Multi-Channel Transfer Learning of Chest X-ray Images for Screening of COVID-19
Abstract
:1. Introduction
2. Materials and Methods
2.1. Dataset and Data Augmentation
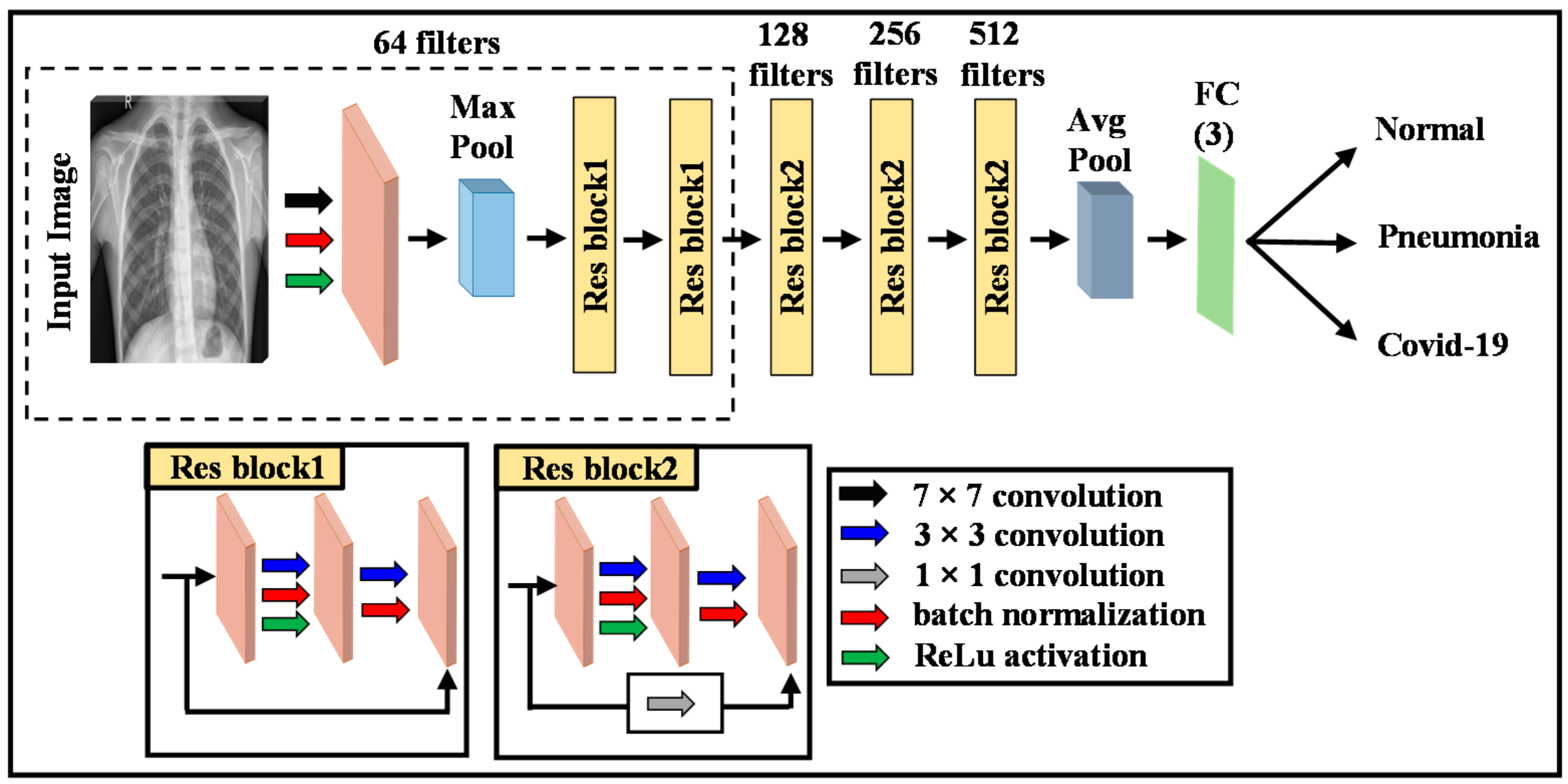
2.2. Deep Learning Model
2.3. Transfer Learning (TL)
2.4. Proposed Methodology
- Build Model a by fine-tuning the pre-trained ResNet model using the Dataset_A, which can classify the normal and diseased images.
- Construct Model b to classify pneumonia and non-pneumonia images based on the pre-trained ResNet model by fine-tuning the Dataset_B.
- Design ResNet-based Model c by fine-tuning the Dataset_C, which can classify the COVID-19 and non-COVID19 images.
- Remove the classification layer of all models to expose activations of their penultimate layers.
- Freeze the weights of Models a, b and c.
- Build ensemble models (Models Bab, Cac, Dbc, and Eabc) by combining Models a + b, a + c, b + c and a + b + c.
- Add a concatenation layer and a classification layer (softmax) into the architecture of the combined models.
- Train (fine-tune) again the combined models using the Dataset_D, which can classify the normal, pneumonia, and COVID-19 images.
3. Results
3.1. Experimental Setup
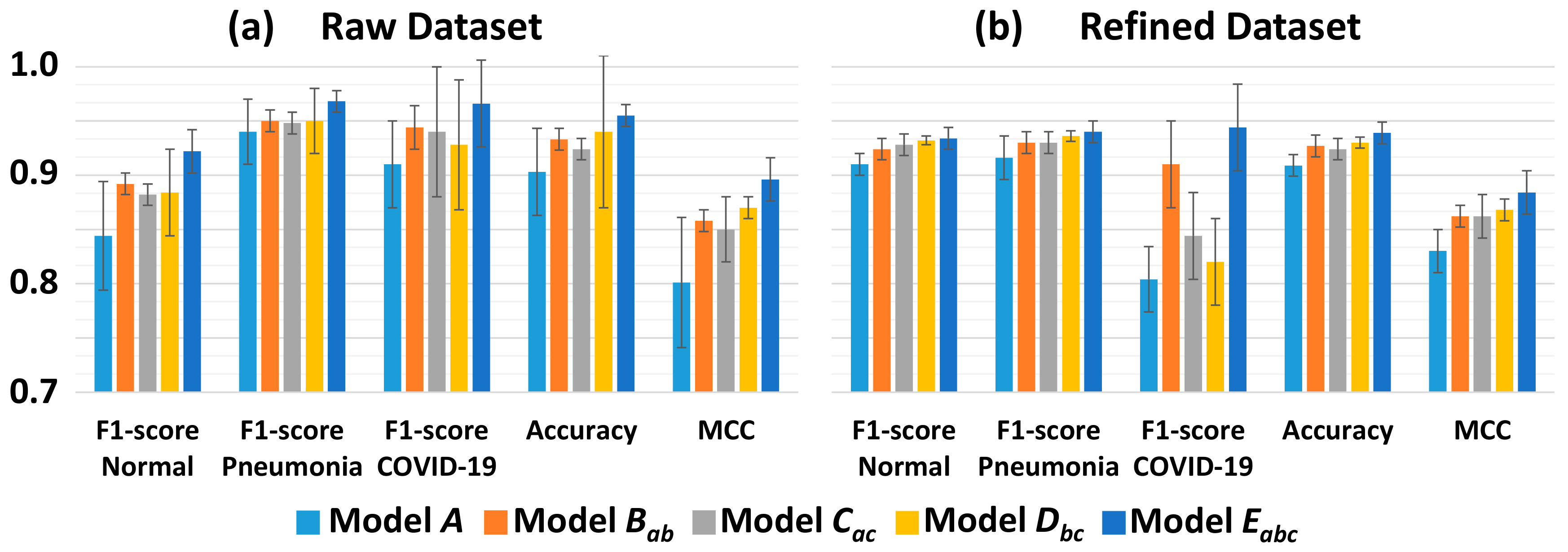
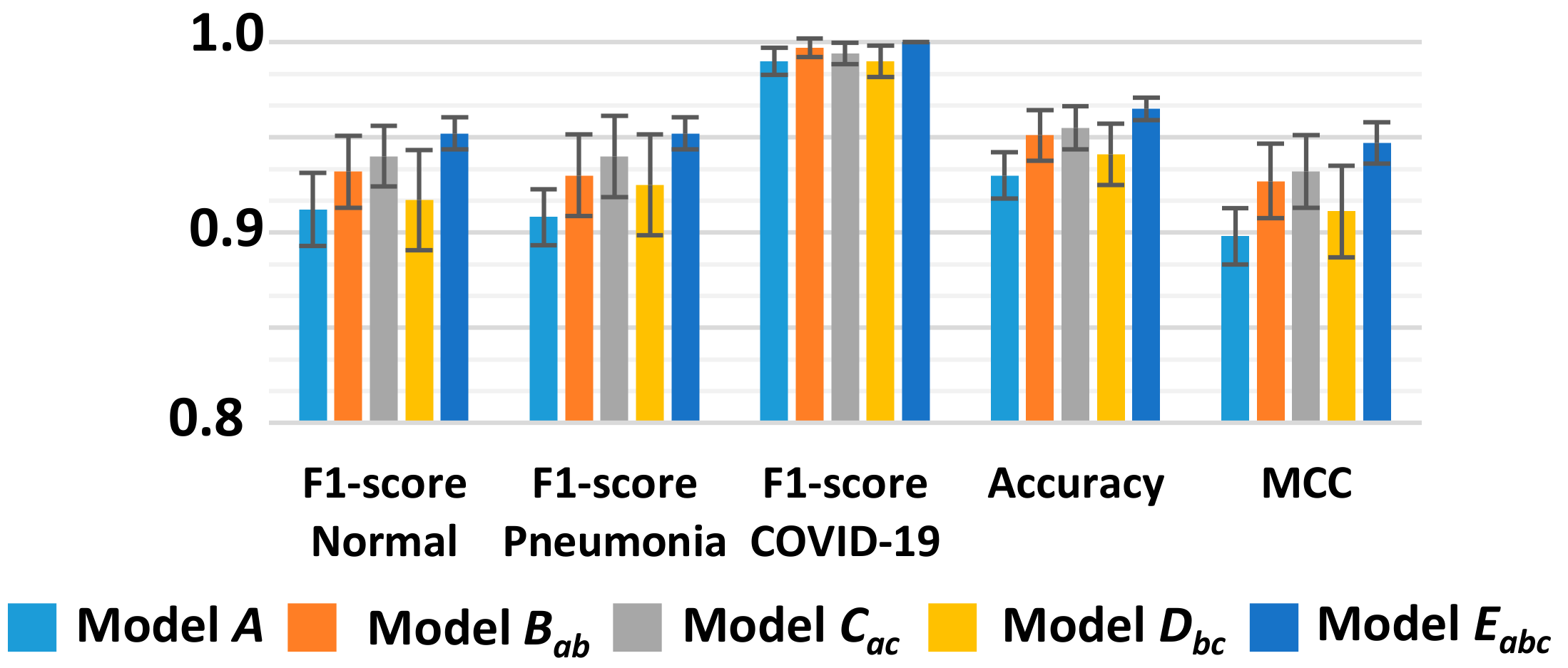
3.2. Classification Performance
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Lai, M.M.C. SARS virus: The beginning of the unraveling of a new coronavirus. J. Biomed. Sci. 2003, 10, 664–675. [Google Scholar] [CrossRef] [PubMed]
- Zumla, A.; Hui, D.S.C.; Perlman, S. Middle East respiratory syndrome. Lancet 2015, 386, 995–1007. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. Coronavirus Disease 2019 (COVID-19): Situation Report, 72; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Chana, J.F.; Yip, C.C.; To, K.K. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel realtime reverse transcription-polymerase chain reaction assay validated in vitro and with clinical specimens. J. Clin. Microbiol. 2020. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Shi, L.; Wang, Y.; Zhang, J.; Huang, L.; Zhang, C.; Liu, S.; Zhao, P.; Liu, H.; Zhu, L.; et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020, 8, 420–422. [Google Scholar] [CrossRef]
- Bai, H.X.; Hsieh, B.; Xiong, Z.; Halsey, K.; Choi, J.W.; Tran, T.M.; Pan, I.; Shi, L.B.; Wang, D.C.; Mei, J.; et al. Performance of radiologists in differentiating COVID-19 from viral pneumonia on chest CT. Radiology 2020. [Google Scholar] [CrossRef] [PubMed]
- Ozturk, T.; Talo, M.; Yildirim, E.A.; Baloglu, U.B.; Yildirim, O.; Acharya, U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020, 121, 103792. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baik, J.W.; Kim, J.Y.; Cho, S.; Choi, S.; Kim, J.; Kim, C. Super Wide-Field Photoacoustic Microscopy of Animals and Humans In Vivo. IEEE Trans. Med. Imaging 2020, 39, 975–984. [Google Scholar] [CrossRef]
- Kim, J.; Kim, J.Y.; Jeon, S.; Baik, J.W.; Cho, S.H.; Kim, C. Super-resolution localization photoacoustic microscopy using intrinsic red blood cells as contrast absorbers. Light. Sci. Appl. 2019, 8, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.; Kim, J.; Lee, D.; Baik, J.W.; Kim, C. Review on practical photoacoustic microscopy. Photoacoustics 2019, 15, 100141. [Google Scholar] [CrossRef]
- Milletari, F.; Ahmadi, S.-A.; Kroll, C.; Plate, A.; Rozanski, V.; Maiostre, J.; Levin, J.; Dietrich, O.; Ertl-Wagner, B.; Bötzel, K.; et al. Hough-CNN: Deep learning for segmentation of deep brain regions in MRI and ultrasound. Comput. Vis. Image Underst. 2017, 164, 92–102. [Google Scholar] [CrossRef] [Green Version]
- Gupta, H.; Jin, K.H.; Nguyen, H.Q.; McCann, M.T.; Unser, M. CNN-Based Projected Gradient Descent for Consistent CT Image Reconstruction. IEEE Trans. Med. Imaging 2018, 37, 1440–1453. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Pan, Y.; Li, M.; Chen, Z.; Tang, L.; Lu, C.; Wang, J. Applications of deep learning to MRI images: A survey. Big Data Min. Anal. 2018, 1, 1–18. [Google Scholar] [CrossRef]
- Pan, S.J.; Yang, Q. A Survey on Transfer Learning. IEEE Trans. Knowl. Data Eng. 2009, 22, 1345–1359. [Google Scholar] [CrossRef]
- Narin, A.; Kaya, C.; Pamuk, Z. Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks. arXiv 2020, arXiv:2003.10849. [Google Scholar]
- Ucar, F.; Korkmaz, D. COVIDiagnosis-Net: Deep Bayes-SqueezeNet based Diagnostic of the Coronavirus Disease 2019 (COVID-19) from X-Ray Images. Med. Hypotheses 2020, 140, 109761. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.I.; Shah, J.L.; Bhat, M.M. CoroNet: A deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput. Methods Progr. Biomed. 2020, 196, 105581. [Google Scholar] [CrossRef]
- Rahimzadeh, M.; Attar, A. A modified deep convolutional neural network for detecting COVID-19 and pneumonia from chest X-ray images based on the concatenation of Xception and ResNet50V2. Inform. Med. Unlocked 2020, 19, 100360. [Google Scholar] [CrossRef]
- Apostolopoulos, I.D.; Mpesiana, T.A. Covid-19: Automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020, 43, 635–640. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Wong, A. COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest radiography images. arXiv 2020, arXiv:2003.09871. [Google Scholar]
- Farooq, M.; Hafeez, A. COVID-ResNet: A Deep Learning Framework for Screening of COVID19 from Radiographs. arXiv 2020, arXiv:2003.14395. [Google Scholar]
- Xuanyang, X.; Yuchang, G.; Shouhong, W.; Xi, L. Computer Aided Detection of SARS Based on Radiographs Data Mining. In Proceedings of the 2005 IEEE Engineering in Medicine and Biology 27th Annual Conference, Shanghai, China, 17–18 January 2006; pp. 7459–7462. [Google Scholar]
- Shan, F.; Gao, Y.; Wang, J.; Shi, W.; Shi, N.; Han, M.; Xue, Z.; Shi, Y. Lung Infection Quantification of COVID-19 in CT Images with Deep Learning. arXiv 2020, arXiv:2003.04655. [Google Scholar]
- Gozes, O.; Frid-Adar, M.; Greenspan, H.; Browning, P.D.; Zhang, H.; Ji, W.; Bernheim, A.; Siegel, E. Rapid ai development cycle for the coronavirus (COVID-19) pandemic: Initial results for automated detection & patient monitoring using deep learning ct image analysis. arXiv 2020, arXiv:2003.05037. [Google Scholar]
- Wang, S.; Kang, B.; Ma, J.; Zeng, X.; Xiao, M.; Guo, J.; Cai, M.; Yang, J.; Li, Y.; Meng, X.; et al. A deep learning algorithm using CT images to screen for Corona Virus Disease (COVID-19). medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Chouhan, V.; Singh, S.K.; Khamparia, A.; Gupta, N.; Tiwari, P.; Moreira, C.; Damasevicius, R.; De Albuquerque, V.H.C. A Novel Transfer Learning Based Approach for Pneumonia Detection in Chest X-ray Images. Appl. Sci. 2020, 10, 559. [Google Scholar] [CrossRef] [Green Version]
- Lakhani, P.; Sundaram, B. Deep Learning at Chest Radiography: Automated Classification of Pulmonary Tuberculosis by Using Convolutional Neural Networks. Radiology 2017, 284, 574–582. [Google Scholar] [CrossRef]
- Fraz, M.M.; Remagnino, P.; Hoppe, A.; Uyyanonvara, B.; Rudnicka, A.R.; Owen, C.G.; Barman, S.A. An Ensemble Classification-Based Approach Applied to Retinal Blood Vessel Segmentation. IEEE Trans. Biomed. Eng. 2012, 59, 2538–2548. [Google Scholar] [CrossRef]
- Radiological Society of North America. RSNA Pneumonia Detection Challenge. 2018. Available online: https://www.rsna.org/en/education/ai-resources-and-training/ai-image-challenge/RSNA-Pneumonia-Detection-Challenge-2018 (accessed on 14 April 2020).
- Kaggle. Available online: https://www.kaggle.com/andrewmvd/convid19-xrays (accessed on 14 April 2020).
- Cohen, J.P.; Morrison, P.; Dao, L. COVID-19 image data collection. arXiv 2020, arXiv:2003.11597. [Google Scholar]
- Shorten, C.; Khoshgoftaar, T.M. A survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Yamashita, R.; Nishio, M.; Do, R.K.G.; Togashi, K. Convolutional neural networks: An overview and application in radiology. Insights Imaging 2018, 9, 611–629. [Google Scholar] [CrossRef] [Green Version]
- Rawat, W.; Wang, Z. Deep convolutional neural networks for image classification: A comprehensive review. Neural Comput. 2017, 29, 2352–2449. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Tharwat, A. Classification assessment methods. Appl. Comput. Inform. 2020. [Google Scholar] [CrossRef]
- Kim, T.-K. T test as a parametric statistic. Korean J. Anesthesiol. 2015, 68, 540–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
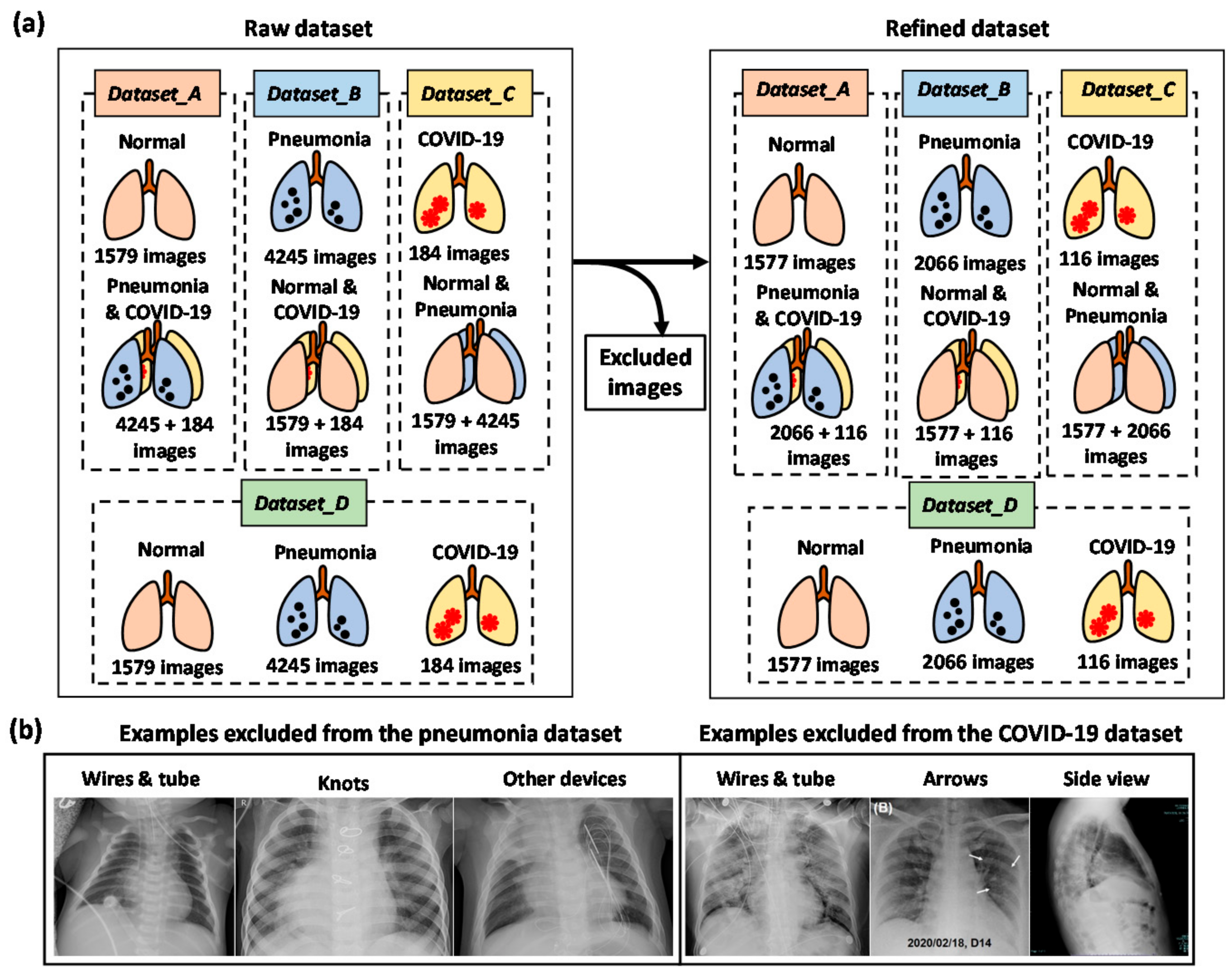

| Dataset | Class | Raw Dataset | Refined Dataset | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | Training | Test | Total | Training | Test | ||||
| Train | Validation | Train | Validation | ||||||
| Dataset_A | Normal | 1579 | 1137 | 284 | 158 * | 1577 | 1136 | 284 | 157 * |
| Pneumonia and COVID-19 | 4429 | 3190 | 797 | 442 (424 * + 18 *) | 2182 | 1572 | 393 | 217 (206 * + 11 *) | |
| Dataset_B | Pneumonia | 4245 | 3057 | 764 | 424 * | 2066 | 1488 | 372 | 206 * |
| Normal and COVID-19 | 1763 | 1270 | 317 | 176 (158 * + 18 *) | 1693 | 1220 | 305 | 168 (157 * + 11 *) | |
| Dataset_C | COVID-19 | 184 | 133 | 33 | 18 * | 116 | 84 | 21 | 11 * |
| Normal and Pneumonia | 5824 | 4194 | 1048 | 582 (158 * + 424 *) | 3643 | 2624 | 656 | 363 (157 * + 206 *) | |
| Dataset_D | Normal | 1579 | 1137 | 284 | 158 * | 1577 | 1136 | 284 | 157 * |
| Pneumonia | 4245 | 3057 | 764 | 424 * | 2066 | 1488 | 372 | 206 * | |
| COVID-19 | 184 | 133 | 33 | 18 * | 116 | 84 | 21 | 11 * | |
| - | Model A (Single ResNet) | Model Bab (a + b) | Model Cac (a + c) | Model Dbc (b + c) | Model Eabc (a + b + c) | ||
|---|---|---|---|---|---|---|---|
| Raw Dataset | Normal | Precision | 0.776 ± 0.11 | 0.830 ± 0.04 | 0.812 ± 0.01 | 0.846 ± 0.10 | 0.874 ± 0.03 |
| Recall | 0.944 ± 0.07 | 0.964 ± 0.02 | 0.980 ± 0.01 | 0.942 ± 0.07 | 0.976 ± 0.01 | ||
| F1-score | 0.844 ±0.05 | 0.892 ± 0.01 | 0.882 ± 0.01 | 0.884 ± 0.04 | 0.922 ± 0.02 | ||
| Pneumonia | Precision | 0.984 ± 0.02 | 0.988 ± 0.01 | 0.996 ± 0.01 | 0.984 ± 0.02 | 0.994 ± 0.004 | |
| Recall | 0.884 ± 0.07 | 0.920 ± 0.02 | 0.902 ± 0.01 | 0.918 ± 0.06 | 0.944 ± 0.01 | ||
| F1-score | 0.940 ± 0.03 | 0.950 ± 0.01 | 0.948 ± 0.01 | 0.950 ± 0.03 | 0.968 ± 0.01 | ||
| COVID-19 | Precision | 0.838 ± 0.07 | 0.896 ± 0.04 | 0.894 ± 0.10 | 0.893 ± 0.12 | 0.940 ± 0.08 | |
| Recall | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | ||
| F1-score | 0.910 ± 0.04 | 0.944 ± 0.02 | 0.940 ± 0.06 | 0.928 ± 0.06 | 0.966± 0.04 | ||
| Accuracy | 0.903 ± 0.04 | 0.933 ± 0.01 | 0.924 ± 0.01 | 0.940 ± 0.07 | 0.955 ± 0.01 | ||
| MCC | 0.801 ± 0.06 | 0.858 ± 0.01 | 0.850 ± 0.03 | 0.870 ± 0.01 | 0.896 ± 0.02 | ||
| Refined Dataset | Normal | Precision | 0.874 ± 0.06 | 0.876 ± 0.02 | 0.884 ± 0.03 | 0.896 ± 0.01 | 0.902 ± 0.03 |
| Recall | 0.950 ± 0.04 | 0.982 ± 0.01 | 0.980 ± 0.01 | 0.970 ± 0.01 | 0.964 ± 0.03 | ||
| F1-score | 0.910 ± 0.01 | 0.924 ± 0.01 | 0.928 ± 0.01 | 0.932 ± 0.004 | 0.934 ± 0.01 | ||
| Pneumonia | Precision | 0.970 ± 0.03 | 0.982 ± 0.01 | 0.984 ± 0.01 | 0.978 ± 0.01 | 0.972 ± 0.02 | |
| Recall | 0.878 ± 0.06 | 0.882 ± 0.02 | 0.880 ± 0.03 | 0.896 ± 0.01 | 0.912 ± 0.03 | ||
| F1-score | 0.916 ± 0.02 | 0.930 ± 0.01 | 0.930 ± 0.01 | 0.936 ± 0.005 | 0.940 ± 0.01 | ||
| COVID-19 | Precision | 0.674 ± 0.05 | 0.834 ± 0.08 | 0.734 ± 0.06 | 0.694 ± 0.06 | 0.896 ± 0.07 | |
| Recall | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | ||
| F1-score | 0.804 ± 0.03 | 0.910 ± 0.04 | 0.844 ± 0.04 | 0.820 ± 0.04 | 0.944 ± 0.04 | ||
| Accuracy | 0.909 ± 0.01 | 0.927 ± 0.01 | 0.924 ± 0.01 | 0.930 ± 0.005 | 0.939 ± 0.01 | ||
| MCC | 0.830 ± 0.02 | 0.862 ± 0.01 | 0.862 ± 0.02 | 0.868 ± 0.01 | 0.884 ± 0.02 | ||
| Models | PPV | A | Eabc | ||
|---|---|---|---|---|---|
| t-Value | p-Value | t-Value | p-Value | ||
| A | 0.898 ± 0.18 | - | - | 4.442 | 0.00001 |
| Bab | 0.915 ± 0.16 | 1.684 | 0.046 | 2.184 | 0.025 |
| Cac | 0.918 ± 0.20 | 1.913 | 0.028 | 2.145 | 0.016 |
| Dbc | 0.918 ± 0.20 | 1.913 | 0.028 | 2.145 | 0.016 |
| Eabc | 0.942 ± 0.16 | 4.442 | 0.00001 | - | - |
| Methods Used In | No. of Images | Model | Accuracy (%) | Precision (%) | Recall (%) |
|---|---|---|---|---|---|
| [7] | 500 Normal, 500 Pneumonia, 125 COVID-19 | DarkCovidNet | 87.02 | 89.96 | 85.35 |
| [17] | 1583 Normal, 4290 Pneumonia, 76 COVID-19 | COVIDiagnosis-Net | 98.26 | 99.35 | 100 |
| [18] | 310 Normal, 330 Pneumonia Bacterial, 327 Pneumonia Viral Images, 284 Covid-19 | CoroNet | 89.6 | 90.0 | 89.92 |
| [19] | 8851 Normal, 6054 Pneumonia, 180 COVID-19 | Xception and ResNet50V2 | 91.4 | 35.27 | 80.53 |
| [20] | 504 Normal, 700 Pneumonia, 224 COVID-19 | VGG-19 | 93.48 | 93.27 | 92.85 |
| [21] | 8066 Normal, 5538 COVID-19(-), 358 COVID-19 | COVID-Net | 93.3 | 98.9 | 91 |
| Proposed | 1579 Normal, 4245 Pneumonia, 184 COVID-19 (raw dataset) | Ensemble 3 ResNet-18 | 95.5 | 94.0 | 100 |
| 1577 Normal, 2066 Pneumonia, 116 COVID-19 (refined dataset) | 93.9 | 89.6 | 100 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Misra, S.; Jeon, S.; Lee, S.; Managuli, R.; Jang, I.-S.; Kim, C. Multi-Channel Transfer Learning of Chest X-ray Images for Screening of COVID-19. Electronics 2020, 9, 1388. https://doi.org/10.3390/electronics9091388
Misra S, Jeon S, Lee S, Managuli R, Jang I-S, Kim C. Multi-Channel Transfer Learning of Chest X-ray Images for Screening of COVID-19. Electronics. 2020; 9(9):1388. https://doi.org/10.3390/electronics9091388
Chicago/Turabian StyleMisra, Sampa, Seungwan Jeon, Seiyon Lee, Ravi Managuli, In-Su Jang, and Chulhong Kim. 2020. "Multi-Channel Transfer Learning of Chest X-ray Images for Screening of COVID-19" Electronics 9, no. 9: 1388. https://doi.org/10.3390/electronics9091388
APA StyleMisra, S., Jeon, S., Lee, S., Managuli, R., Jang, I.-S., & Kim, C. (2020). Multi-Channel Transfer Learning of Chest X-ray Images for Screening of COVID-19. Electronics, 9(9), 1388. https://doi.org/10.3390/electronics9091388






