Integrating Machine Learning in Clinical Practice for Characterizing the Malignancy of Solitary Pulmonary Nodules in PET/CT Screening
Abstract
1. Introduction
2. Materials and Methods
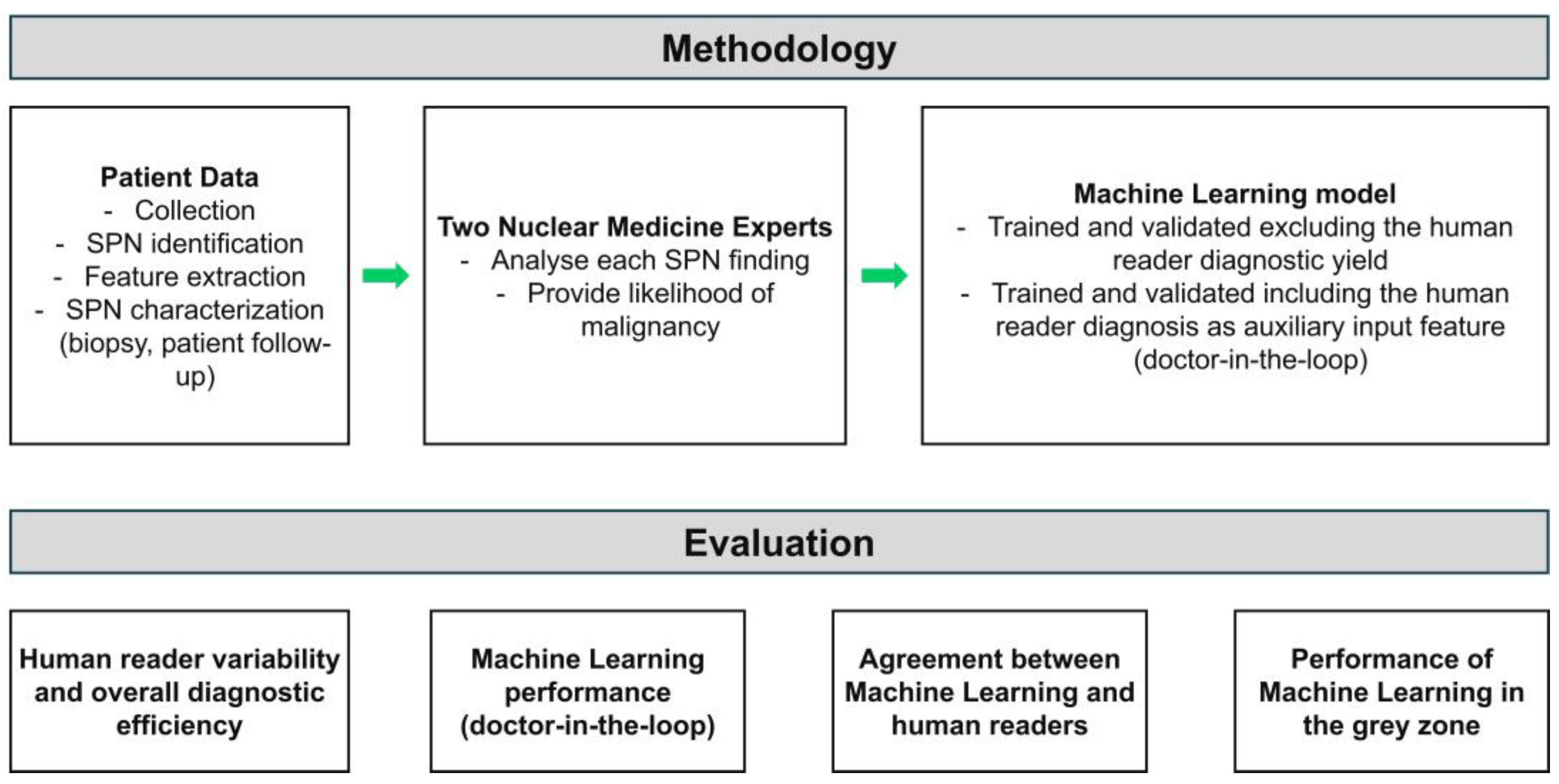
2.1. Research Methodology
2.2. Patient Data
2.3. Data Preprocessing
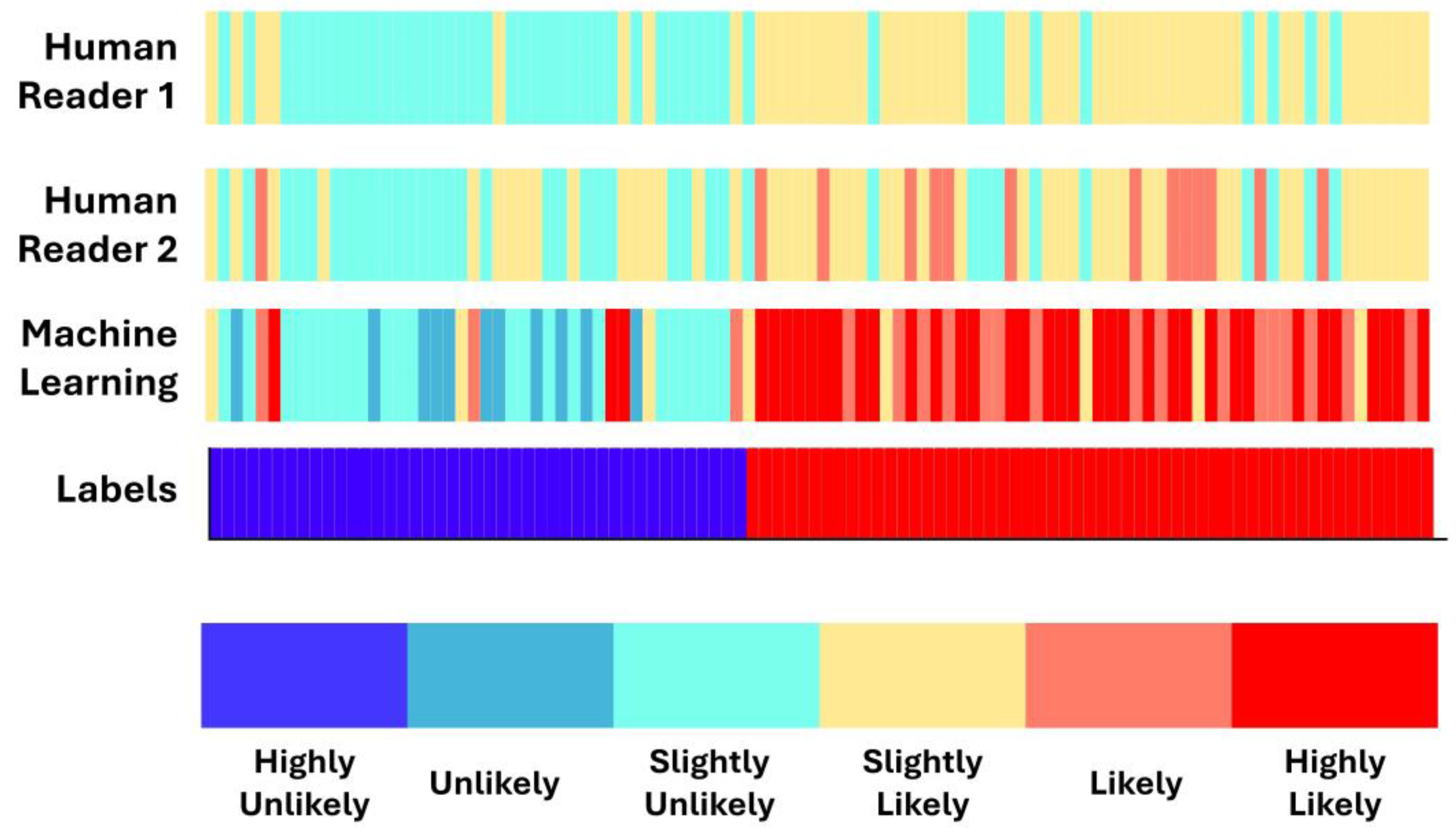
2.4. Expert Probability Scores
- Highly unlikely to be malignant;
- Unlikely to be malignant;
- Possibly not malignant;
- Possibly malignant;
- Likely to be malignant;
- Highly likely to be malignant.
2.5. Machine Learning
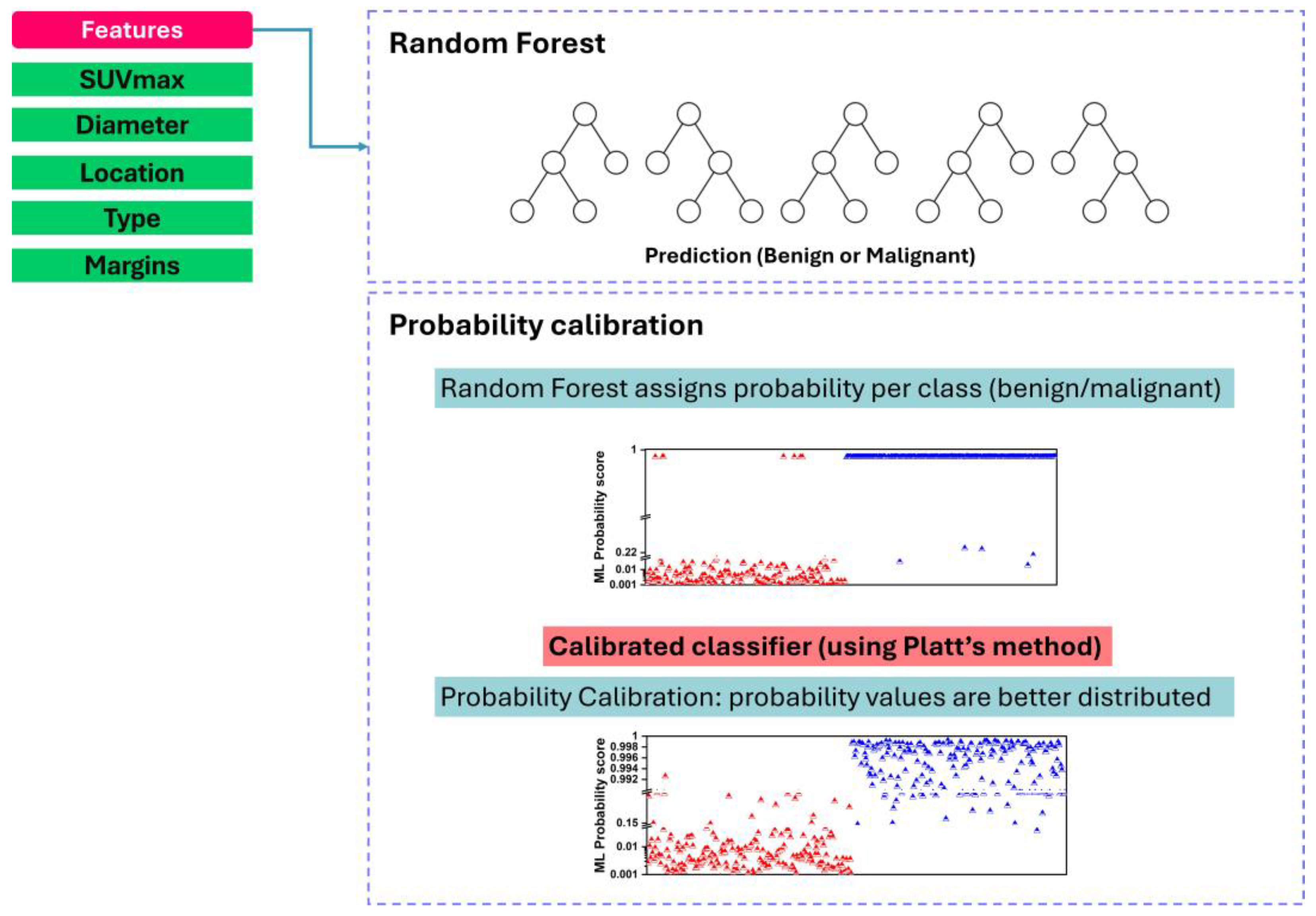
2.5.1. Random Forest Algorithm
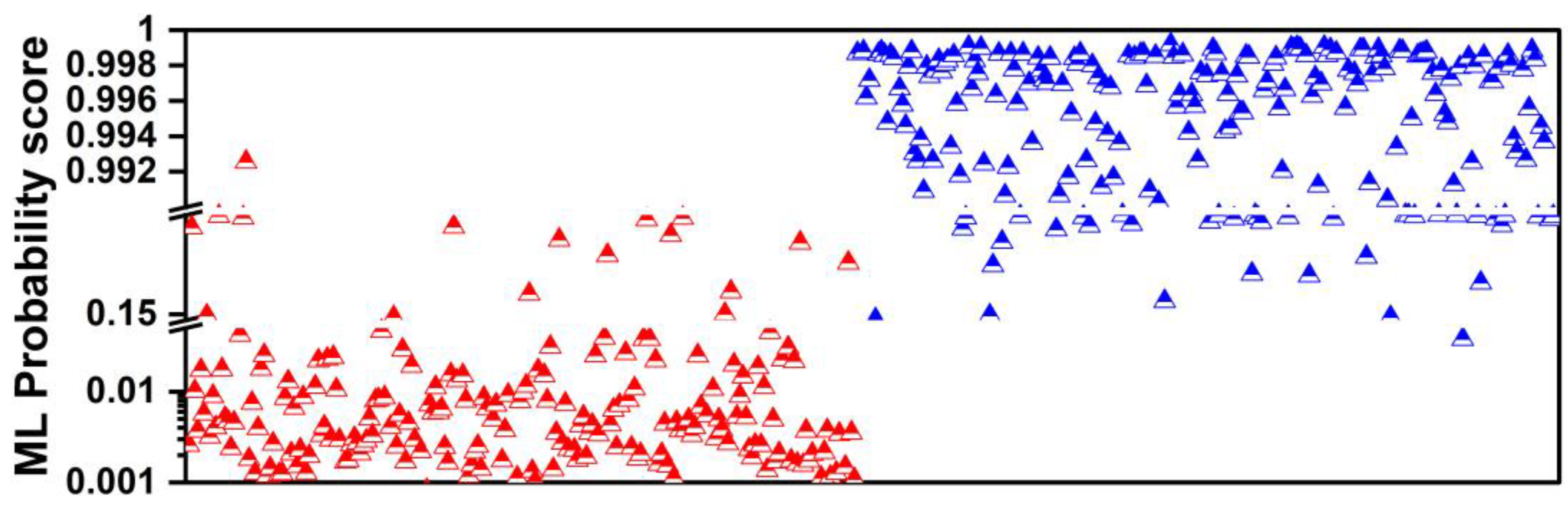
2.5.2. Probability Calibration
- Splitting the dataset into ten roughly equal-sized folds;
- Iterating through each fold as the validation set while training the Random Forest classifier on the remaining nine folds;
- Generating predicted probabilities for the validation set using the trained RF classifier;
- Applying probability calibration (e.g., sigmoid calibration) to the predicted probabilities of the validation set, ensuring calibration within each fold;
- Repeating steps 2–4 for each fold to obtain calibrated probabilities for the entire dataset.
3. Results
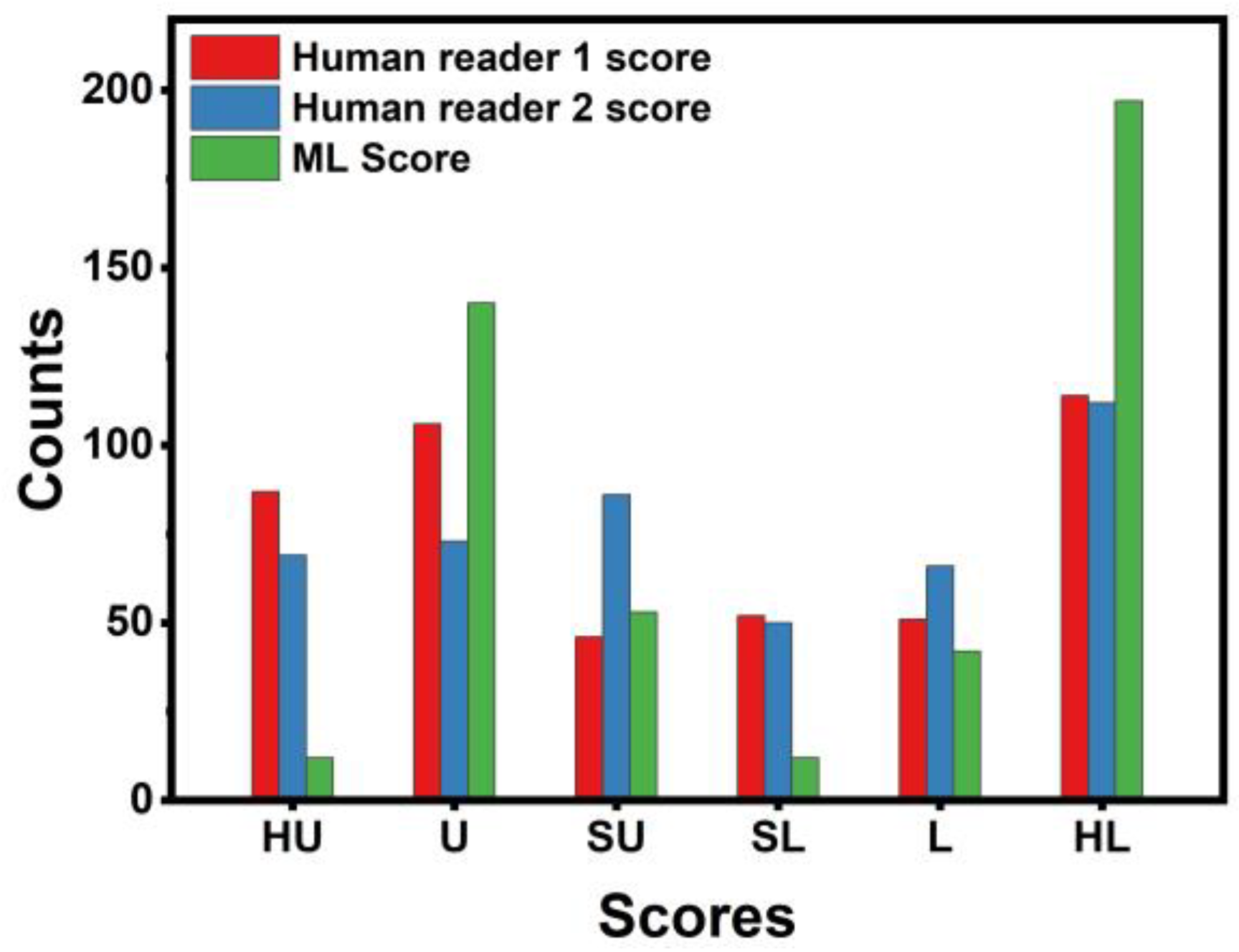
3.1. Human Experts’ Diagnostic Yield
3.2. Performance of Machine Learning without Expert’s Verdict
3.3. Performance of Machine Learning When Using Human Readers’ Diagnostic Yield as Additional Input
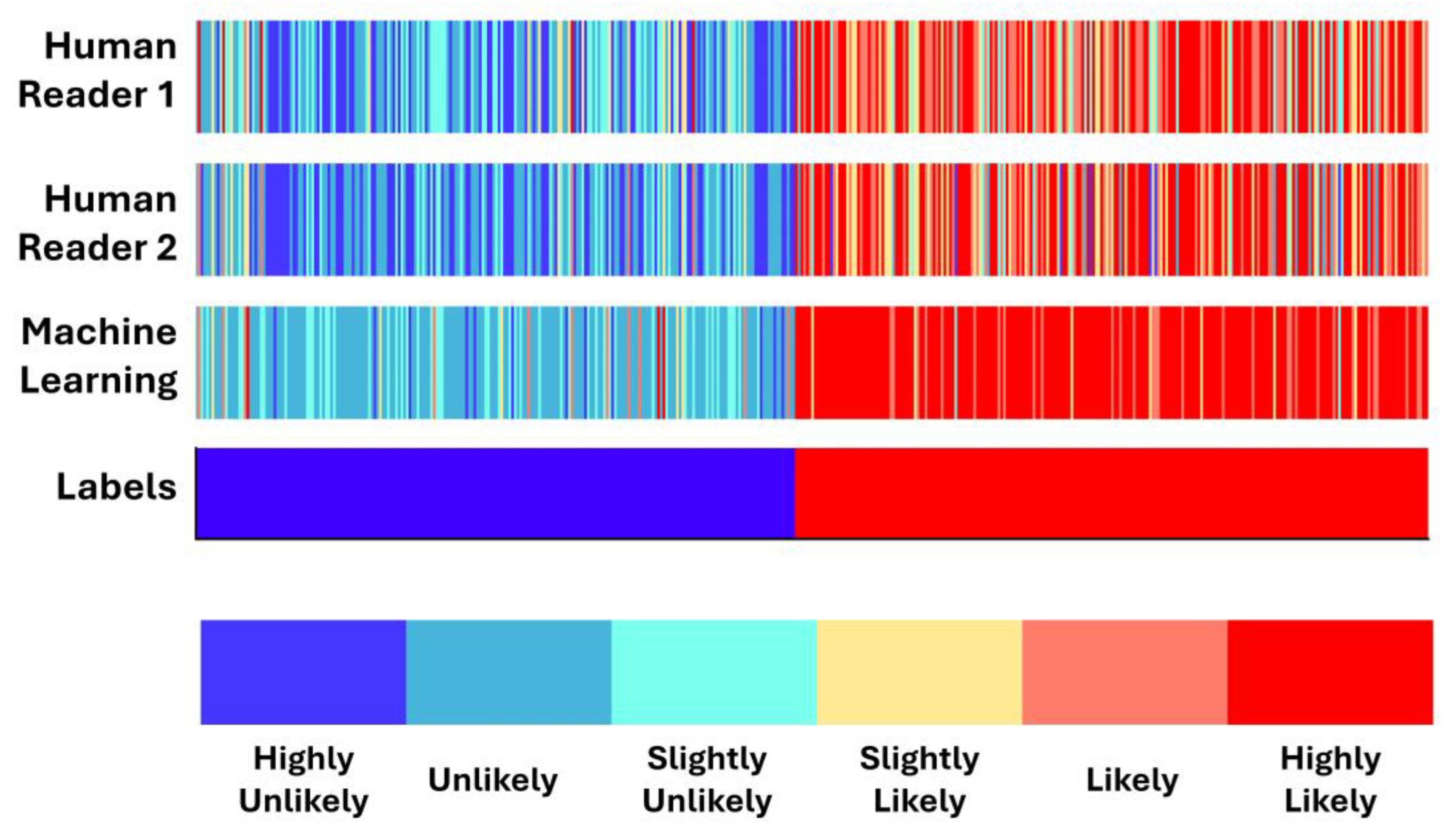
3.4. Concordance between Machine Learning and Human Readers
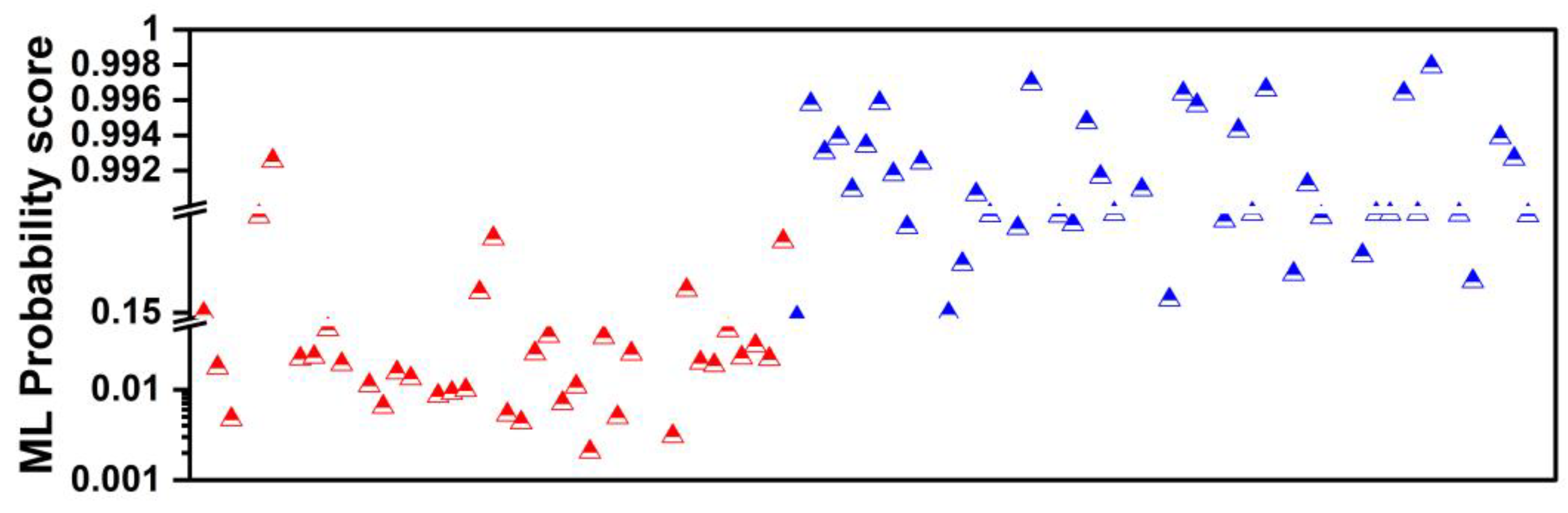
3.5. Performance of Machine Learning in Ambiguous Cases
3.6. Comparison with the Literature
4. Discussion
- (a)
- Human readers exhibited variability when providing their assessments. The latter was reflected in the different performance scores (first reader: ~90% accuracy; second reader: ~88% accuracy).
- (b)
- Integrating both readers’ diagnostic scores in the training features of the ML model resulted in improving the diagnostic efficiency of the ML model by ~3%. The latter underlines the importance of the synergistic contribution of ML and human judgement in discriminating between benign and malignant SPNs.
- (c)
- (d)
- ML performed better than the best human reader in challenging instances (i.e., SPNs with probability grades of Slightly Unlikely and Slightly Likely). ML produced 2 FNs, whilst the best human reader had 11 FNs. The latter is reflected in the overall performance of ML in the grey zones (~90% accuracy, 80% sensitivity, and 81.39% specificity). Hence, ML could be particularly useful in such cases where diagnostic yields are challenged.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alexander, M.; Kim, S.Y.; Cheng, H. Update 2020: Management of Non-Small Cell Lung Cancer. Lung 2020, 198, 897–907. [Google Scholar] [CrossRef] [PubMed]
- Cruickshank, A.; Stieler, G.; Ameer, F. Evaluation of the Solitary Pulmonary Nodule. Intern. Med. J. 2019, 49, 306–315. [Google Scholar] [CrossRef] [PubMed]
- Mazzone, P.J.; Lam, L. Evaluating the Patient with a Pulmonary Nodule: A Review. JAMA 2022, 327, 264. [Google Scholar] [CrossRef] [PubMed]
- Senent-Valero, M.; Librero, J.; Pastor-Valero, M. Solitary Pulmonary Nodule Malignancy Predictive Models Applicable to Routine Clinical Practice: A Systematic Review. Syst. Rev. 2021, 10, 308. [Google Scholar] [CrossRef] [PubMed]
- Snoeckx, A.; Reyntiens, P.; Desbuquoit, D.; Spinhoven, M.J.; Van Schil, P.E.; Van Meerbeeck, J.P.; Parizel, P.M. Evaluation of the Solitary Pulmonary Nodule: Size Matters, but Do Not Ignore the Power of Morphology. Insights Imaging 2018, 9, 73–86. [Google Scholar] [CrossRef] [PubMed]
- Groheux, D.; Quere, G.; Blanc, E.; Lemarignier, C.; Vercellino, L.; De Margerie-Mellon, C.; Merlet, P.; Querellou, S. FDG PET-CT for Solitary Pulmonary Nodule and Lung Cancer: Literature Review. Diagn. Interv. Imaging 2016, 97, 1003–1017. [Google Scholar] [CrossRef] [PubMed]
- Basso Dias, A.; Zanon, M.; Altmayer, S.; Sartori Pacini, G.; Henz Concatto, N.; Watte, G.; Garcez, A.; Mohammed, T.-L.; Verma, N.; Medeiros, T.; et al. Fluorine 18–FDG PET/CT and Diffusion-Weighted MRI for Malignant versus Benign Pulmonary Lesions: A Meta-Analysis. Radiology 2019, 290, 525–534. [Google Scholar] [CrossRef]
- Wang, H.; Zhu, H.; Ding, L.; Yang, K. A Diagnostic Classification of Lung Nodules Using Multiple-Scale Residual Network. Sci. Rep. 2023, 13, 11322. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, Y.; Han, J.; Lin, Q.; Zhao, L.; Li, Q.; Zhao, J.; Li, H.; Wang, Y.; Hu, C. A Machine Learning-Based PET/CT Model for Automatic Diagnosis of Early-Stage Lung Cancer. Front. Oncol. 2023, 13, 1192908. [Google Scholar] [CrossRef]
- Astaraki, M.; Zakko, Y.; Toma Dasu, I.; Smedby, Ö.; Wang, C. Benign-Malignant Pulmonary Nodule Classification in Low-Dose CT with Convolutional Features. Phys. Med. 2021, 83, 146–153. [Google Scholar] [CrossRef]
- Apostolopoulos, I.D.; Pintelas, E.G.; Livieris, I.E.; Apostolopoulos, D.J.; Papathanasiou, N.D.; Pintelas, P.E.; Panayiotakis, G.S. Automatic classification of solitary pulmonary nodules in PET/CT imaging employing transfer learning techniques. Med. Biol. Eng. Comput. 2021, 59, 1299–1310. [Google Scholar] [CrossRef] [PubMed]
- Apostolopoulos, I.D.; Apostolopoulos, D.J.; Panayiotakis, G.S. Solitary Pulmonary Nodule Malignancy Classification Utilising 3D Features and Semi-Supervised Deep Learning. In Proceedings of the 2022 13th International Conference on Information, Intelligence, Corfu, Greece, 18–20 July 2022, Systems & Applications (IISA); IEEE: Piscataway, NJ, USA, 2022; pp. 1–6. [Google Scholar]
- Lin, C.-Y.; Guo, S.-M.; Lien, J.-J.J.; Lin, W.-T.; Liu, Y.-S.; Lai, C.-H.; Hsu, I.-L.; Chang, C.-C.; Tseng, Y.-L. Combined Model Integrating Deep Learning, Radiomics, and Clinical Data to Classify Lung Nodules at Chest CT. Radiol. Med. 2023, 129, 56–69. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-J.; Hua, K.-L.; Hsu, C.-H.; Cheng, W.-H.; Hidayati, S.C. Computer-Aided Classification of Lung Nodules on Computed Tomography Images via Deep Learning Technique. OncoTargets Ther. 2015, 2015, 2015–2022. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Cao, H.; Song, E.; Ma, G.; Xu, X.; Jin, R.; Liu, C.; Hung, C.-C. Multi-Model Ensemble Learning Architecture Based on 3D CNN for Lung Nodule Malignancy Suspiciousness Classification. J. Digit. Imaging 2020, 33, 1242–1256. [Google Scholar] [CrossRef] [PubMed]
- Samaras, A.-D.; Moustakidis, S.; Apostolopoulos, I.D.; Papandrianos, N.; Papageorgiou, E. Classification Models for Assessing Coronary Artery Disease Instances Using Clinical and Biometric Data: An Explainable Man-in-the-Loop Approach. Sci. Rep. 2023, 13, 6668. [Google Scholar] [CrossRef] [PubMed]
- Setio, A.A.A.; Traverso, A.; De Bel, T.; Berens, M.S.N.; Bogaard, C.V.D.; Cerello, P.; Chen, H.; Dou, Q.; Fantacci, M.E.; Geurts, B.; et al. Validation, Comparison, and Combination of Algorithms for Automatic Detection of Pulmonary Nodules in Computed Tomography Images: The LUNA16 Challenge. Med. Image Anal. 2017, 42, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Shi, F.; Chen, B.; Cao, Q.; Wei, Y.; Zhou, Q.; Zhang, R.; Zhou, Y.; Yang, W.; Wang, X.; Fan, R.; et al. Semi-Supervised Deep Transfer Learning for Benign-Malignant Diagnosis of Pulmonary Nodules in Chest CT Images. IEEE Trans. Med. Imaging 2022, 41, 771–781. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Sollee, J.; Luo, Y.-H.; Reddy, A.; Zhong, Z.; Wu, J.; Mammarappallil, J.; Healey, T.; Cheng, G.; Azzoli, C.; et al. Prediction of Lung Malignancy Progression and Survival with Machine Learning Based on Pre-Treatment FDG-PET/CT. eBioMedicine 2022, 82, 104127. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.-J.; Choi, D.; Choi, J.Y.; Hyun, S.H. Performance Evaluation of a Deep Learning System for Differential Diagnosis of Lung Cancer with Conventional CT and FDG PET/CT Using Transfer Learning and Metadata. Clin. Nucl. Med. 2021, 46, 635–640. [Google Scholar] [CrossRef]
- Salihoğlu, Y.S.; Uslu Erdemir, R.; Aydur Püren, B.; Özdemir, S.; Uyulan, Ç.; Ergüzel, T.T.; Tekin, H.O. Diagnostic Performance of Machine Learning Models Based on 18F-FDG PET/CT Radiomic Features in the Classification of Solitary Pulmonary Nodules. Mol. Imaging Radionucl. Ther. 2022, 31, 82–88. [Google Scholar] [CrossRef]
- Chae, K.J.; Jin, G.Y.; Ko, S.B.; Wang, Y.; Zhang, H.; Choi, E.J.; Choi, H. Deep Learning for the Classification of Small (≤2 Cm) Pulmonary Nodules on CT Imaging: A Preliminary Study. Acad. Radiol. 2020, 27, e55–e63. [Google Scholar] [CrossRef] [PubMed]
- Svetnik, V.; Liaw, A.; Tong, C.; Culberson, J.C.; Sheridan, R.P.; Feuston, B.P. Random forest: A classification and regression tool for compound classification and QSAR modeling. J. Chem. Inf. Comput. Sci. 2003, 43, 1947–1958. [Google Scholar] [CrossRef]
- Ho, T.K. The Random Subspace Method for Constructing Decision Forests. IEEE Trans. Pattern Anal. Mach. Intell. 1998, 20, 832–844. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and Regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Apostolopoulos, I.D.; Papathanasiou, N.D.; Panayiotakis, G.S. Classification of Lung Nodule Malignancy in Computed Tomography Imaging Utilising Generative Adversarial Networks and Semi-Supervised Transfer Learning. Biocybern. Biomed. Eng. 2021, 41, 1243–1257. [Google Scholar] [CrossRef]
- Chen, S.; Harmon, S.; Perk, T.; Li, X.; Chen, M.; Li, Y.; Jeraj, R. Diagnostic Classification of Solitary Pulmonary Nodules Using Dual Time 18F-FDG PET/CT Image Texture Features in Granuloma-Endemic Regions. Sci. Rep. 2017, 7, 9370. [Google Scholar] [CrossRef]
- Weir-McCall, J.R.; Debruyn, E.; Harris, S.; Qureshi, N.R.; Rintoul, R.C.; Gleeson, F.V.; Gilbert, F.J.; Lucy Brindle, A.B.; Callister, M.; Clegg, A.; et al. Diagnostic Accuracy of a Convolutional Neural Network Assessment of Solitary Pulmonary Nodules Compared with PET with CT Imaging and Dynamic Contrast-Enhanced CT Imaging Using Unenhanced and Contrast-Enhanced CT Imaging. Chest 2023, 163, 444–454. [Google Scholar] [CrossRef]
| Clinical Characteristics | Frequency |
|---|---|
| No. of participants | 456 |
| Age (mean ± sd) | 66 ± 8 |
| Sex (male/female) | 69% male/31% female |
| Total benign SPNs | 222 (48.6%) |
| Total malignant SPNs | 234 (51.4%) |
| Feature Name | Type of Feature | Type of Values | Potential Values |
|---|---|---|---|
| SUVmax | FDG Uptake | Numeric | |
| Diameter | SPN Feature | Numeric | 0.6 to 3 cm |
| Location | SPN Feature | Categorical | Left Lower Lobe (LLL) Lingula Middle Right Upper Lobe (RUL) Right Lower Lobe (RLR) |
| Type | SPN Feature | Categorical | Solid Semi-solid Ground-class |
| Margins | SPN Feature | Categorical | Well-defined Lobulated Spiculated Ill-defined |
| TP | FP | TN | FN | ACC | SEN | SPE | |
|---|---|---|---|---|---|---|---|
| Human Reader 1 | 203 | 14 | 208 | 31 | 0.9013 | 0.8675 | 0.9369 |
| Human Reader 2 | 203 | 25 | 197 | 31 | 0.8771 | 0.8675 | 0.8873 |
| ACC | SEN | SPE | |
|---|---|---|---|
| ML without human diagnosis as input feature | 0.9295 (CI 95%: 0.9287–0.9303) | 0.9171 (CI 95%: 0.9156–0.9186) | 0.9426 (CI 95%: 0.9402–0.9449) |
| ML with human diagnosis as input feature | 0.9539 (CI 95%: 0.9529–0.9549) | 0.9688 (CI 95%: 0.967–0.9707) | 0.9383 (CI 95%: 0.9373–0.9393) |
| Metric | Score |
|---|---|
| Accuracy | 0.9539 (CI 95%: 0.9529–0.9549) |
| Sensitivity | 0.9688 (CI 95%: 0.967–0.9707) |
| Specificity | 0.9383 (CI 95%: 0.9373–0.9393) |
| F1 | 0.9557 (CI 95%: 0.9547–0.9567) |
| AUC | 0.992 (CI 95%: 0.9919–0.9921) |
| Kappa | 0.9078 (CI 95%: 0.9058–0.9098) |
| TP | 227 |
| TN | 208 |
| FP | 14 |
| FN | 7 |
| FPR | 0.0617 (CI 95%: 0.0607–0.0627) |
| FNR | 0.0312 (CI 95%: 0.0293–0.033) |
| PPV | 0.943 (CI 95%: 0.9422–0.9439) |
| NPV | 0.9152 (CI 95%: 0.9139–0.9165) |
| TP | FP | TN | FN | ACC | SEN | SPE | |
|---|---|---|---|---|---|---|---|
| Human Reader 1 | 44 | 8 | 35 | 11 | 0.8061 | 0.8 | 0.8139 |
| ML | 53 | 8 | 35 | 2 | 0.8979 | 0.9636 | 0.8139 |
| Study | Data Type | Test Data Size | Results |
|---|---|---|---|
| [12] | CT image | 897 | ACC: 90.85% SEN: 94.76% SPE: 82.05% |
| [26] | CT image | 1113 | ACC: 92.07% SEN: 89.35% SPE: 94.80% |
| [11] | CT image | 112 | ACC: 94% SEN: 92% SPE: 94.50% |
| [10] | CT image | 1297 | AUC: 0.936 |
| [22] | CT image | 208 | AUC: 0.85 |
| [15] | CT image | 252 | ACC: 90.6% SEN: 83.7% SPE: 93.9% |
| [8] | CT image | 2119 | ACC: 85.23% SEN: 92.79% SPE: 72.89% AUC: 0.9275 |
| [21] | PET/CT image | 48 | AUC: 0.81 SEN: 88% SPE: 86% |
| [19] | PET + CT images | 1168 | ACC: 79% AUC: 0.876 |
| [20] | PET + CT + Clinical | 105 | ACC: 85% SEN: 86% SPE: 33% |
| [27] | PET image | 86 | ACC: 86% SEN: 64% SPE: 91% |
| [28] | CT + PET images | 270 | SEN: 72% SPE: 82% |
| [18] | CT image | 1175 | ACC: 74.5% AUC: 0.795 |
| [16] | Clinical data | 452 | ACC: 86.54% |
| [17] | CT image + Clinical data | 227 | AUC: 88.2% |
| [18] | Clinical data | 200 | ACC: 75.6% F1: 72.2% AUC: 82% |
| [15] | CT image | 552 | ACC: 84.1% AUC: 90.3% |
| [19] | PET/CT image | 55 | SEN: 85.2% SPE: 82.1% |
| [21] | PET/CT image + Radiomics | 106 | AUC: 82% |
| This study | CT and PET image features | 456 | ACC: 0.9539 (CI 95%: 0.9529–0.9549) SEN: 0.9688 (CI 95%: 0.967–0.9707) SPE: 0.9383 (CI 95%: 0.9373–0.9393) AUC: 0.992 (CI 95%: 0.9919–0.9921) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Apostolopoulos, I.D.; Papathanasiou, N.D.; Apostolopoulos, D.J.; Papandrianos, N.; Papageorgiou, E.I. Integrating Machine Learning in Clinical Practice for Characterizing the Malignancy of Solitary Pulmonary Nodules in PET/CT Screening. Diseases 2024, 12, 115. https://doi.org/10.3390/diseases12060115
Apostolopoulos ID, Papathanasiou ND, Apostolopoulos DJ, Papandrianos N, Papageorgiou EI. Integrating Machine Learning in Clinical Practice for Characterizing the Malignancy of Solitary Pulmonary Nodules in PET/CT Screening. Diseases. 2024; 12(6):115. https://doi.org/10.3390/diseases12060115
Chicago/Turabian StyleApostolopoulos, Ioannis D., Nikolaos D. Papathanasiou, Dimitris J. Apostolopoulos, Nikolaos Papandrianos, and Elpiniki I. Papageorgiou. 2024. "Integrating Machine Learning in Clinical Practice for Characterizing the Malignancy of Solitary Pulmonary Nodules in PET/CT Screening" Diseases 12, no. 6: 115. https://doi.org/10.3390/diseases12060115
APA StyleApostolopoulos, I. D., Papathanasiou, N. D., Apostolopoulos, D. J., Papandrianos, N., & Papageorgiou, E. I. (2024). Integrating Machine Learning in Clinical Practice for Characterizing the Malignancy of Solitary Pulmonary Nodules in PET/CT Screening. Diseases, 12(6), 115. https://doi.org/10.3390/diseases12060115







