistances [134].
The cobas® MTB test (Roche Molecular Diagnostics, Pleasanton, CA, USA) targets 16S rRNA and esx genes through qPCR (LoD = 8.8 CFU/mL in raw sputum). It is intended for use on the cobas® 5800/6800/8800 Systems and is an automated, qualitative in vitro diagnostic test. The test can detect MTBC DNA in various human respiratory specimens, including sputum and bronchoalveolar lavage samples [135]. Additionally, the cobas® MTB-RIF/INH test (Roche Molecular Diagnostics, Pleasanton, CA, USA), also conducted on the cobas® 5800/6800/8800 Systems, is an automated PCR test designed as a reflex test with cobas® MTB. It identifies RIF and INH resistance mutations in M. tuberculosis, analyzing eighteen RIF-resistance-associated mutations in the rpoB gene and seven INH-resistance-associated mutations in the katG gene and inhA gene promotor region. The LoD in raw sputum is 182 CFU/mL for RIF-resistant M. tuberculosis and 27.5 CFU/mL for INH-resistant M. tuberculosis [137].
The aforementioned Abbott RealTime MTB and Abbott RealTime MTB RIF/INH, BD MAX™ MDR-TB, FluoroType® MTBDR and FluoroType® MTB, and cobas® MTB and cobas MTB-RIF/INH have been classified as Moderate complexity automated (MC-a) NAATs for detection of TB and resistance to RIF and INH [69]. Overall, WHO prioritizes using mWRDs, including MC-aNAATs, as the initial diagnostic test for suspected TB. The use of molecular approaches, in addition to culture methods, offers several advantages, including faster results, improved sensitivity, rifampicin resistance detection, and, ultimately, quicker initiation of appropriate treatment [69]. All the NAATs presented in Table 1 (except for Truenat® MTB) detect DNA from MTBC but do not differentiate the subspecies, especially Mycobacterium bovis, which is a significant cause of zoonotic TB. This is particularly important as the zoonotic transmission of TB, especially that involving M. bovis, has gained increasing attention globally [143,144,145]. Therefore, because these methods do not differentiate between these two species, crucial information about the zoonotic aspect of TB may be overlooked, and a traditional culture-based method must be performed [125,128,129].
4.1.2. Line Probe Assays (LPAs)
Line-probe assays (LPAs) represent a category of DNA strip-based tests that detect the presence of MTBC and mutations associated with drug resistance. The interpretation of the results involves analyzing a band pattern on the strips, where immobilized probes are bound to MTBC amplicons. The probes are specifically designed to target prevalent mutations linked to resistance against both first- and second-line anti-TB drugs, as well as distinct MTBC wild-type DNA sequences. Two commonly used LPAs, GenoType® MTBDRplus and GenoType® MTBDRsl, are recommended by the WHO [146].
GenoType® MTBDRplus (VER 2.0; Hain Lifescience GmbH, Nehren, Germany) enables the identification of RIF resistance by detecting significant mutations in the rpoB gene. Moreover, it assesses INH resistance through the katG gene and the promoter region of the inhA gene. Benefits include testing from patient specimens or cultures and obtaining results in five hours (compared to 1 to 2 months required by conventional methods) [147]. In a study conducted by Moga et al. [148], GenoType® MTBDRplus VER 2.0 achieved a sensitivity of ~94.3% to detect INH resistance among MDR-TB isolates and a specificity of 100%. Similarly, Stephen et al. [149] reported 100% sensitivity and specificity to detect resistance to both INH and RIF. Furthermore, Tan et al. [150] reported that the test displayed a sensitivity of ~92.7% and a specificity of ~94.5% in diagnosing TB, while Meaza et al. [151] reported a sensitivity and specificity of ~96.4% and 100%, respectively, to detect MDR-TB.
GenoType® MTBDRsl (VER 1.0 and VER 2.0; Hain Lifescience GmbH, Nehren, Germany) assists in detecting extensively drug-resistant tuberculosis (XDR-TB) by identifying MTBC and its resistance to fluoroquinolones, aminoglycosides/cyclic peptides, second-line injectable drugs (KAN: kanamycin; AMK: amikacin; CAP: capreomycin) and ethambutol (only VER 1.0) [152]. According to Bouzouita et al. [153], GenoType® MTBDRsl 2.0 presented sensitivities of approximately 92.8% and 80% to detect resistance to fluoroquinolones and second-line injectable drugs (KAN, AMK, and CAP), respectively. Notably, specificity was 100% for the drugs mentioned above.
Detecting MDR-TB is a notable strength of LPAs. Given that MDR-TB poses a substantial public health risk, timely identification is essential for effectively treating and preventing the spread of the disease [154]. Another advantage is the ability of LPAs to detect specific genetic mutations associated with drug resistance. This allows for personalized treatment plans, as different M. tuberculosis strains may respond differently to different drugs [146].
However, drawbacks include the potential for false results, reliance on skilled personnel, and higher costs compared to traditional methods. Despite challenges, line probe assays are crucial in managing MDR-TB due to their ability to identify drug resistance-associated genetic markers [146].
4.2. Sequencing
Several studies highlight the impact of sequencing on diagnosing infectious diseases [155,156,157]. In 2018, WHO published one of the first guides concerning the use of whole genome sequencing (WGS) as a tool to study MTBC, particularly the mutations related to drug resistance. The document discussed the use of the sequencing platforms Illumina MiSeqTM (Illumina Inc., San Diego, CA, USA), Ion Personal Genome Machine® (Thermo Fisher Scientific, Inc., Waltham, MA, USA), Nanopore MinION® (Oxford Nanopore Technologies, Oxford, UK), and the GeneReader system (Qiagen, Hilden, Germany) [158]. Similarly, in October 2023, WHO published a document to help laboratories implement next-generation sequencing (NGS), also known as high throughput sequencing (HTS), for TB bacteria characterization, focusing on drug resistance mutations to complement existing TB surveillance systems [159]. The study by Vogel et al. [160] demonstrated the technical and financial aspects of implementing WGS in a National Reference Laboratory in Kyrgyzstan. The authors highlighted that implementing WGS for TB diagnostics involves challenges such as higher sequencing costs, extended procurement and capacity building timelines, early consideration of infrastructure requirements, tailored solutions for quality assurance, careful planning for transitioning WGS to routine diagnostics, and the necessity of ongoing support by experienced experts for sustainable success. A review by Ness, DiNardo, and Farhat [161] presents sequencing platforms for NGS of M. tuberculosis, as well as applications of targeted HTS in the context of TB.
4.3. MALDI-TOF MS
Regarding mass spectrometry, Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS) is revolutionizing clinical microbiology by rapidly and accurately identifying microorganisms in various samples, potentially improving patient outcomes, and reducing hospital stays [162]. Alcolea-Medina et al. [163] successfully established a fast and affordable method (MALDI-TOF) for identifying mycobacteria species in hospitals. It achieved 85% accuracy compared to existing methods, making it an attractive tool for clinical use.
Some studies present the use of nucleotide MALDI-TOF-MS and MALDI-TOF-MS as promising rapid tools for detecting drug resistance in M. tuberculosis [164,165,166], as well as the simultaneous detection of MTBC and mutations related to drug resistance [167,168,169], although these techniques cannot identify de novo drug resistance mutations [164] nor detect mutations related to novel resistance mechanisms [166].
MALDI-TOF MS is a rapid and cost-effective method for microbial identification. While molecular-based assays like PCR offer high specificity and sensitivity, they are generally more expensive and time-consuming. In laboratories equipped with MALDI-TOF MS, it is often the preferred method due to its ease of use and speed [163].
Although a pure culture enhances the specificity and sensitivity of the technique, some studies have shown that MALDI-TOF MS can be performed on raw specimens such as sputum and bronchoalveolar lavage fluid [167,168,170].
4.4. Biosensors
Lastly, biosensors are versatile biomedical diagnostic tools using targeted molecules to detect analytes [171] that usually consist of a biological sensing element along with a physicochemical transducer and a processor [172]. Notably, in TB diagnosis, DNA electrochemical biosensors, usually targeting the IS6110, demonstrate exceptional sensitivity, holding potential for drug-resistance identification [173,174]. In addition to DNA, biosensors can target antigens such as CFP-10/ESAT-6, MPT64, and Ag85, as well as antibodies, and cytokines [172]. Furthermore, biosensors are categorized based on their sensing technologies. In addition to electrochemical biosensors, there are other types, such as surface plasmon resonance (SPR), optical, mechanical, and quartz crystal microbalance (QCM)-based biosensors [175].
5. Immunological Approaches
The interaction between M. tuberculosis and the host involves a complex interplay of immune responses, influencing infection outcomes [176]. In TB diagnosis, immunology relies on important tools to detect adaptive immune responses in humans, such as interferon-gamma release assays (IGRAs) and tuberculin skin tests (TST), which are recommended by WHO for detecting LTBI [177]. However, no gold-standard technique exists for diagnosing this form of infection, and neither TST nor IGRAs are entirely accurate in predicting disease risk. Positive results from these tests should be considered alongside other clinical factors [178].
A comprehensive review by McIntyre et al. [179] highlighted that no serological tests have yet met WHO criteria for TB diagnosis, underscoring a gap in the understanding of the role of antibodies in TB immunity. Despite ongoing research, this incomplete knowledge complicates the development of effective vaccines, diagnostic tools, and treatments. Recent advances include the introduction of novel M. tuberculosis antigen-based skin tests (TBSTs) since 2022, which aim to enhance diagnostic accuracy alongside IGRAs and TSTs [180]. Additionally, a review by Melkie et al. [181] concluded that antibody tests remain insufficiently reliable for routine TB diagnosis, underscoring the need for further research and development in this area.
5.1. Interferon-Gamma Release Assays (IGRAs)
Interferon-gamma release assays (IGRAs) are blood-based tests that measure the production of interferon-gamma (IFN-γ) in response to M. tuberculosis antigens [182]. WHO recommends both IGRAs and TST for detecting LTBI [182], but IGRAs offer several advantages over TST, including a single-visit requirement and no influence by prior Bacille Calmette–Guérin (BCG) vaccination [183].
Despite their advantages, IGRAs are more expensive and require specialized laboratory equipment, which may not be readily available in low- and middle-income countries [183]. Furthermore, IGRAs yield indeterminate results in approximately 1 in 26 tests, particularly for immunocompromised individuals and young children [184].
Currently, three WHO-recommended IGRAs include:
- T-SPOT®.TB (T-Spot; Oxford Immunotec Ltd., Oxford, UK): uses the enzyme-linked immunospot (ELISPOT) method to count M. tuberculosis-sensitized T cells [185];
- QuantiFERON®-TB Gold Plus (QFT-Plus; Qiagen, Hilden, Germany): a fourth-generation assay that measures the cell-mediated immune response to two specific M. tuberculosis antigens—Early Secreted Antigenic Target 6 (ESAT-6) and Culture Filtrate Protein 10 (CFP-10)—using an ELISA-based approach [186];
- WANTAI TB-IGRA (Beijing Wantai Biological Pharmacy Enterprise Co Ltd., Beijing, China): an ELISA-based IGRA test similar to QFT-Plus, using a recombinant fusion protein of CFP-10 and ESAT-6 antigens [187].
Despite not being usually recommended for the diagnosis of active TB [156], several studies have explored the potential of IGRAs for this purpose [188,189,190,191,192].
Three new IGRAs—(1) Advansure TB IGRA (LG Chem, Seoul, Republic of Korea), (2) Lioferon TB/LTBI (LIONEX Diagnostics & Therapeutics GmbH, Braunschweig, Germany), and (3) Quantiferon-Diasorin (Stillwater, MN, USA)—are under review for potential WHO policy recommendations [93].
5.2. Tuberculin Skin Test (TST)
Tuberculin skin testing (TST), also known as the Mantoux test, involves the intradermal injection of 0.1 mL of tuberculin-purified protein derivative (PPD) into the forearm, creating a 6 to 10 mm wheal [193]. This wheal typically disappears within 15 to 20 min as the liquid is absorbed. The test is evaluated by a healthcare worker 48 to 72 h later by measuring the diameter of the induration (the raised, hardened area) in millimeters, which results from a delayed hypersensitivity reaction. The measurement excludes any surrounding erythema (redness) [194]. A result is considered positive for individuals devoid of risk factors for TB if the induration measures ≥15 mm; however, in immunocompromised patients or those receiving immunosuppressive treatments, a cut-off of ≥5 mm is used [194]. BCG vaccination significantly impacts TST specificity [183]. Nevertheless, BCG history does not interfere with TST results in children over three years old. For those under three, for whom BCG may cause false positives, using IGRAs is recommended. In situations in which IGRAs are unavailable or inconclusive, ignoring the BCG vaccination history is advised [195].
Despite limitations, TST remains a valuable tool for LTBI detection due to its cost-effectiveness and field applicability. However, it requires a cold chain for PPD storage and transportation, two healthcare visits, and specific training in intradermal injection, reading, and interpretation [183]. Although PPD requires a cold chain for storage and transportation [183], a study developed by Maes et al. [196] shows that tuberculin is not as heat labile as commonly believed. This finding is crucial since it suggests that tuberculin may be more resilient to temperature fluctuations than previously thought, emphasizing the importance of proper handling while offering reassurance regarding its viability even if cold chain is compromised. In general, TST continues to be clinically significant in both low and high TB-endemic regions until more advanced and widely available tests become accessible [197].
5.3. Mycobacterium Tuberculosis Antigen-Based Skin Tests (TBSTs)
Emerging as alternatives to traditional TST, TBSTs like Cy-Tb™ (Serum Institute of India, Pune, India), Diaskintest® (Generium, Moscow, Russian Federation) and C-TST (Anhui Zhifei Longcom, Chongqing, China) offer improved specificity and sensitivity for TB diagnosis [180]. The tests involve intradermal injections with M. tuberculosis antigen-based reagents and assess induration 48–72 h later. Cy-Tb™ employs a unique test dose (0.1 mL containing 0.05 μg each of recombinant dimer ESAT-6 (rdESAT-6) and recombinant CFP-10 (rCFP-10), boasting high accuracy (73.9% sensitivity, 99.3% specificity) [180,198]. Diaskintest® utilizes recombinant proteins (CFP-10 and ESAT-6), with hyperallergic reactions like blistering and necrosis considered rare [180], achieving approximate sensitivity and specificity of 91.18% and 99.15%, respectively [199]. Moreover, C-TST, formerly EC-Test, uses a similar antigen and assessment method [180], reaching 90.6% and 88.2% of sensitivity and specificity, respectively [193]. These TBSTs represent a promising advancement in TB diagnosis, offering improved accuracy and potentially reducing limitations associated with the traditional TST.
Regarding safety, a systematic review by Hamada et al. [200] found that these TBSTs showed safety profiles similar to TSTs, with mainly mild injection site reactions. This suggests their potential as alternatives, especially considering their accuracy near IGRA tests.
Despite their ease of administration and favorable safety profile, novel TBSTs have not yet surpassed the use of TST and IGRAs in clinical practice [200,201]. However, further data regarding TBSTs are needed, particularly for pregnant women [200].
6. Ongoing Research
Recent studies showcase promising advancements in TB diagnostics:
- Cepheid MTB-HR cartridge: This fingerstick blood test identifies a three-gene transcriptomic signature, achieving a sensitivity of 59.8% in distinguishing TB from non-TB cases. Combined with other methods, it identified 71.2% of confirmed TB cases [202].
- Immuno-affinity LC-MS (ILM) assay: This novel approach quantifies peptides from HIV-1 and M. tuberculosis proteins, achieving high sensitivity and specificity for both infections [203]. Additionally, it can differentiate treatment responders from non-responders, providing valuable insights for integrated TB and HIV management [203].
- CAPTURE-XT technology: This “lab-on-a-chip” platform uses dielectrophoresis to isolate M. tuberculosis from sputum, enabling efficient bacterial purification for subsequent molecular confirmation. It demonstrated high concordance with culture diagnosis, highlighting its potential as a robust sample preparation tool [204].
- Electronic nose (EN): Ketchanji Mougang et al. [205] conducted a study in Douala, Cameroon, assessing an EN for diagnosing PTB in a clinical setting. The EN utilizes eleven quartz microbalance sensors modified with metalloporphyrins and corroles to detect volatile organic compounds (VOCs) present in exhaled breath samples collected using a specialized breath sampler. Breath samples were segregated into alveolar and non-alveolar fractions, with analysis focusing exclusively on the alveolar portion to minimize external contaminants. The sensors detect changes in frequency resulting from interactions with VOCs, which exhibit unique patterns associated with TB. The EN demonstrated an accuracy of 88.0%, with a sensitivity of 90.8% and specificity of 85.7%, effectively distinguishing between PTB patients and healthy controls. Notably, the sensitivity of the EN was comparable to TB-LAMP and CXR, surpassing SSM.
These studies offer hope for improved TB diagnosis, particularly in challenging settings. Further research and development are crucial to translate these technologies into practical applications, contributing to global efforts for TB elimination.
Tests Undergoing WHO Policy Review
Various innovative diagnostic tools for TB have emerged, and several of these promising technologies are currently undergoing rigorous evaluation by the WHO to assess their suitability for incorporation into global TB diagnostic guidelines [93].
Concerning the culture-based drug susceptibility testing, Sensititre™ Mycobacterium tuberculosis MYCOTBI AST Plate (Thermo Fisher Scientific, Inc., Waltham, MA, USA) is a manual semiquantitative test whose results can be interpreted visually or with the aid of the ThermoScientific™ Sensititre™ Vizion™ System [206]. The test is based on microbroth dilution, testing 12 drugs: the first-line antibiotics rifampicin, rifabutin, isoniazid, and ethambutol, as well as the second-line antibiotics ofloxacin, moxifloxacin, amikacin, kanamycin, streptomycin, para-aminosalicylic acid, ethionamide, and cycloserine. Minimum inhibitory concentration (MIC) results can be obtained from 7 to 21 days [206].
Fujifilm SILVAMP TB LAM test (FujiLAM; Fujifilm, Tokyo, Japan), like the Abbott LF-LAM, detects lipoarabinomannan in urine samples [207], with 70% and 93% of sensitivity and specificity to detect TB in adults, and 51% and 87% for children [208]. This new point-of-care test has been considered suitable to detect PTB as well as extrapulmonary forms of the disease in patients with HIV [209] and is easy to be performed by any healthcare worker [210].
The three IGRAs currently undergoing WHO policy review involve (1) StandardTM E TB-Feron ELISA (SD Biosensor, Gyeonggi-do, Republic of Korea), (2) STANDARDTM F TB-Feron FIA (SD Biosensor, Gyeonggi-do, Republic of Korea), and (3) VIDAS® TB-IGRA (bioMérieux, France) [93]. The first is not influenced by previous BCG vaccination; results are available in approximately 100 min and present 98.03% and 98.55% sensitivity and specificity, respectively [211]. Concerning the efficacy of StandardTM E TB-Feron ELISA compared to QFT-Plus, accordance of 92% between the tests has been reported by Yoo et al. [212] and 94% by Jung et al. [213]. STANDARDTM F TB-Feron FIA, in turn, quantifies the IFN-γ in the blood samples through a fluorescent immunoassay (FIA) technique, delivering results within 15 min [214]. In South America, Saint-Pierre et al. [215] reported a sensitivity of 88.59% and a specificity of 92.5% for this test. Lastly, VIDAS® TB-IGRA exhibits sensitivity and specificity of 97.5% and 97.6%, respectively, with results available within 17 h (estimative for one patient) [216]. Petruccioli et al. [217] reported that the test is able to detect the IFN-γ response in CD4+/CD8+ T-cells for both TB and LTBI. Diagbouga et al. [218] also discussed that the test is promising for both active and latent TB.
Low complexity automated NAATs undergoing WHO policy review include the all-in-one cartridges STANDARD™ M10 MDR-TB (SD Biosensor, Gyeonggi-do, Republic of Korea) and IRON-qPCR™ RFIA Kit (Bioneer, Daejeon, Republic of Korea) [93]. STANDARD™ M10 MDR-TB performs the simultaneous detection of M. tuberculosis and resistance to RIF and INH from sputum samples based on qPCR technology, with a turnaround time of 80 min [219]. IRON-qPCR™ RFIA Kit is undergoing a clinical trial estimated to be completed in 2024. The test detects M. tuberculosis and mutations related to RIF, INH, fluoroquinolones, and aminoglycosides resistance, which is relevant since there are no WHO-endorsed tests to detect resistance to first- and second-line drugs to treat TB [220].
Finally, the DeepChek® Assay 13-Plex KB Drug Susceptibility Testing (ABL SA, Luxembourg, Luxembourg) assesses TB drug resistance through NGS. The test detects resistance-associated mutations in specific M. tuberculosis-targeted genes by sequencing. The key steps involve DNA extraction, multiplex PCR, NGS, and data analysis [221].
7. Final Remarks
In order to summarize the methods discussed in this review, an overview of the advantages and drawbacks of these methods, as well as the main diagnostics approaches used to diagnose pulmonary tuberculosis, can be visualized in Table 2 and Figure 1, respectively.

Table 2.
Overall advantages and disadvantages of the tests currently used to diagnose tuberculosis addressed in this review.
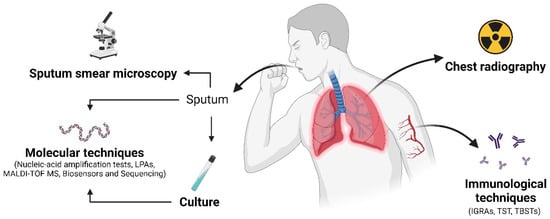
Figure 1.
This figure illustrates relevant diagnostic techniques employed for pulmonary tuberculosis diagnosis. Sputum is collected and examined under a microscope for acid-fast bacilli (AFB). Chest radiography assesses lung abnormalities such as infiltrates, cavities, or pleural effusions. Molecular techniques detect the genetic material of MTBC using methods like NAATs, LAMP, MALDI-TOF MS, biosensors, and sequencing. Immunological tests, such as IGRAs, TST, and TBSTs, measure the body’s immunity in response to mycobacterial proteins. Culture involves growing the mycobacteria in a laboratory for identification and further antimicrobial susceptibility testing. These techniques collectively aid in the diagnosis of pulmonary tuberculosis. Created with BioRender.com.
Finally, although the diagnosis of extrapulmonary TB is not the focus of this review, it is important to highlight the clinical relevance of the assessment of adenosine deaminase (ADA), which has been extensively used to diagnose these forms of tuberculosis [222,223,224]. ADA, an enzyme found in some types of leukocytes and crucial for purine metabolism, is associated with intracellular infections [225]. Elevated pleural fluid ADA levels are a useful marker for diagnosing tuberculous pleurisy (TPE), especially in high TB burden areas, though they can also be high in other conditions. Low ADA levels can help exclude TPE, prompting further investigation to identify the cause of pleural effusion [224]. ADA is vital for regulating immune, neurological, and vascular functions and aids in lymphocyte development. Elevated serum ADA can indicate various conditions that stimulate the immune system, including TB. However, ADA levels alone are not reliable for differentiating PTB from other lung infections [226].
8. Conclusions
Tuberculosis remains an important cause of death among infectious diseases, with granulomas as the hallmark of its pathophysiology. Since a wide range of the population is estimated to be infected with M. tuberculosis, exhibiting no symptoms, the infection can become active upon a series of factors, including the interaction between the pathogen and the host immune system. An important consequence of tuberculosis reactivation is the significant risk of transmitting M. tuberculosis to other individuals, which can amplify the spread of the disease within the community.
This review addresses the multiple approaches to diagnosing tuberculosis, focusing on pulmonary tuberculosis. Despite the availability of several molecular testing techniques, they are not accessible in various settings, especially in low- and middle-income countries. Here, culture-based methods play a critical role. The culture of M. tuberculosis not only remains the gold standard for diagnosis but also allows for the characterization of the pathogen, including drug susceptibility profile. This is crucial for ensuring effective treatment regimens and controlling the spread of drug-resistant strains. In many resource-limited settings, culture-based methods offer a vital approach to diagnosis where advanced molecular tests may not be feasible.
When feasible, the scenario of tuberculosis diagnosis can be improved with molecular testing without neglecting culture-based methods and SSM, thus improving specific identification of the etiological agent and the drug susceptibility testing, along with the use of decentralized and multi-disease testing (especially M. tuberculosis/HIV coinfection). Importantly, the tests should be affordable and favor non-sputum samples, such as oral swabs and urine. These samples are preferable since sputum samples are more difficult to obtain, making them a critical consideration for improving accessibility and convenience in testing [227].
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/diseases12090202/s1, Table S1: Some examples of tests developed for the diagnosis of non-tuberculous mycobacteria (NTM).
Author Contributions
All authors listed have made a substantial intellectual contribution to the study. Conceptualization, writing—original draft preparation, review and editing, G.B.-G., M.R.E.P. and S.F.Y.-O.; writing—original draft preparation, J.M.d.S., B.T.F., L.F.A.S., G.F.C., I.M.d.C., P.H.G.B., G.S.-R. and M.P.; writing—review and editing, E.R.T. and L.M.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, Financial Code 01).
Acknowledgments
G.B.-G., J.M.d.S., B.T.F., L.F.A.S., G.F.C., P.H.G.B. and G.S.-R. were funded by a graduate scholarship from CAPES. I.M.d.C. was funded by a graduate scholarship from Fundação Araucária-PR. S.F.Y.-O. was funded by a research fellowship from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, 309260/2022-1).
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Gagneux, S. Ecology and evolution of Mycobacterium tuberculosis. Nat. Rev. Microbiol. 2018, 16, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Pai, M.; Behr, M.A.; Dowdy, D.; Dheda, K.; Divangahi, M.; Boehme, C.C.; Ginsberg, A.; Swaminathan, S.; Spigelman, M.; Getahun, H.; et al. Tuberculosis. Nat. Rev. Dis. Primers 2016, 2, 16076. [Google Scholar] [CrossRef]
- World Health Organization. Global Tuberculosis Report 2023. 2023. Available online: https://www.who.int/teams/global-tuberculosis-programme/tb-reports/global-tuberculosis-report-2023 (accessed on 23 June 2024).
- Silva, S.; Arinaminpathy, N.; Atun, R.; Goosby, E.; Reid, M. Economic impact of tuberculosis mortality in 120 countries and the cost of not achieving the Sustainable Development Goals tuberculosis targets: A full-income analysis. Lancet Glob. Health 2021, 9, e1372–e1379. [Google Scholar] [CrossRef] [PubMed]
- Bhalla, A.S.; Goyal, A.; Guleria, R.; Gupta, A.K. Chest tuberculosis: Radiological review and imaging recommendations. Indian J. Radiol. Imaging 2015, 25, 213–225. [Google Scholar] [CrossRef]
- Natarajan, A.; Beena, P.M.; Devnikar, A.V.; Mali, S. A systemic review on tuberculosis. Indian J. Tuberc. 2020, 67, 295–311. [Google Scholar] [CrossRef]
- Sharma, S.K.; Mohan, A.; Kohli, M. Extrapulmonary tuberculosis. Expert Rev. Respir. Med. 2021, 15, 931–948. [Google Scholar] [CrossRef] [PubMed]
- Htet, K.K.K.; Chongsuvivatwong, V.; Aung, S.T. Sensitivity and specificity of tuberculosis signs and symptoms screening and adjunct role of social pathology characteristics in predicting bacteriologically confirmed tuberculosis in Myanmar. Trop. Med. Health 2021, 49, 3. [Google Scholar] [CrossRef]
- Knechel, N.A. Tuberculosis: Pathophysiology, clinical features, and diagnosis. Crit. Care Nurse 2009, 29, 34–44. [Google Scholar] [CrossRef]
- World Health Organization. Guidelines on the Management of Latent Tuberculosis Infection. 2015. Available online: https://www.who.int/publications/i/item/9789241548908 (accessed on 23 June 2024).
- Kiazyk, S.; Ball, T.B. Latent tuberculosis infection: An overview. Can. Commun. Dis. Rep. 2017, 43, 62–66. [Google Scholar] [CrossRef]
- Turner, R.D.; Bothamley, G.H. Cough and the transmission of tuberculosis. J. Infect. Dis. 2015, 211, 1367–1372. [Google Scholar] [CrossRef]
- Turner, R.D.; Chiu, C.; Churchyard, G.J.; Esmail, H.; Lewinsohn, D.M.; Gandhi, N.R.; Fennelly, K.P. Tuberculosis Infectiousness and Host Susceptibility. J. Infect. Dis. 2017, 216, S636–S643. [Google Scholar] [CrossRef] [PubMed]
- Imtiaz, S.; Shield, K.D.; Roerecke, M.; Samokhvalov, A.V.; Lönnroth, K.; Rehm, J. Alcohol consumption as a risk factor for tuberculosis: Meta-analyses and burden of disease. Eur. Respir. J. 2017, 50, 1700216. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, N.; Bhattarai, R.B.; Basnet, R.; Joshi, L.R.; Tinkari, B.S.; Thapa, A.; Joshi, B. Prevalence and associated risk factors for tuberculosis among people living with HIV in Nepal. PLoS ONE 2022, 17, e0262720. [Google Scholar] [CrossRef] [PubMed]
- Gelaw, Y.; Getaneh, Z.; Melku, M. Anemia as a risk factor for tuberculosis: A systematic review and meta-analysis. Environ. Health Prev. Med. 2021, 26, 13. [Google Scholar] [CrossRef]
- Chen, Z.; Liu, Q.; Song, R.; Zhang, W.; Wang, T.; Lian, Z.; Sun, X.; Liu, Y. The association of glycemic level and prevalence of tuberculosis: A meta-analysis. BMC Endocr. Disord. 2021, 16, 123. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, R.; Akugizibwe, P.; Siegfried, N.; Rees, D. The association between silica exposure, silicosis and tuberculosis: A systematic review and meta-analysis. BMC Public Health 2021, 21, 953. [Google Scholar] [CrossRef]
- Obore, N.; Kawuki, J.; Guan, J.; Papabathini, S.S.; Wang, L. Association between indoor air pollution, tobacco smoke and tuberculosis: An updated systematic review and meta-analysis. Public Health 2020, 187, 24–35. [Google Scholar] [CrossRef]
- Dias, M.; Gaio, R.; Sousa, P.; Abranches, M.; Gomes, M.; Oliveira, O.; Correia-Neves, M.; Ferreira, E.; Duarte, R. Tuberculosis among the homeless: Should we change the strategy? Int. J. Tuberc. Lung Dis. 2017, 21, 327–332. [Google Scholar] [CrossRef]
- Velen, K.; Charalambous, S. Tuberculosis in prisons: An unintended sentence? Lancet Public Health 2021, 6, e263–e264. [Google Scholar] [CrossRef]
- Cronan, M.R. In the Thick of It: Formation of the Tuberculous Granuloma and Its Effects on Host and Therapeutic Responses. Front. Immunol. 2022, 13, 820134. [Google Scholar] [CrossRef]
- Alsayed, S.S.; Gunosewoyo, H. Tuberculosis: Pathogenesis, current treatment regimens and new drug targets. Int. J. Mol. Sci. 2023, 24, 5202. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Chen, Y.; Chen, J.; Xu, K.; Xu, F.; Shi, J. The relationship between previous pulmonary tuberculosis and risk of lung cancer in the future. Infect. Agents Cancer 2022, 17, 20. [Google Scholar] [CrossRef]
- Hwang, S.Y.; Kim, J.Y.; Lee, H.S.; Lee, S.; Kim, D.; Kim, S.; Hyun, J.H.; Shin, J.I.; Lee, K.H.; Han, S.H.; et al. Pulmonary Tuberculosis and Risk of Lung Cancer: A Systematic Review and Meta-Analysis. J. Clin. Med. 2022, 11, 765. [Google Scholar] [CrossRef] [PubMed]
- Ho, L.J.; Yang, H.Y.; Chung, C.H.; Chang, W.C.; Yang, S.S.; Sun, C.A.; Chien, W.C.; Su, R.Y. Increased risk of secondary lung cancer in patients with tuberculosis: A nationwide, population-based cohort study. PLoS ONE 2021, 16, e0250531. [Google Scholar] [CrossRef]
- MacGregor-Fairlie, M.; Wilkinson, S.; Besra, G.S.; Goldberg Oppenheimer, P. Tuberculosis diagnostics: Overcoming ancient challenges with modern solutions. Emerg. Top. Life Sci. 2020, 4, 423–436. [Google Scholar] [CrossRef]
- Ahmed, I.; Tiberi, S.; Farooqi, J.; Jabeen, K.; Yeboah-Manu, D.; Migliori, G.B.; Hasan, R. Non-tuberculous mycobacterial infections-A neglected and emerging problem. Int. J. Infect. Dis. 2020, 92, S46–S50. [Google Scholar] [CrossRef] [PubMed]
- Ying, C.; Zhang, L.; Jin, X.; Zhu, D.; Wu, W. Advances in diagnosis and treatment of non-tuberculous mycobacterial lung disease. Diagn. Microbiol. Infect. Dis. 2024, 109, 116254. [Google Scholar] [CrossRef]
- Chindam, A.; Vengaldas, S.; Srigiri, V.R.; Syed, U.; Kilaru, H.; Chenimilla, N.P.; Kilaru, S.C.; Patil, E. Challenges of diagnosing and treating non-tuberculous mycobacterial pulmonary disease [NTM-PD]: A case series. J. Clin. Tuberc. Other Mycobact. Dis. 2021, 25, 100271. [Google Scholar] [CrossRef]
- Zhu, L.; Jiang, G.; Wang, S.; Wang, C.; Li, Q.; Yu, H.; Zhou, Y.; Zhao, B.; Huang, H.; Xing, W.; et al. Biochip system for rapid and accurate identification of mycobacterial species from isolates and sputum. J. Clin. Microbiol. 2010, 48, 3654–3660. [Google Scholar] [CrossRef]
- Ramos, A.; Carvalho, T.; Ribeiro, M.; Guimarães, J.T. Capilia™ TB-Neo assay: A new tool for rapid distinction between tuberculous and non-tuberculous mycobacteria. Int. J. Tuberc. Lung. Dis. 2016, 20, 753–756. [Google Scholar] [CrossRef]
- Chae, H.; Han, S.J.; Kim, S.Y.; Ki, C.S.; Huh, H.J.; Yong, D.; Koh, W.J.; Shin, S.J. Development of a One-Step Multiplex PCR Assay for Differential Detection of Major Mycobacterium Species. J. Clin. Microbiol. 2017, 55, 2736–2751. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, K.; Garima, K.; Narang, A.; Bhattacharyya, K.; Vishnoi, E.; Singh, R.K.; Chaudhry, A.; Prasad, R.; Bose, M.; Varma-Basil, M. Rv1458c: A new diagnostic marker for identification of Mycobacterium tuberculosis complex in a novel duplex PCR assay. J. Med. Microbiol. 2017, 66, 371–376. [Google Scholar] [CrossRef]
- Singh, S.; Sharma, M.; Modi, M.; Sharma, A.; Singh, R.; Ray, P.; Sharma, K. High-resolution melting curve analysis of heat shock protein 65 for identification of mycobacterial isolates. Int. J. Tuberc. Lung. Dis. 2018, 22, 1511–1513. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Shu, Q.; Zhao, Z.; Fan, J.; Lyon, C.J.; Zelazny, A.M.; Hu, Y. Antigen 85B peptidomic analysis allows species-specific mycobacterial identification. Clin. Proteom. 2018, 15, 1. [Google Scholar] [CrossRef] [PubMed]
- Chuensirikulchai, K.; Laopajon, W.; Phunpae, P.; Apiratmateekul, N.; Surinkaew, S.; Tayapiwatana, C.; Pata, S.; Kasinrerk, W. Sandwich antibody-based biosensor system for identification of Mycobacterium tuberculosis complex and nontuberculous mycobacteria. J. Immunoass. Immunochem. 2019, 40, 590–604. [Google Scholar] [CrossRef] [PubMed]
- Wood, H.N.; Sidders, A.E.; Brumsey, L.E.; Morozkin, E.S.; Gerasimova, Y.V.; Rohde, K.H. Species Typing of Nontuberculous Mycobacteria by Use of Deoxyribozyme Sensors. Clin. Chem. 2019, 65, 333–341. [Google Scholar] [CrossRef]
- Shin, S.; Yoo, I.Y.; Shim, H.J.; Kang, O.K.; Jhun, B.W.; Koh, W.J.; Huh, H.J.; Lee, N.Y. Diagnostic Performance of the GENEDIA MTB/NTM Detection Kit for Detecting Mycobacterium tuberculosis and Nontuberculous Mycobacteria with Sputum Specimens. Ann. Lab. Med. 2020, 40, 169–173. [Google Scholar] [CrossRef]
- Xiao, G.; Zhang, S.; Liang, Z.; Li, G.; Fang, M.; Liu, Y.; Zhang, J.; Ou, M.; He, X.; Zhang, T.; et al. Identification of Mycobacterium abscessus species and subspecies using the Cas12a/sgRNA-based nucleic acid detection platform. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 551–558. [Google Scholar] [CrossRef]
- Li, B.; Zhu, C.; Sun, L.; Dong, H.; Sun, Y.; Cao, S.; Zhen, L.; Qi, Q.; Zhang, Q.; Mo, T.; et al. Performance evaluation and clinical validation of optimized nucleotide MALDI-TOF-MS for mycobacterial identification. Front. Cell. Infect. Microbiol. 2022, 12, 1079184. [Google Scholar] [CrossRef]
- Fukushima, K.; Matsumoto, Y.; Matsuki, T.; Saito, H.; Motooka, D.; Komukai, S.; Fukui, E.; Yamuchi, J.; Nitta, T.; Niitsu, T.; et al. MGIT-seq for the Identification of Nontuberculous Mycobacteria and Drug Resistance: A Prospective Study. J. Clin. Microbiol. 2023, 61, e0162622. [Google Scholar] [CrossRef]
- Uwamino, Y.; Aono, A.; Tomita, Y.; Morimoto, K.; Kawashima, M.; Kamata, H.; Sasaki, Y.; Nagai, H.; Hasegawa, N.; Mitarai, S. Diagnostic Utility of a Mycobacterium Multiplex PCR Detection Panel for Tuberculosis and Nontuberculous Mycobacterial Infections. Microbiol. Spectr. 2023, 11, e0516222. [Google Scholar] [CrossRef]
- Wang, J.; Xu, H.; Wang, X.; Lan, J. Rapid diagnosis of non-tuberculous mycobacterial pulmonary diseases by metagenomic next-generation sequencing in non-referral hospitals. Front. Cell. Infect. Microbiol. 2023, 12, 1083497. [Google Scholar] [CrossRef]
- Wu, T.; Shen, C.; Zhao, Z.; Lyu, M.; Bai, H.; Hu, X.; Zhao, J.; Zhang, R.; Qian, K.; Xu, G.; et al. Integrating Paper-Based Microfluidics and Lateral Flow Strip into Nucleic Acid Amplification Device toward Rapid, Low-Cost, and Visual Diagnosis of Multiple Mycobacteria. Small Methods 2024, 11, e2400095. [Google Scholar] [CrossRef] [PubMed]
- Dong, B.; He, Z.; Li, Y.; Xu, X.; Wang, C.; Zeng, J. Improved Conventional and New Approaches in the Diagnosis of Tuberculosis. Front. Microbiol. 2022, 13, 924410. [Google Scholar] [CrossRef]
- Filardo, T.D.; Feng, P.J.; Pratt, R.H.; Price, S.F.; Self, J.L. Tuberculosis—United States, 2021. MMWR Morb. Mortal. Wkly. 2022, 71, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.A.; Leite, A.; Soares, P.; Duarte, R.; Nunes, C. Delayed diagnosis of active pulmonary tuberculosis—Potential risk factors for patient and healthcare delays in Portugal. BMC Public Health 2021, 21, 2178. [Google Scholar] [CrossRef] [PubMed]
- Kraef, C.; Bentzon, A.; Panteleev, A.; Skrahina, A.; Bolokadze, N.; Tetradov, S.; Podlasin, R.; Karpov, I.; Borodulina, E.; Denisova, E.; et al. Delayed diagnosis of tuberculosis in persons living with HIV in Eastern Europe: Associated factors and effect on mortality-a multicentre prospective cohort study. BMC Infect. Dis. 2021, 21, 1038. [Google Scholar] [CrossRef] [PubMed]
- Ershova, J.V.; Volchenkov, G.V.; Somova, T.R.; Kuznetsova, T.A.; Kaunetis, N.V.; Kaminski, D.; Demikhova, O.V.; Chernousova, L.N.; Vasilyeva, I.A.; Kerr, E.M.; et al. Impact of GeneXpert MTB/RIF® on treatment initiation and outcomes of RIF-resistant and RIF-susceptible TB patients in Vladimir TB dispensary, Russia. BMC Infect. Dis. 2020, 20, 543. [Google Scholar] [CrossRef]
- Raja, R.; Sreeramulu, P.N.; Dave, P.; Srinivasan, D. GeneXpert assay–A cutting-edge tool for rapid tissue diagnosis of tuberculous lymphadenitis. J. Clin. Tuberc. Other Mycobact. Dis. 2020, 21, 100204. [Google Scholar] [CrossRef]
- World Health Organization. Implementing the End TB Strategy: The Essentials, 2022 Update. 2022. Available online: https://www.who.int/publications/i/item/9789240065093 (accessed on 23 June 2024).
- World Health Organization. Global Tuberculosis Report 2022. 2022. Available online: https://www.who.int/teams/global-tuberculosis-programme/tb-reports/global-tuberculosis-report-2022 (accessed on 23 June 2024).
- Campelo, T.A.; Cardoso de Sousa, P.R.; Nogueira, L.L.; Frota, C.C.; Zuquim Antas, P.R. Revisiting the methods for detecting Mycobacterium tuberculosis: What has the new millennium brought thus far? Access Microbiol. 2021, 3, 000245. [Google Scholar] [CrossRef]
- Seki, M.; Kim, C.K.; Hayakawa, S.; Mitarai, S. Recent advances in tuberculosis diagnostics in resource-limited settings. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 1405–1410. [Google Scholar] [CrossRef] [PubMed]
- Figueredo, L.J.A.; Miranda, S.S.; Santos, L.B.D.; Manso, C.G.G.; Soares, V.M.; Alves, S.; Vater, M.C.; Kritski, A.L.; Carvalho, W.D.S.; Pádua, C.M.; et al. Cost analysis of smear microscopy and the Xpert assay for tuberculosis diagnosis: Average turnaround time. Rev. Soc. Bras. Med. Trop. 2020, 53, e20200314. [Google Scholar] [CrossRef]
- Vilchèze, C.; Kremer, L. Acid-Fast Positive and Acid-Fast Negative Mycobacterium tuberculosis: The Koch Paradox. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef] [PubMed]
- Steingart, K.R.; Henry, M.; Ng, V.; Hopewell, P.C.; Ramsay, A.; Cunningham, J.; Urbanczik, R.; Perkins, M.; Aziz, M.A.; Pai, M. Fluorescence versus conventional sputum smear microscopy for tuberculosis: A systematic review. Lancet Infect. Dis. 2006, 6, 570–581. [Google Scholar] [CrossRef]
- Desikan, P. Sputum smear microscopy in tuberculosis: Is it still relevant? Indian J. Med. Res. 2013, 137, 442–444. [Google Scholar]
- Chopra, S.; Mahajan, S.; Singh, Y.; Chopra, M. Comparative evaluation of Ziehl-Neelsen staining and Kinyoun’s staining in the diagnosis of clinically suspected cases of tuberculosis. IP Int. J. Med. Microbiol. Trop. Dis. 2022, 8, 149–153. [Google Scholar] [CrossRef]
- Zheng, R.; Xu, F.; Huang, X.; Wang, J.; Feng, Y.; Huang, J.; Qin, L. Evaluation of Aptamer Fluorescence Microscopy in the Diagnosis of Pulmonary Tuberculosis. Microbiol. Spectr. 2022, 10, e0260221. [Google Scholar] [CrossRef]
- Whitelaw, A.; Peter, J.; Sohn, H.; Viljoen, D.; Theron, G.; Badri, M.; Davids, V.; Pai, M.; Dheda, K. Comparative cost and performance of light-emitting diode microscopy in HIV-tuberculosis-co-infected patients. Eur. Respir. J. 2011, 38, 1393–1397. [Google Scholar] [CrossRef] [PubMed]
- Steingart, K.R.; Ng, V.; Henry, M.; Hopewell, P.C.; Ramsay, A.; Cunningham, J.; Urbanczik, R.; Perkins, M.D.; Aziz, M.A.; Pai, M. Sputum processing methods to improve the sensitivity of smear microscopy for tuberculosis: A systematic review. Lancet Infect. Dis. 2006, 6, 664–674. [Google Scholar] [CrossRef]
- Fu, H.T.; Tu, H.Z.; Lee, H.S.; Lin, Y.E.; Lin, C.W. Evaluation of an AI-Based TB AFB Smear Screening System for Laboratory Diagnosis on Routine Practice. Sensors 2022, 22, 8497. [Google Scholar] [CrossRef]
- Kotei, E.; Thirunavukarasu, R. Computational techniques for the automated detection of Mycobacterium tuberculosis from digitized sputum smear microscopic images: A systematic review. Prog. Biophys. Mol. Biol. 2022, 171, 4–16. [Google Scholar] [CrossRef] [PubMed]
- Zingue, D.; Weber, P.; Soltani, F.; Raoult, D.; Drancourt, M. Automatic microscopic detection of mycobacteria in sputum: A proof-of-concept. Sci. Rep. 2018, 8, 11308. [Google Scholar] [CrossRef]
- Panicker, R.O.; Kalmady, K.S.; Rajan, J.; Sabu, M.K. Automatic detection of tuberculosis bacilli from microscopic sputum smear images using deep learning methods. Biocybern. Biomed. Eng. 2018, 38, 691–699. [Google Scholar] [CrossRef]
- Van Deun, A.; Tahseen, S.; Affolabi, D.; Hossain, M.A.; Joloba, M.L.; Angra, P.K.; Ridderhof, J.C.; de Jong, B.C.; Rieder, H.L. Sputum smear microscopy in the Xpert® MTB/RIF era. Int. J. Tuberc. Lung. Dis. 2019, 23, 12–18. [Google Scholar] [CrossRef]
- World Health Organization. Operational Handbook on Tuberculosis. Module 3: Diagnosis—Rapid Diagnostics for Tuberculosis Detention, 2021 Update. 2021. Available online: https://www.who.int/publications/i/item/9789240030589 (accessed on 23 June 2024).
- Das, P.K.; Ganguly, S.B.; Mandal, B. Sputum smear microscopy in tuberculosis: It is still relevant in the era of molecular diagnosis when seen from the public health perspective. Biomed. Biotechnol. Res. J. 2019, 3, 77–79. [Google Scholar] [CrossRef]
- Sohn, H.; Kasaie, P.; Kendall, E.; Gomez, G.B.; Vassall, A.; Pai, M.; Dowdy, D. Informing decision-making for universal access to quality tuberculosis diagnosis in India: An economic-epidemiological model. BMC Med. 2019, 17, 155. [Google Scholar] [CrossRef] [PubMed]
- Maheshwari, P.; Chauhan, K.; Kadam, R.; Pujani, A.; Kaur, M.; Chitalia, M.; Dabas, H.; Perkins, M.D.; Boehme, C.C.; Denkinger, C.M.; et al. Market assessment of tuberculosis diagnostics in India in 2013. Int. J. Tuberc. Lung. Dis. 2016, 20, 304–313. [Google Scholar] [CrossRef]
- Kik, S.V.; Denkinger, C.M.; Chedore, P.; Pai, M. Replacing smear microscopy for the diagnosis of tuberculosis: What is the market potential? Eur. Respir. J. 2014, 43, 1793–1796. [Google Scholar] [CrossRef] [PubMed]
- Nachiappan, A.C.; Rahbar, K.; Shi, X.; Guy, E.S.; Mortani Barbosa, E.J., Jr.; Shroff, G.S.; Ocazionez, D.; Schlesinger, A.E.; Katz, S.I.; Hammer, M.M. Pulmonary Tuberculosis: Role of Radiology in Diagnosis and Management. Radiographics 2017, 37, 52–72. [Google Scholar] [CrossRef]
- Pinto, L.M.; Pai, M.; Dheda, K.; Schwartzman, K.; Menzies, D.; Steingart, K.R. Scoring systems using chest radiographic features for the diagnosis of pulmonary tuberculosis in adults: A systematic review. Eur. Respir. J. 2013, 42, 480–494. [Google Scholar] [CrossRef]
- World Health Organization. Chest Radiography in Tuberculosis Detection: Summary of Current WHO Recommendations and Guidance on Programmatic Approaches. 2016. Available online: https://www.who.int/publications/i/item/9789241511506 (accessed on 23 June 2024).
- Van’t Hoog, A.; Viney, K.; Biermann, O.; Yang, B.; Leeflang, M.M.; Langendam, M.W. Symptom- and chest-radiography screening for active pulmonary tuberculosis in HIV-negative adults and adults with unknown HIV status. Cochrane Database Syst. Rev. 2022, 3, CD010890. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.C.; Tan, Q.; Becerra, M.C.; Calderon, R.; Chiang, S.S.; Contreras, C.; Lecca, L.; Jimenez, J.; Perez-Velez, C.M.; Roya-Pabon, C.L.; et al. The Contribution of Chest Radiography to the Clinical Management of Children Exposed to Tuberculosis. Am. J. Respir. Crit. Care Med. 2022, 206, 892–900. [Google Scholar] [CrossRef]
- Nel, M.; Franckling-Smith, Z.; Pillay, T.; Andronikou, S.; Zar, H.J. Chest Imaging for Pulmonary TB-An Update. Pathogens 2022, 11, 161. [Google Scholar] [CrossRef] [PubMed]
- Feyisa, D.W.; Ayano, Y.M.; Debelee, T.G.; Schwenker, F. Weak Localization of Radiographic Manifestations in Pulmonary Tuberculosis from Chest X-ray: A Systematic Review. Sensors 2023, 23, 6781. [Google Scholar] [CrossRef]
- Meghji, J.; Simpson, H.; Squire, S.B.; Mortimer, K. A Systematic Review of the Prevalence and Pattern of Imaging Defined Post-TB Lung Disease. PLoS ONE 2016, 11, e0161176. [Google Scholar] [CrossRef]
- Park, M.; Lee, Y.; Kim, S.; Kim, Y.J.; Kim, S.Y.; Kim, Y.; Kim, H.M. Distinguishing nontuberculous mycobacterial lung disease and Mycobacterium tuberculosis lung disease on X-ray images using deep transfer learning. BMC Infect. Dis. 2023, 23, 32. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ding, W.; Mo, Y.; Shi, D.; Zhang, S.; Zhong, L.; Wang, K.; Wang, J.; Huang, C.; Zhang, S.; et al. Distinguishing nontuberculous mycobacteria from Mycobacterium tuberculosis lung disease from CT images using a deep learning framework. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 4293–4306. [Google Scholar] [CrossRef]
- Mohammed, H.; Oljira, L.; Roba, K.T.; Ngadaya, E.; Tesfaye, D.; Manyazewal, T.; Yimer, G. Impact of early chest radiography on delay in pulmonary tuberculosis case notification in Ethiopia. Int. J. Mycobacteriol. 2021, 10, 364–372. [Google Scholar] [CrossRef]
- Kulkarni, S.; Jha, S. Artificial intelligence, radiology, and tuberculosis: A review. Acad. Radiol. 2019, 27, 71–75. [Google Scholar] [CrossRef]
- Harris, M.; Qi, A.; Jeagal, L.; Torabi, N.; Menzies, D.; Korobitsyn, A.; Pai, M.; Nathavitharana, R.R.; Ahmad Khan, F. A systematic review of the diagnostic accuracy of artificial intelligence-based computer programs to analyze chest x-rays for pulmonary tuberculosis. PLoS ONE 2019, 14, e0221339. [Google Scholar] [CrossRef]
- World Health Organization. Rapid Communication on Systematic Screening for Tuberculosis. 2020. Available online: https://www.who.int/publications/i/item/9789240016552 (accessed on 23 June 2024).
- Nijiati, M.; Ma, J.; Hu, C.; Tuersun, A.; Abulizi, A.; Kelimu, A.; Zhang, D.; Li, G.; Zou, X. Artificial Intelligence Assisting the Early Detection of Active Pulmonary Tuberculosis from Chest X-Rays: A Population-Based Study. Front. Mol. Biosci. 2022, 9, 874475. [Google Scholar] [CrossRef]
- Heo, S.J.; Kim, Y.; Yun, S.; Lim, S.S.; Kim, J.; Nam, C.M.; Park, E.C.; Jung, I.; Yoon, J.H. Deep Learning Algorithms with Demographic Information Help to Detect Tuberculosis in Chest Radiographs in Annual Workers’ Health Examination Data. Int. J. Environ. Res. Public Health 2019, 16, 250. [Google Scholar] [CrossRef] [PubMed]
- Rahman, T.; Khandakar, A.; Kadir, M.A.; Islam, K.R.; Islam, K.F.; Mazhar, R.; Hamid, T.; Islam, M.T.; Mahbub, Z.B.; Ayari, M.A.; et al. Reliable tuberculosis detection using chest X-ray with deep learning, segmentation and visualization. IEEE Access 2020, 8, 191586–191601. [Google Scholar] [CrossRef]
- Qin, Z.Z.; Barrett, R.; Ahmed, S.; Sarker, M.S.; Paul, K.; Adel, A.S.S.; Banu, S.; Creswell, J. Comparing different versions of computer-aided detection products when reading chest X-rays for tuberculosis. PLoS Digit. Health 2022, 1, e0000067. [Google Scholar] [CrossRef] [PubMed]
- Geric, C.; Qin, Z.Z.; Denkinger, C.M.; Kik, S.V.; Marais, B.; Anjos, A.; David, P.M.; Ahmad Khan, F.; Trajman, A. The rise of artificial intelligence reading of chest X-rays for enhanced TB diagnosis and elimination. Int. J. Tuberc. Lung Dis. 2023, 27, 367–372. [Google Scholar] [CrossRef]
- World Health Organization. Global Tuberculosis Report 2023. TB Research and Innovation. 2023. Available online: https://www.who.int/teams/global-tuberculosis-programme/tb-reports/global-tuberculosis-report-2023/tb-research-and-innovation (accessed on 26 December 2023).
- Xu, Y.; Wang, G.; Xu, M. Biohazard levels and biosafety protection for Mycobacterium tuberculosis strains with different virulence. Biosaf. Health 2020, 2, 135–141. [Google Scholar] [CrossRef]
- Drancourt, M.; Carrieri, P.; Gévaudan, M.J.; Raoult, D. Blood agar and Mycobacterium tuberculosis: The end of a dogma. J. Clin. Microbiol. 2003, 41, 1710–1711. [Google Scholar] [CrossRef]
- Stop TB Partnership. GLI Mycobacteriology Laboratory Manual. 2014. Available online: https://stoptb.org/wg/gli/assets/documents/gli_mycobacteriology_lab_manual_web.pdf (accessed on 21 August 2024).
- Kudoh, S.; Kudoh, T. A simple technique for culturing tubercle bacilli. Bull. World Health Organ. 1974, 51, 71–82. [Google Scholar]
- Franco-Sotomayor, G.; Rivera-Olivero, I.A.; Leon-Benitez, M.; Uruchima-Campoverde, S.E.; Cardenas-Franco, G.; Perdomo-Castro, M.E.; Cardenas-Franco, C.S.; Ortega-Vivanco, J.; Abad-Ruiz, A.S.; de Waard, J.H.; et al. Fast, Simple, and Cheap: The Kudoh-Ogawa Swab Method as an Alternative to the Petroff-Lowenstein-Jensen Method for Culturing of Mycobacterium tuberculosis. J. Clin. Microbiol. 2020, 58, e01424-19. [Google Scholar] [CrossRef]
- Costa, R.R.D.; Silva, S.F.D.; Fochat, R.C.; Macedo, R.L.; Pereira, T.V.; Silva, M.R.; Pinto, C.P.G.; Leite, I.C.G. Comparison between Ogawa-Kudoh and modified Petroff techniques for mycobacteria cultivation in the diagnosis of pulmonary tuberculosis. Einstein (São Paulo) 2018, 16, eAO4214. [Google Scholar] [CrossRef]
- Jaspe, R.C.; Rojas, Y.M.; Flores, L.A.; Sofia Toro, E.; Takiff, H.; de Waard, J.H. Evaluation of the Kudoh swab method for the culturing of Mycobacterium tuberculosis in rural areas. Trop. Med. Int. Health 2009, 14, 468–471. [Google Scholar] [CrossRef] [PubMed]
- Jobarteh, T.; Out, J.; Gitteh, E.; Mendy, F.S.; Faal-Jawara, T.I.; Ofori-Anyinam, B.; Sarr, B.; Riley, A.J.; Ayorinde, A.; de Jong, B.C.; et al. The use of Kudoh method for culture of Mycobacterium tuberculosis and Mycobacterium africanum in The Gambia. PLoS ONE 2024, 19, e0300042. [Google Scholar] [CrossRef]
- Madeira, C.M.; Azam, K.I.; Sato, D.N.; Khosa, C.; Bhatt, N.; Viegas, S.O. Evaluation of the Ogawa-Kudoh method for tuberculosis isolation in two health units in Mozambique. Afr. J. Lab. Med. 2020, 9, 929. [Google Scholar] [CrossRef] [PubMed]
- American Society for Microbiology. How TB Diagnostics Have Evolved Since the Second Century. 2021. Available online: https://asm.org/Articles/2021/March/How-TB-Diagnostics-Have-Evolved-Since-the-Second-C (accessed on 15 January 2024).
- Banaei, N.; Kincaid, E.Z.; Lin, S.Y.; Desmond, E.; Jacobs, W.R., Jr.; Ernst, J.D. Lipoprotein processing is essential for resistance of Mycobacterium tuberculosis to malachite green. Antimicrob. Agents Chemother. 2009, 53, 3799–3802. [Google Scholar] [CrossRef]
- Sigma Aldrich. Product M0178. Middlebrook 7H9 Broth Base. Available online: https://www.sigmaaldrich.com/BR/pt/product/sial/m0178?utm_source=google&utm_medium=cpc&utm_campaign=19329107722&utm_content=141938264102&gclid=CjwKCAiA-vOsBhAAEiwAIWR0TU6ck_CPWtIy4WGvax-giogdjnsUXVIiDWGXxJltDfyeCc_KXuKI8xoCFTAQAvD_BwE (accessed on 15 January 2024).
- Franzblau, S.G.; DeGroote, M.A.; Cho, S.H.; Andries, K.; Nuermberger, E.; Orme, I.M.; Mdluli, K.; Angulo-Barturen, I.; Dick, T.; Dartois, V.; et al. Comprehensive analysis of methods used for the evaluation of compounds against Mycobacterium tuberculosis. Tuberculosis 2012, 92, 453–488. [Google Scholar] [CrossRef] [PubMed]
- Roquet-Banères, F.; Alcaraz, M.; Hamela, C.; Abendroth, J.; Edwards, T.E.; Kremer, L. In vitro and in vivo efficacy of NITD-916 against Mycobacterium fortuitum. Antimicrob. Agents Chemother. 2023, 67, e0160722. [Google Scholar] [CrossRef]
- Aguilera-Correa, J.J.; Boudehen, Y.M.; Kremer, L. Characterization of Mycobacterium abscessus colony-biofilms based on bi-dimensional images. Antimicrob. Agents Chemother. 2023, 67, e00402-23. [Google Scholar] [CrossRef]
- Pedro, H.D.S.P.; Coelho, A.G.V.; Nardi, S.M.T.; Vilela, G.; Silva, J.O.; Nascimento, A.C.C.; Galle, L.C.; Aily, D.C.G.; Ferro e Silva, R.R.; Shikama, M.L.M.; et al. Performance of liquid culture MGIT after implementation in a network of public laboratories of Sao Paulo state. Rev. Inst. Adolfo Lutz 2017, 76, e1727. [Google Scholar]
- Palomino, J.C.; Martin, A.; Von Groll, A.; Portaels, F. Rapid culture-based methods for drug-resistance detection in Mycobacterium tuberculosis. J. Microbiol. Methods 2008, 75, 161–166. [Google Scholar] [CrossRef]
- Ma, Y.; Fan, J.; Li, S.; Dong, L.; Li, Y.; Wang, F.; Huo, F.; Pang, Y.; Qin, S. Comparison of Lowenstein-Jensen medium and MGIT culture system for recovery of Mycobacterium tuberculosis from abscess samples. Diagn. Microbiol. Infect. Dis. 2020, 96, 114969. [Google Scholar] [CrossRef]
- Kumari, P.; Thakur, J.K.; Kumar, P.; Kumar, R.; Parekh, D. Comparison of LJ Medium and BACTEC MGIT 960 Culture System for the Diagnosis of Tuberculosis. J. Clin. Diagn. Res. 2020, 14, DC09–DC13. [Google Scholar] [CrossRef]
- Kumar, H.; Singh, V.A.; Mehta, S.; Nagpal, S.; Bala, R.; Biswas, D. Comparative Evaluation of Conventional Media with Bactec MGIT 960 for Detection of Mycobacterium tuberculosis in Clinically Suspected Cases of Pulmonary and Extra-Pulmonary Tuberculosis. Indian J. Public Health 2020, 11, 818–822. [Google Scholar] [CrossRef]
- Salam, A.A.; Rehman, S.; Munir, M.K.; Iqbal, R.; Saeed, S.S.; Khan, S.U. Importance of Ziehl-Neelsen smear and culture on Lowenstein Jensen medium in diagnosis of pulmonary tuberculosis. Pak. J. Chest Med. 2014, 20, 1–5. [Google Scholar]
- Gopalaswamy, R.; Shanmugam, S.; Mondal, R.; Subbian, S. Of tuberculosis and non-tuberculous mycobacterial infections—A comparative analysis of epidemiology, diagnosis and treatment. J. Biomed. Sci. 2020, 27, 74. [Google Scholar] [CrossRef]
- MacLean, E.; Kohli, M.; Weber, S.F.; Suresh, A.; Schumacher, S.G.; Denkinger, C.M.; Pai, M. Advances in Molecular Diagnosis of Tuberculosis. J. Clin. Microbiol. 2020, 58, e01582-19. [Google Scholar] [CrossRef]
- Acharya, B.; Acharya, A.; Gautam, S.; Ghimire, S.P.; Mishra, G.; Parajuli, N.; Sapkota, B. Advances in diagnosis of Tuberculosis: An update into molecular diagnosis of Mycobacterium tuberculosis. Mol. Biol. Rep. 2020, 47, 4065–4075. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Manual for Selection of Molecular WHO-Recommended Rapid Diagnostic Tests for Detection of Tuberculosis and Drug-Resistant Tuberculosis. 2022. Available online: https://www.who.int/publications/i/item/9789240042575 (accessed on 23 June 2024).
- Nagai, K.; Horita, N.; Yamamoto, M.; Tsukahara, T.; Nagakura, H.; Tashiro, K.; Shibata, Y.; Watanabe, H.; Nakashima, K.; Ushio, R.; et al. Diagnostic test accuracy of loop-mediated isothermal amplification assay for Mycobacterium tuberculosis: Systematic review and meta-analysis. Sci. Rep. 2016, 6, 39090. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. The Use of Loop-Mediated Isothermal Amplification (TB-LAMP) for the Diagnosis of Pulmonary Tuberculosis: Policy Guidance. 2016. Available online: https://iris.who.int/bitstream/handle/10665/249154/9789241511186-eng.pdf?sequence=1 (accessed on 23 June 2024).
- Hain Lifescience. FluoroType® MTB. Available online: https://www.hain-lifescience.de/en/products/microbiology/mycobacteria/tuberculosis/fluorotype-mtb.html (accessed on 2 December 2023).
- World Health Organization. A2.4 Information Sheet: Practical Considerations for Implementation of the Bruker-Hain Lifesciences FluoroType MTB and FluoroType MTBDR. Available online: https://tbksp.org/en/node/1709 (accessed on 13 December 2023).
- Hofmann-Thiel, S.; Hoffmann, H. Evaluation of Fluorotype MTB for detection of Mycobacterium tuberculosis complex DNA in clinical specimens from a low-incidence country. BMC Infect. Dis. 2014, 14, 59. [Google Scholar] [CrossRef]
- Dippenaar, A.; Derendinger, B.; Dolby, T.; Beylis, N.; van Helden, P.D.; Theron, G.; Warren, R.M.; de Vos, M. Diagnostic accuracy of the FluoroType MTB and MTBDR VER 2.0 assays for the centralized high-throughput detection of Mycobacterium tuberculosis complex DNA and isoniazid and rifampicin resistance. Clin. Microbiol. Infect. 2021, 27, 1351.e1–1351.e4. [Google Scholar] [CrossRef]
- Cepheid. Xpert MTB/RIF Assay: Package Insert (Rev. G). 2020. Available online: https://www.cepheid.com/content/dam/www-cepheid-com/documents/package-insert-files/Xpert-MTB-RIF-PORTUGUESE-Package-Insert-301-1404-PT-Rev-G.pdf (accessed on 9 December 2023).
- Li, S.; Liu, B.; Peng, M.; Chen, M.; Yin, W.; Tang, H.; Luo, Y.; Hu, P.; Ren, H. Diagnostic accuracy of Xpert MTB/RIF for tuberculosis detection in different regions with different endemic burden: A systematic review and meta-analysis. PLoS ONE 2017, 12, e0180725. [Google Scholar] [CrossRef]
- Kaswala, C.; Schmiedel, Y.; Kundu, D.; George, M.M.; Dayanand, D.; Devasagayam, E.; Manesh, S.A.M.; Kumar, S.S.; Michael, J.S.; Ninan, M.M.; et al. Accuracy of Xpert MTB/RIF Ultra for the diagnosis of tuberculosis in adult patients: A retrospective cohort study. Int. J. Infect. Dis. 2022, 122, 566–568. [Google Scholar] [CrossRef] [PubMed]
- Molbio Diagnostics. Truenat™ MTB Package Insert [PDF Document]. Available online: https://www.molbiodiagnostics.com/uploads/product_download/20231107.150156~MTB-pack-insert-V-08.pdf (accessed on 23 June 2024).
- Molbio Diagnostic. Truenat™ MTB Plus Package Insert [PDF Document]. Available online: https://www.molbiodiagnostics.com/uploads/product_download/20190927.152146~Truenat-MTB-Plus-packinsert.pdf (accessed on 23 June 2024).
- Abbott. RealTime MTB Assay. Molecular Diagnostics. Available online: https://www.molecular.abbott/int/en/products/infectious-disease/realtime-mtb (accessed on 14 December 2023).
- Chen, J.H.; She, K.K.; Kwong, T.C.; Wong, O.Y.; Siu, G.K.; Leung, C.C.; Chang, K.C.; Tam, C.M.; Ho, P.L.; Cheng, V.C.; et al. Performance of the new automated Abbott RealTime MTB assay for rapid detection of Mycobacterium tuberculosis complex in respiratory specimens. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 1827–1832. [Google Scholar] [CrossRef]
- Abbott. RealTime MTB RIF/INH. Molecular Diagnostics. Available online: https://www.molecular.abbott/int/en/products/infectious-disease/realtime-mtb-rif-inh-resistance (accessed on 23 December 2023).
- BD MAX™ MDR-TB Assay Package Insert; BD Life Sciences: Sparks, MD, USA, 2019; Available online: https://static.bd.com/documents/eifu/P0228_ZMG_H_SD_P0228.pdf (accessed on 14 December 2023).
- Hain Lifescience. FluoroType® MTBDR VER 2.0—Your Test System for True MDR-TB Testing. Available online: https://www.hain-lifescience.de/en/products/microbiology/mycobacteria/tuberculosis/fluorotype-mtbdr.html (accessed on 14 December 2023).
- Cobas® MTB. Roche Diagnostics. Available online: https://diagnostics.roche.com/global/en/products/params/cobas-mtb.html (accessed on 14 December 2023).
- World Health Organization. Information Sheet: Practical Consideration for Implementation of the Roche Cobas MTB and Cobas MTB-RIF/INH Assays. 2021. Available online: https://www.stoptb.org/file/10477/download (accessed on 23 December 2023).
- Cobas® MTB-RIF/INH. Roche Diagnostics. Available online: https://diagnostics.roche.com/global/en/products/params/cobas-mtb-rif-inh.html (accessed on 23 December 2023).
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef]
- Tomita, N.; Mori, Y.; Kanda, H.; Notomi, T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 2008, 3, 877–882. [Google Scholar] [CrossRef]
- Mori, Y.; Nagamine, K.; Tomita, N.; Notomi, T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 2001, 289, 150–154. [Google Scholar] [CrossRef]
- World Health Organization. WHO Operational Handbook on Tuberculosis: Module 3: Diagnosis: Rapid Diagnostics for Tuberculosis Detection, 3rd ed.; WHO: Geneva, Switzerland, 2024; Available online: https://iris.who.int/bitstream/handle/10665/376155/9789240089501-eng.pdf?sequence=1 (accessed on 23 June 2024).
- Hain Lifescience. FluoroType® Mycobacteria VER 1.0—Differentiate Nontuberculous Mycobacteria. Available online: https://www.hain-lifescience.de/en/products/microbiology/mycobacteria/ntm/fluorotype-mycobacteria.html (accessed on 20 August 2024).
- Khairullah, A.R.; Moses, I.B.; Kusala, M.K.J.; Tyasningsih, W.; Ayuti, S.R.; Rantam, F.A.; Fauziah, I.; Silaen, O.S.M.; Puspitasari, Y.; Aryaloka, S.; et al. Unveiling insights into bovine tuberculosis: A comprehensive review. Open. Vet. J. 2024, 14, 1330–1344. [Google Scholar] [CrossRef]
- Kasir, D.; Osman, N.; Awik, A.; El Ratel, I.; Rafei, R.; Al Kassaa, I.; El Safadi, D.; Salma, R.; El Omari, K.; Cummings, K.J.; et al. Zoonotic Tuberculosis: A Neglected Disease in the Middle East and North Africa (MENA) Region. Diseases 2023, 11, 39. [Google Scholar] [CrossRef]
- de Macedo Couto, R.; Santana, G.O.; Ranzani, O.T.; Waldman, E.A. One Health and surveillance of zoonotic tuberculosis in selected low-income, middle-income and high-income countries: A systematic review. PLoS Negl. Trop. Dis. 2022, 16, e0010428. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Line Probe Assays for Detection of Drug-Resistant Tuberculosis: Interpretation and Reporting Manual for Laboratory Staff and Clinicians. 2022. Available online: https://www.who.int/publications/i/item/9789240046665 (accessed on 23 June 2024).
- Hain Lifescience. GenoType MTBDRplus. Available online: https://www.hain-lifescience.de/en/products/microbiology/mycobacteria/tuberculosis/genotype-mtbdrplus.html (accessed on 3 January 2024).
- Moga, S.; Bobosha, K.; Fikadu, D.; Zerihun, B.; Diriba, G.; Amare, M.; Kempker, R.R.; Blumberg, H.M.; Abebe, T. Diagnostic performance of the GenoType MTBDRplus VER 2.0 line probe assay for the detection of isoniazid resistant Mycobacterium tuberculosis in Ethiopia. PLoS ONE 2023, 18, e0284737. [Google Scholar] [CrossRef] [PubMed]
- Stephen, S.; Muzhizhizhi, D.; Dhibi, N.; Chidemo, T.; Samaneka, W.; Matubu, T.A.; Hakim, J.G.; Chirenje, Z.M. Validation of the GenoType® MTBDRplus Ver 2.0 assay for detection of rifampicin and isoniazid resistance in Mycobacterium tuberculosis complex isolates at UZCHS-CTRC TB research laboratory. Int. J. Mycobacteriol. 2019, 8, 83–88. [Google Scholar] [CrossRef]
- Tan, Y.; Li, Q.; Wang, Q.; Sun, H.; Chen, J.; Cai, X.; Yao, Y.; Bao, X.; Wang, C.; Liu, Y.; et al. Evaluation of the MTBDRplus 2.0 assay for the detection of multidrug resistance among persons with presumptive pulmonary TB in China. Sci. Rep. 2017, 7, 3364. [Google Scholar] [CrossRef] [PubMed]
- Meaza, A.; Kebede, A.; Yaregal, Z.; Dagne, Z.; Moga, S.; Yenew, B.; Diriba, G.; Molalign, H.; Tadesse, M.; Adisse, D.; et al. Evaluation of genotype MTBDRplus VER 2.0 line probe assay for the detection of MDR-TB in smear positive and negative sputum samples. BMC Infect. Dis. 2017, 17, 280. [Google Scholar] [CrossRef] [PubMed]
- Hain Lifescience. GenoType MTBDRsl. Available online: https://www.hain-lifescience.de/en/products/microbiology/mycobacteria/tuberculosis/genotype-mtbdrsl.html (accessed on 3 January 2024).
- Bouzouita, I.; Draoui, H.; Cabibbe, A.M.; Essalah, L.; Bejaoui, S.; Trovato, A.; Messadi, F.; Cirillo, D.M.; Slim-Saidi, L. Performance of the GenoType MTBDRsl V 2.0 for detecting second-line drugs resistance of Mycobacterium tuberculosis isolates in Tunisia. Res. Microbiol. 2021, 172, 103816. [Google Scholar] [CrossRef]
- Nathavitharana, R.R.; Cudahy, P.G.; Schumacher, S.G.; Steingart, K.R.; Pai, M.; Denkinger, C.M. Accuracy of line probe assays for the diagnosis of pulmonary and multidrug-resistant tuberculosis: A systematic review and meta-analysis. Eur. Respir. J. 2017, 49, 1601075. [Google Scholar] [CrossRef]
- Miller, S.; Chiu, C. The Role of Metagenomics and Next-Generation Sequencing in Infectious Disease Diagnosis. Clin. Chem. 2021, 68, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Di Resta, C.; Galbiati, S.; Carrera, P.; Ferrari, M. Next-generation sequencing approach for the diagnosis of human diseases: Open challenges and new opportunities. EJIFCC 2018, 29, 4–14. [Google Scholar]
- Lecuit, M.; Eloit, M. The diagnosis of infectious diseases by whole genome next generation sequencing: A new era is opening. Front. Cell. Infect. Microbiol. 2014, 4, 25. [Google Scholar] [CrossRef]
- World Health Organization. The Use of Next-Generation Sequencing Technologies for the Detection of Mutations Associated with Drug Resistance in Mycobacterium tuberculosis Complex: Technical Guide. 2018. Available online: https://apps.who.int/iris/handle/10665/274443 (accessed on 23 December 2023).
- World Health Organization. The Use of Next-Generation Sequencing for the Surveillance of Drug-Resistant Tuberculosis: An Implementation Manual. 2023. Available online: https://www.who.int/publications/i/item/9789240078079 (accessed on 23 December 2023).
- Vogel, M.; Utpatel, C.; Corbett, C.; Kohl, T.A.; Iskakova, A.; Ahmedov, S.; Antonenka, U.; Dreyer, V.; Ibrahimova, A.; Kamarli, C.; et al. Implementation of whole genome sequencing for tuberculosis diagnostics in a low-middle income, high MDR-TB burden country. Sci. Rep. 2021, 11, 15333. [Google Scholar] [CrossRef]
- Ness, T.E.; DiNardo, A.; Farhat, M.R. High Throughput Sequencing for Clinical Tuberculosis: An Overview. Pathogens 2022, 11, 1343. [Google Scholar] [CrossRef]
- Tsuchida, S.; Umemura, H.; Nakayama, T. Current Status of Matrix-Assisted Laser Desorption/Ionization-Time-of-Flight Mass Spectrometry (MALDI-TOF MS) in Clinical Diagnostic Microbiology. Molecules 2020, 25, 4775. [Google Scholar] [CrossRef]
- Alcolea-Medina, A.; Fernandez, M.T.C.; Montiel, N.; García, M.P.L.; Sevilla, C.D.; North, N.; Lirola, M.J.M.; Wilks, M. An improved simple method for the identification of Mycobacteria by MALDI-TOF MS (Matrix-Assisted Laser Desorption- Ionization mass spectrometry). Sci. Rep. 2019, 9, 20216. [Google Scholar] [CrossRef] [PubMed]
- Ou, X.; Song, Z.; Zhao, B.; Pei, S.; Teng, C.; Zheng, H.; He, W.; Xing, R.; Wang, Y.; Wang, S.; et al. Diagnostic efficacy of an optimized nucleotide MALDI-TOF-MS assay for anti-tuberculosis drug resistance detection. Eur. J. Clin. Microbiol. Infect. Dis. 2024, 43, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Tan, G.; Yang, J.; Guo, Y.; Huang, C.; Sha, W.; Yu, F. Prediction of Mycobacterium tuberculosis drug resistance by nucleotide MALDI-TOF-MS. Int. J. Infect. Dis. 2022, 121, 47–54. [Google Scholar] [CrossRef]
- Shi, J.; He, G.; Ning, H.; Wu, L.; Wu, Z.; Ye, X.; Qiu, C.; Jiang, X. Application of matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) in the detection of drug resistance of Mycobacterium tuberculosis in re-treated patients. Tuberculosis 2022, 135, 102209. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, Q.; Xu, B.; Lin, Y.; Yang, X.; Tong, J.; Huang, C. Clinical performance of nucleotide MALDI-TOF-MS in the rapid diagnosis of pulmonary tuberculosis and drug resistance. Tuberculosis 2023, 143, 102411. [Google Scholar] [CrossRef]
- Su, K.Y.; Yan, B.S.; Chiu, H.C.; Yu, C.J.; Chang, S.Y.; Jou, R.; Liu, J.L.; Hsueh, P.R.; Yu, S.L. Rapid Sputum Multiplex Detection of the M. tuberculosis Complex (MTBC) and Resistance Mutations for Eight Antibiotics by Nucleotide MALDI-TOF MS. Sci. Rep. 2017, 7, 41486. [Google Scholar] [CrossRef]
- Ceyssens, P.J.; Soetaert, K.; Timke, M.; Van den Bossche, A.; Sparbier, K.; De Cremer, K.; Kostrzewa, M.; Hendrickx, M.; Mathys, V. Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry for Combined Species Identification and Drug Sensitivity Testing in Mycobacteria. J. Clin. Microbiol. 2017, 55, 624–634. [Google Scholar] [CrossRef]
- Jiang, L.; Xin, J.; Liang, L.; Xia, M.; Li, J.; Tong, J.; Huang, C.; Li, T. Enhanced diagnosis of pulmonary tuberculosis through nucleotide MALDI-TOF MS analysis of BALF: A retrospective clinical study. Sci. Rep. 2024, 14, 18416. [Google Scholar] [CrossRef] [PubMed]
- Bhalla, N.; Jolly, P.; Formisano, N.; Estrela, P. Introduction to biosensors. Essays Biochem. 2016, 60, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Fan, S.; Ma, Y.; Chen, H.; Xu, J.F.; Pi, J.; Wang, W.; Chen, G. Current progress of functional nanobiosensors for potential tuberculosis diagnosis: The novel way for TB control? Front. Bioeng. Biotechnol. 2022, 10, 1036678. [Google Scholar] [CrossRef]
- Rizi, K.S.; Hatamluyi, B.; Rezayi, M.; Meshkat, Z.; Sankian, M.; Ghazvini, K.; Farsiani, H.; Aryan, E. Response surface methodology optimized electrochemical DNA biosensor based on HAPNPTs/PPY/MWCNTs nanocomposite for detecting Mycobacterium tuberculosis. Talanta 2021, 226, 122099. [Google Scholar] [CrossRef] [PubMed]
- Golichenari, B.; Nosrati, R.; Farokhi-Fard, A.; Faal Maleki, M.; Gheibi Hayat, S.M.; Ghazvini, K.; Vaziri, F.; Behravan, J. Electrochemical-based biosensors for detection of Mycobacterium tuberculosis and tuberculosis biomarkers. Crit. Rev. Biotechnol. 2019, 39, 1056–1077. [Google Scholar] [CrossRef]
- Mobed, A.; Darvishi, M.; Kohansal, F.; Dehfooli, F.M.; Alipourfard, I.; Tahavvori, A.; Ghazi, F. Biosensors; nanomaterial-based methods in diagnosing of Mycobacterium tuberculosis. J. Clin. Tuberc. Other Mycobact. Dis. 2023, 34, 100412. [Google Scholar] [CrossRef]
- de Martino, M.; Lodi, L.; Galli, L.; Chiappini, E. Immune Response to Mycobacterium tuberculosis: A Narrative Review. Front. Pediatr. 2019, 7, 350. [Google Scholar] [CrossRef] [PubMed]
- Scriba, T.J.; Coussens, A.K.; Fletcher, H.A. Human Immunology of Tuberculosis. Microbiol. Spectr. 2017, 5, TBTB2-0016. [Google Scholar] [CrossRef] [PubMed]
- Trajman, A.; Steffen, R.E.; Menzies, D. Interferon-Gamma Release Assays versus Tuberculin Skin Testing for the Diagnosis of Latent Tuberculosis Infection: An Overview of the Evidence. Pulm. Med. 2013, 2013, 601737. [Google Scholar] [CrossRef]
- McIntyre, S.; Warner, J.; Rush, C.; Vanderven, H.A. Antibodies as clinical tools for tuberculosis. Front. Immunol. 2023, 14, 1278947. [Google Scholar] [CrossRef]
- World Health Organization. WHO Operational Handbook on Tuberculosis. Module 3: Diagnosis. Tests for Tuberculosis Infection. 2022. Available online: https://www.who.int/publications/i/item/9789240058347 (accessed on 23 January 2024).
- Melkie, S.T.; Arias, L.; Farroni, C.; Jankovic Makek, M.; Goletti, D.; Vilaplana, C. The role of antibodies in tuberculosis diagnosis, prophylaxis and therapy: A review from the ESGMYC study group. Eur. Respir. Rev. 2022, 31, 210218. [Google Scholar] [CrossRef]
- World Health Organization. Use of Alternative Interferon-Gamma Release Assays for the Diagnosis of TB Infection: WHO Policy Statement. 2022. Available online: https://www.who.int/publications/i/item/9789240042346 (accessed on 25 January 2024).
- World Health Organization. WHO Consolidated Guidelines on Tuberculosis. Module 1: Prevention—Tuberculosis Preventive Treatment. 2020. Available online: https://www.who.int/publications/i/item/9789240001503 (accessed on 26 January 2024).
- Zhou, G.; Luo, Q.; Luo, S.; Chen, H.; Cai, S.; Guo, X.; He, J.; Xia, Y.; Li, H.; Zhou, Y.; et al. Indeterminate results of interferon gamma release assays in the screening of latent tuberculosis infection: A systematic review and meta-analysis. Front. Immunol. 2023, 14, 1170579. [Google Scholar] [CrossRef]
- Oxford Immunotec. T-SPOT.TB [Package Insert]. Available online: https://www.oxfordimmunotec.com/international/wp-content/uploads/sites/3/Final-File-PI-TB-US-V6.pdf (accessed on 16 November 2023).
- Qiagen. QuantiFERON-TB Gold Plus (QFT-Plus) Package Insert 01/2023. 2023. Available online: https://www.qiagen.com/us/resources/download.aspx?id=ac068fc7-a994-4443-ac7c-dda43ce2bc5e&lang=en (accessed on 16 November 2023).
- WANTAI TB-IGRA Package Insert; Beijing Wantai Biological Pharmacy Enterprise Co., Ltd.: Beijing, China, 2022; Available online: https://www.ystwt.cn/wp-content/uploads/2018/04/Wantai-TB-IGRA.pdf (accessed on 16 November 2023).
- Sun, Y.; Yao, X.; Ni, Y.; Peng, Y.; Shi, G. Diagnostic Efficacy of T-SPOT.TB for Active Tuberculosis in Adult: A Retrospective Study. Infect. Drug Resist. 2022, 15, 7077–7093. [Google Scholar] [CrossRef]
- Luo, Y.; Tang, G.; Yuan, X.; Lin, Q.; Mao, L.; Song, H.; Xue, Y.; Wu, S.; Ouyang, R.; Hou, H.; et al. Combination of Blood Routine Examination and T-SPOT.TB Assay for Distinguishing Between Active Tuberculosis and Latent Tuberculosis Infection. Front. Cell Infect. Microbiol. 2021, 1, 575650. [Google Scholar] [CrossRef]
- Nguyen, D.T.; Phan, H.; Trinh, T.; Nguyen, H.; Doan, H.; Pham, N.; Nguyen, H.; Nguyen, H.; Nguyen, H.V.; Le, H.V.; et al. Sensitivity and characteristics associated with positive QuantiFERON-TB Gold-Plus assay in children with confirmed tuberculosis. PLoS ONE 2019, 14, e0213304. [Google Scholar] [CrossRef] [PubMed]
- Takasaki, J.; Manabe, T.; Morino, E.; Muto, Y.; Hashimoto, M.; Iikura, M.; Izumi, S.; Sugiyama, H.; Kudo, K. Sensitivity and specificity of QuantiFERON-TB Gold Plus compared with QuantiFERON-TB Gold In-Tube and T-SPOT.TB on active tuberculosis in Japan. J. Infect. Chemother. 2018, 24, 188–192. [Google Scholar] [CrossRef]
- Zhang, L.; Shi, X.; Zhang, Y.; Zhang, Y.; Huo, F.; Zhou, B.; Deng, G.; Liu, X. Analysis of Factors Influencing Diagnostic Accuracy of T-SPOT.TB for Active Tuberculosis in Clinical Practice. Sci. Rep. 2017, 7, 7764. [Google Scholar] [CrossRef] [PubMed]
- Hamada, Y.; Cirillo, D.M.; Matteelli, A.; Penn-Nicholson, A.; Rangaka, M.X.; Ruhwald, M. Tests for tuberculosis infection: Landscape analysis. Eur. Respir. J. 2021, 58, 2100167. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Tuberculin Skin Testing. CDC. 2020. Available online: https://www.cdc.gov/tb/publications/factsheets/testing/Tuberculin_Skin_Testing_Information_for_Health_Care_Providers.pdf (accessed on 16 November 2023).
- Piñeiro, R.; Mellado, M.J.; Cilleruelo, M.J.; García-Ascaso, M.; Medina-Claros, A.; García-Hortelano, M. Tuberculin skin test in bacille Calmette-Guérin-vaccinated children: How should we interpret the results? Eur. J. Pediatr. 2012, 171, 1625–1632. [Google Scholar] [CrossRef]
- Maes, M.; Giménez, J.F.; D’Alessandro, A.; De Waard, J.H. The stability of human, bovine and avian tuberculin purified protein derivative (PPD). J. Infect. Dev. Ctries. 2011, 5, 781–785. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gualano, G.; Mencarini, P.; Lauria, F.N.; Palmieri, F.; Mfinanga, S.; Mwaba, P.; Chakaya, J.; Zumla, A.; Ippolito, G. Tuberculin skin test—Outdated or still useful for Latent TB infection screening? Int. J. Infect. Dis. 2019, 80, S20–S22. [Google Scholar] [CrossRef]
- Cy-Tb Package Insert; Serum Institute: Pune, India, 2022.
- Goscé, L.; Allel, K.; Hamada, Y.; Korobitsyn, A.; Ismail, N.; Bashir, S.; Denkinger, C.M.; Abubakar, I.; White, P.J.; Rangaka, M.X. Economic evaluation of novel Mycobacterium tuberculosis specific antigen-based skin tests for detection of TB infection: A modelling study. PLoS Glob. Public Health 2023, 3, e0002573. [Google Scholar] [CrossRef]
- Hamada, Y.; Kontsevaya, I.; Surkova, E.; Wang, T.T.; Wan-Hsin, L.; Matveev, A.; Ziganshina, L.E.; Denkinger, C.M.; Korobitsyn, A.; Ismail, N.; et al. A Systematic Review on the Safety of Mycobacterium tuberculosis-Specific Antigen-Based Skin Tests for Tuberculosis Infection Compared with Tuberculin Skin Tests. Open Forum Infect. Dis. 2023, 10, ofad228. [Google Scholar] [CrossRef]
- To, K.W.; Zhang, R.; Lee, S.S. Is the new tuberculous antigen-based skin test ready for use as an alternative to tuberculin skin test/interferon-gamma release assay for tuberculous diagnosis? A narrative review. Int. J. Infect. Dis. 2024, 141, 106992. [Google Scholar] [CrossRef] [PubMed]
- Olbrich, L.; Verghese, V.P.; Franckling-Smith, Z.; Sabi, I.; Ntinginya, N.E.; Mfinanga, A.; Banze, D.; Viegas, S.; Khosa, C.; Semphere, R.; et al. Diagnostic accuracy of a three-gene Mycobacterium tuberculosis host response cartridge using fingerstick blood for childhood tuberculosis: A multicentre prospective study in low-income and middle-income countries. Lancet Infect. Dis. 2024, 24, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Lyon, C.J.; LaCourse, S.M.; Zheng, W.; Stern, J.; Escudero, J.N.; Murithi, W.B.; Njagi, L.; John-Stewart, G.; Hawn, T.R.; et al. Sensitive Blood-Based Detection of HIV-1 and Mycobacterium tuberculosis Peptides for Disease Diagnosis by Immuno-Affinity Liquid Chromatography-Tandem Mass Spectrometry: A Method Development and Proof-of-Concept Study. Clin. Chem. 2023, 69, 1409–1419. [Google Scholar] [CrossRef]
- Moore, C.M.; Dhillon, J.; Flynn, R.; Gizynski, K.; Adams, C.; Morgan, G.; McGurk, D.; Boada, E.; Shabestary, S.; Peat, J.; et al. A Novel Microfluidic Dielectrophoresis Technology to Enable Rapid Diagnosis of Mycobacterium tuberculosis in Clinical Samples. J. Mol. Diagn. 2023, 25, 513–523. [Google Scholar] [CrossRef]
- Ketchanji Mougang, Y.C.; Endale Mangamba, L.M.; Capuano, R.; Ciccacci, F.; Catini, A.; Paolesse, R.; Mbatchou Ngahane, H.B.; Palombi, L.; Di Natale, C. On-Field Test of Tuberculosis Diagnosis through Exhaled Breath Analysis with a Gas Sensor Array. Biosensors 2023, 13, 570. [Google Scholar] [CrossRef]
- Thermo Fisher Scientific. Sensititre™ Mycobacterium tuberculosis MYCOTBI AST Plate. Available online: https://www.thermofisher.com/order/catalog/product/MYCOTBI (accessed on 12 January 2024).
- Olbrich, L.; Khambati, N.; Bijker, E.M.; Ruhwald, M.; Heinrich, N.; Song, R. FujiLAM for the diagnosis of childhood tuberculosis: A systematic review. BMJ Paediatr. Open 2022, 6, e001447. [Google Scholar] [CrossRef]
- Li, Z.; Tong, X.; Liu, S.; Yue, J.; Fan, H. The Value of FujiLAM in the Diagnosis of Tuberculosis: A Systematic Review and Meta-Analysis. Front. Public Health 2021, 9, 757133. [Google Scholar] [CrossRef]
- Kerkhoff, A.D.; Sossen, B.; Schutz, C.; Reipold, E.I.; Trollip, A.; Moreau, E.; Schumacher, S.G.; Burton, R.; Ward, A.; Nicol, M.P.; et al. Diagnostic sensitivity of SILVAMP TB-LAM (FujiLAM) point-of-care urine assay for extra-pulmonary tuberculosis in people living with HIV. Eur. Respir. J. 2020, 55, 1901259. [Google Scholar] [CrossRef] [PubMed]
- Chenai Mathabire Rücker, S.; Lissouba, P.; Akinyi, M.; Vicent Lubega, A.; Stewart, R.; Tamayo Antabak, N.; Taremwa Mugisha, I.; Ohler, L.; Macuácua, H.; Atieno, M.; et al. Feasibility and acceptability of using the novel urine-based FujiLAM test to detect tuberculosis: A multi-country mixed-methods study. J. Clin. Tuberc. Other Mycobact. Dis. 2022, 27, 100316. [Google Scholar] [CrossRef]
- SD Biosensor. STANDARD E TB-Feron ELISA. Available online: https://www.sdbiosensor.com/product/product_view?product_no=113 (accessed on 10 January 2024).
- Yoo, I.Y.; Lee, J.; Choi, A.R.; Jun, Y.H.; Lee, H.Y.; Kang, J.Y.; Park, Y.-J. Comparative Evaluation of Standard E TB-Feron ELISA and QuantiFERON-TB Gold Plus Assays in Patients with Tuberculosis and Healthcare Workers. Diagnostics 2021, 11, 1659. [Google Scholar] [CrossRef]
- Jung, J.; Jhun, B.W.; Jeong, M.; Yoon, S.J.; Huh, H.J.; Jung, C.W.; Kim, K.; Park, J.B.; Kim, D.J.; Huh, W.; et al. Is the New Interferon-Gamma Releasing Assay Beneficial for the Diagnosis of Latent and Active Mycobacterium tuberculosis Infections in Tertiary Care Setting? J. Clin. Med. 2021, 10, 1376. [Google Scholar] [CrossRef]
- SD Biosensor. STANDARD F TB-Feron FIA (IFN-Gamma). Available online: https://www.sdbiosensor.com/product/product_view?product_no=167# (accessed on 10 January 2024).
- Saint-Pierre, G.; Conei, D.; Cantillana, P.; Raijmakers, M.; Vera, A.; Gutiérrez, D.; Kennedy, C.; Peralta, P.; Ramonda, P. Comparison of Two Tuberculosis Infection Tests in a South American Tertiary Hospital: STANDARD F TB-Feron FIA vs. QIAreachTM QuantiFERON-TB. Diagnostics 2023, 13, 1162. [Google Scholar] [CrossRef] [PubMed]
- bioMérieux. VIDAS® TB-IGRA. bioMérieux Diagnostics. Available online: https://www.biomerieux-diagnostics.com/vidasr-tb-igra (accessed on 10 January 2024).
- Petruccioli, E.; Farroni, C.; Cuzzi, G.; Vanini, V.; Palmieri, F.; Vittozzi, P.; Goletti, D. VIDAS® TB-IGRA reagents induce a CD4+ and CD8+ T-cell IFN-γ response for both TB infection and active TB. Int. J. Tuberc. Lung Dis. 2022, 26, 65–68. [Google Scholar] [CrossRef] [PubMed]
- Diagbouga, S.; Djibougou, A.D.; Pease, C.; Alcaide, A.; Berthoux, A.; Bruiners, N.; Cirillo, D.M.; Combary, A.; Falchero, N.; Handler, D.; et al. Preliminary diagnostic performance of the VIDAS® TB-IGRA for the detection of Mycobacterium tuberculosis infection and disease. medRxiv 2022. [Google Scholar] [CrossRef]
- SD Biosensor. STANDARD M M10 MDR-TB. Available online: https://www.sdbiosensor.com/product/product_view?product_no=23007 (accessed on 12 January 2024).
- ClinicalTrials.gov. Phase II: Multicentre Clinical Study to Assess the Performance of the Bioneer IRONqPCR ™ RFIA Kit for INH-, RIF-, FQ- and AG-Resistance Detection (NCT05117788). 2022. Available online: https://clinicaltrials.gov/study/NCT05117788#study-overview (accessed on 12 January 2024).
- ABL SA. DEEPCHEK® ASSAY 13-PLEX KB DRUG SUSCEPTIBILITY TESTING (CE-IVD). Available online: https://www.ablsa.com/laboratory-applications/bacteriochek-tb-software/ (accessed on 17 January 2024).
- Zhou, R.; Qiu, X.; Ying, J.; Yue, Y.; Ruan, T.; Yu, L.; Liu, Q.; Sun, X.; Wang, S.; Qu, Y.; et al. Diagnostic performance of adenosine deaminase for abdominal tuberculosis: A systematic review and meta-analysis. Front. Public Health. 2022, 10, 938544. [Google Scholar] [CrossRef]
- Gopalaswamy, R.; Dusthackeer, V.A.; Kannayan, S.; Subbian, S. Extrapulmonary tuberculosis—An update on the diagnosis, treatment and drug resistance. J. Respir. 2021, 1, 141–164. [Google Scholar] [CrossRef]
- Aggarwal, A.N.; Agarwal, R.; Sehgal, I.S.; Dhooria, S. Adenosine deaminase for diagnosis of tuberculous pleural effusion: A systematic review and meta-analysis. PLoS ONE 2019, 14, e0213728. [Google Scholar] [CrossRef]
- Gao, L.; Wang, W.; Zhang, Y.; Hu, X.; An, J.; Li, Y.; Chen, M.; Shen, Y. Adenosine deaminase-based measurement in the differential diagnosis of pleural effusion: A multicenter retrospective study. Ther. Adv. Respir. Dis. 2023, 17, 17534666231155747. [Google Scholar] [CrossRef]
- Salmanzadeh, S.; Tavakkol, H.; Bavieh, K.; Alavi, S.M. Diagnostic Value of Serum Adenosine Deaminase (ADA) Level for Pulmonary Tuberculosis. Jundishapur J. Microbiol. 2015, 8, e21760. [Google Scholar] [CrossRef]
- Pai, M.; Dewan, P.K.; Swaminathan, S. Transforming tuberculosis diagnosis. Nat. Microbiol. 2023, 8, 756–759. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).