1. Introduction
The electrocardiogram (ECG) is one of the standard clinical methods of diagnosis for cardiac-related diseases. By means of the ECG, the electrical activity of the heart is recorded and analyzed. This technology is inexpensive, easy to use and clinically established. The main drawback of the ECG is the use of adhesive electrodes that can provoke allergic reactions, especially if applied during a relatively long period. Another issue is the reduced comfort for patients, mainly due to the cables attached to the electrodes.
To overcome these issues, various unobtrusive ECG systems (uECG) have been developed. One realization method is capacitive ECG (cECG), which has been built into everyday objects such as chairs [
1,
2], car seats [
3], beds [
4,
5,
6,
7] and clothing [
8]. The principle is to use ECG electrodes incorporated in these objects that do not require conductive contact to the human body, but acquire the physiological surface potentials with capacitive coupling. Similar unobtrusive systems are dry ECGs; these have conductive contact to the skin but do not use any electrolyte gel or adhesive material [
9,
10,
11,
12]. However, the uECG requires a more complicated electronic setup [
13,
14]. In many cases, an operational amplifier is used as a buffer located directly at the electrode that drives the electrode cable and reduces interference (active electrode). Nevertheless, these systems have their drawbacks, e.g., they are especially prone to movement artifacts. So far, uECG are foremost used in laboratory settings or clinical studies.
For the cECG in particular, several hardware-based approaches have been proposed to improve the quality, but they have also increased the complexity of the circuits [
14,
15,
16]. Furthermore, the morphology of the cECG differs slightly from that of the standard ECG, which is important for clinical diagnosis. When comparing the shape of the ECGs, a study using a cECG chair showed that some features could be reliably extracted, including the heart rate and QRS duration, as well as QT and PQ length. However, other features of the ECG were distorted, i.e., (1) depression of the ST segment; (2) flattening of the R-peak; (3) depression of the S wave below the equipotential line; (4) a rounding of the QRS complex [
17,
18].
Previously, our group described the development of a dry 12-lead ECG T-shirt with active electrodes [
19]. It was found that the accuracy of the system related to extracting the beat-to-beat intervals was very high, whereas some morphological features of the ECG waves (especially of the T wave) were not in agreement with the reference ECG [
19].
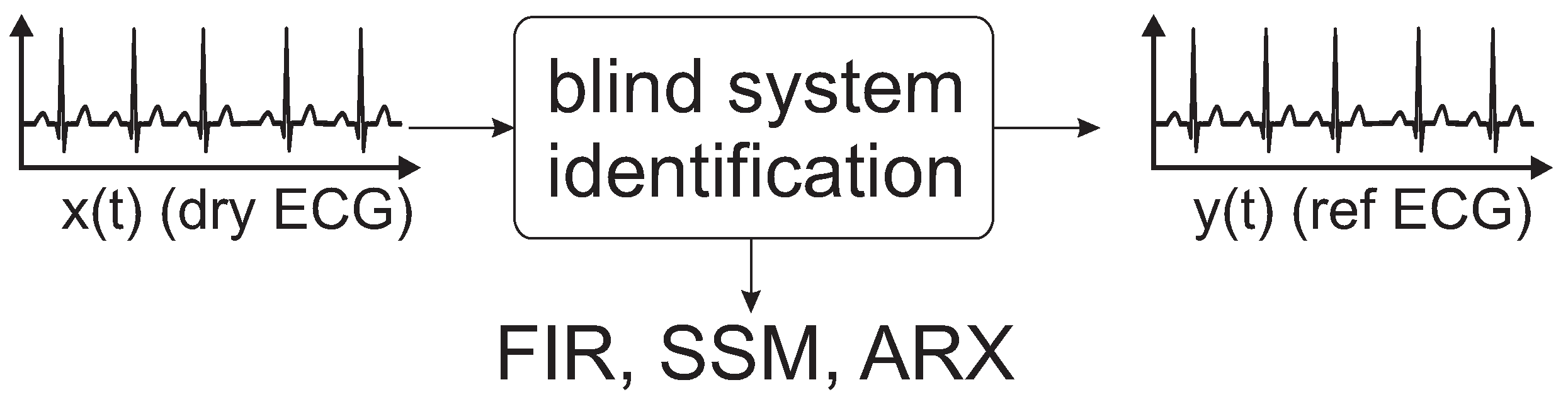
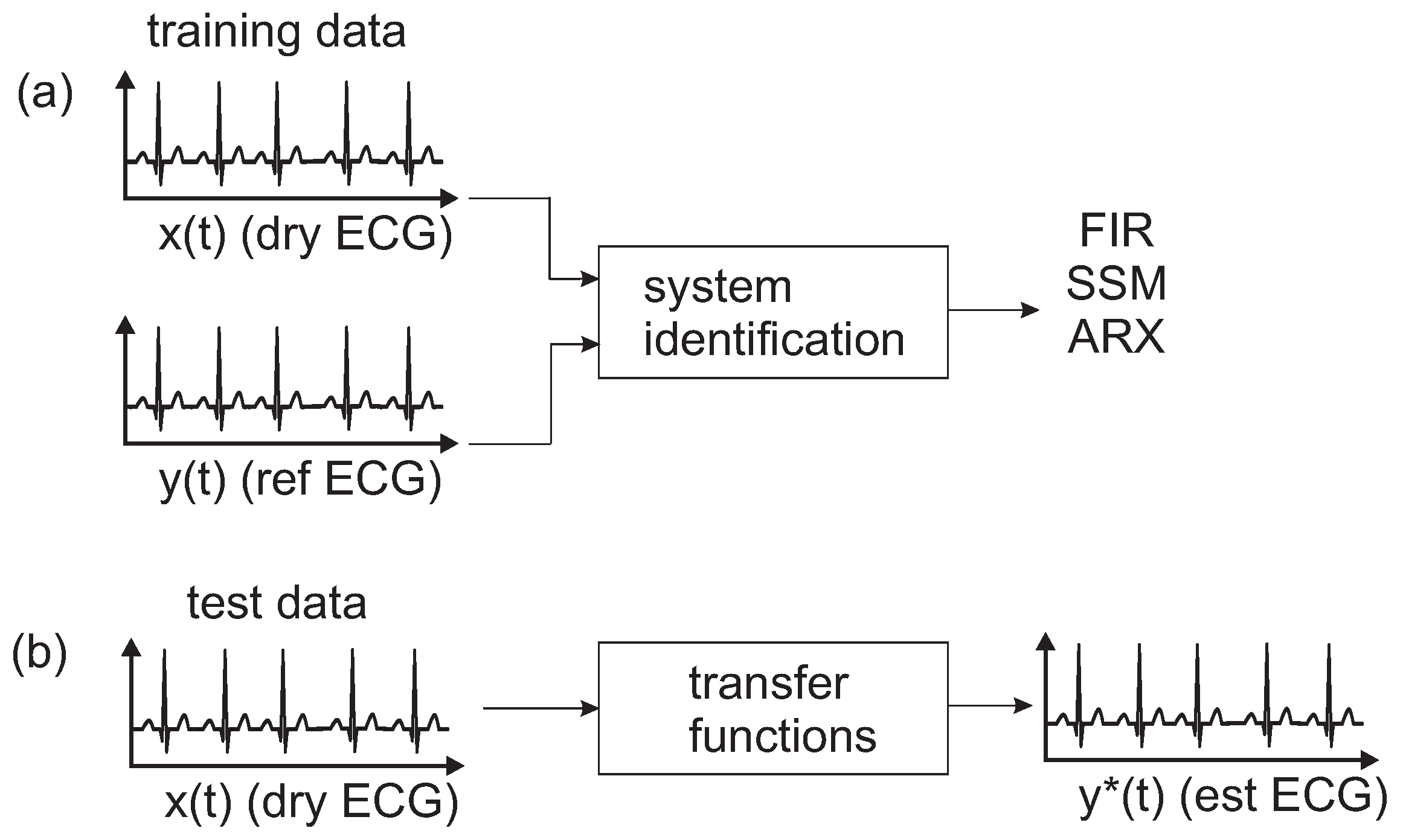
The aim of this study is to present different models and transfer functions to correct and compensate a uECG for deformations that arise due to the recording setup. After a short calibration phase using a uECG (dry-contact) and a reference ECG, models are trained that can be applied for correction of the subsequent uECG acquisition. The approach performs system identification using an finite impulse response (FIR) model, a state space model (SSM) and an autoregressive with exogenous terms (ARX).
Section 2 introduces the study, presents an equivalent system model for a uECG, and describes different methods for system identification.
Section 3 gives the results,
Section 4 presents the discussion and
Section 5 gives the conclusions.
4. Discussion
This paper presents different approaches to determine a model usable for correction and compensation of capacitive coupling occurring in uECG signals. The approach is to blindly perform system identification using an FIR model, a SSM and an ARX model. These models were trained on short excerpts of five heartbeats and were later validated on test data.
The goal was to evaluate different methods to correct deformations in a uECG using system identification. After a short calibration phase using a reference ECG and uECG, a correction method was applied to the following uECG acquisition. The models do not take into account changes in the electrode-skin interface, e.g., loss of electrode contact, but take into account the changes in ECG morphology due to the ECG acquisition method.
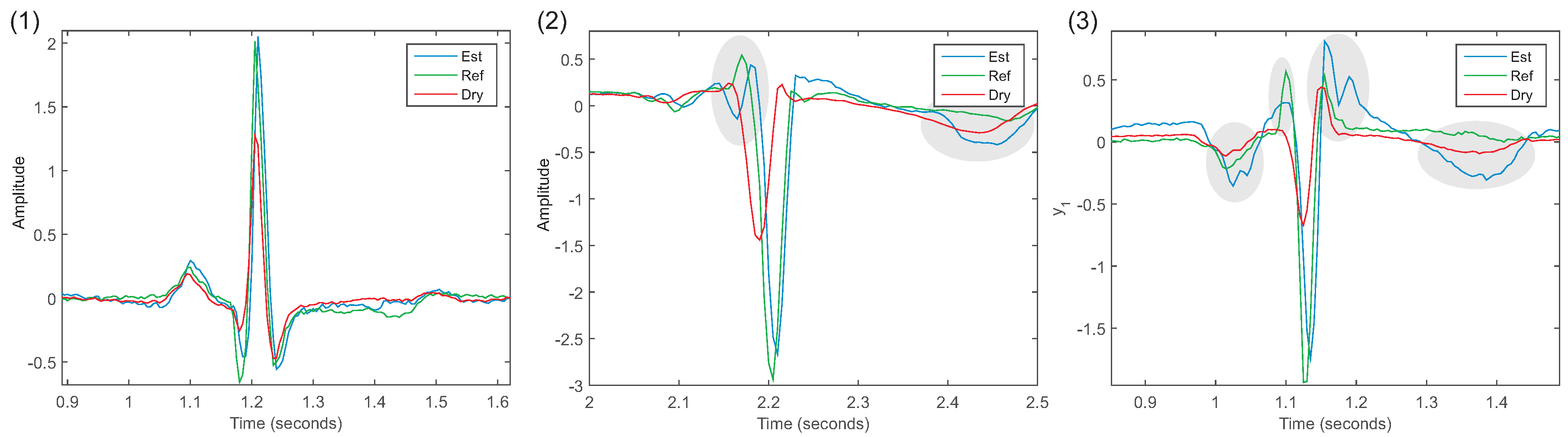
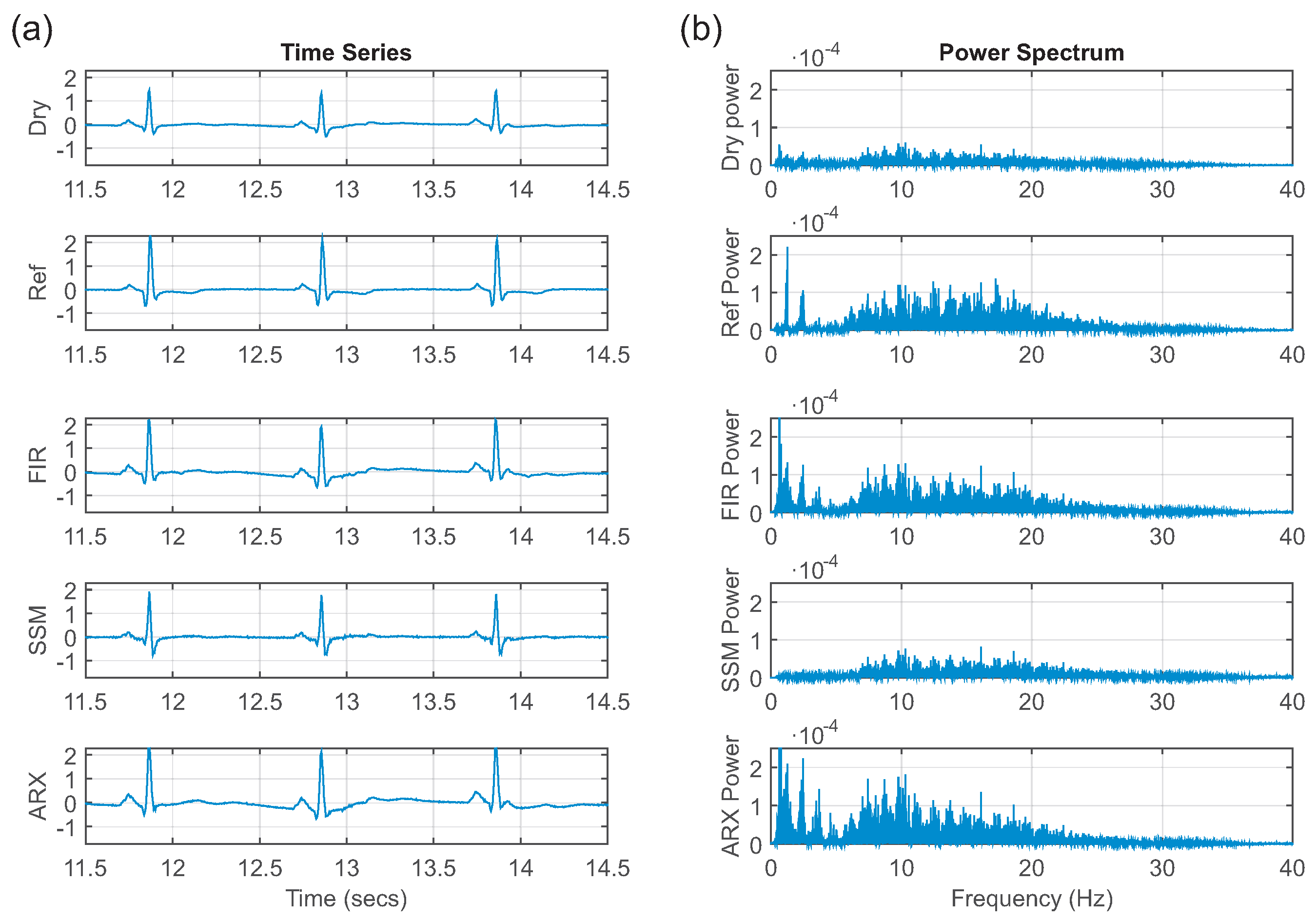
The use of FIR filters proved to be a feasible correction method. For each lead and each subject, an individual FIR filter was estimated. The estimated ECG was in better agreement compared to the reference ECG, which was supported by the high correlation coefficients (
= 0.84 ± 0.10,
= 0.65 ± 0.24,
= 0.88 ± 0.04). The improvement of the correlation of the ECGs was consistent with our findings in [
23]; analysis of the spectrum also supported this statement. Regarding morphology, the flattening of the R-peaks was successfully corrected (
= 0.10 ± 0.10,
= 0.14 ± 0.27,
= 0.03 ± 0.02). The other ECG waves (P, Q, S and T) were comparable with the uECG. To summarize: FIR filters had the best properties for correcting the uECG recordings. It is possible to tune the models by selecting different filter lengths (FIR) or model orders. We assume that the discrepancy between aVL and the other leads was probably due to an unknown acquisition error. Several sources of acquisition disturbances exist, the prominent are movement and high-frequency noise [
25]. We assume that our problems were due to insufficient electrode contact to the skin, leading to more movement artifacts or baseline wander. Furthermore, the setting for the study was in an office space with computers and electronic devices that lead to high-frequency noise.
For the SSM and the ARX, in most experiments, the correlation coefficient did not indicate a better agreement with the reference ECG than with the uECG. Furthermore, a few of the estimated models were unstable for the SSM model (aVR subject 4, aVL subject 3, aVF subject 3) and the ARX model (aVR subject 1, 5; aVL subject 3, aVF subject 3). For future applications, each model needs to be checked for stability before it is used for correction. The values of the cross-spectrum indicated a slightly better agreement with the reference ECG for aVL and a much higher agreement with aVF than the uECG. In the analysis of the ECG waves, the flattening of the R-peak was corrected by both models. The MSE of the other waves indicated inconsistent results that showed no significant improvement compared with the uECG.
In future studies, additional issues of unobtrusive ECGs that are related to artifacts (based on motion or electric discharge) also need to be addressed. However, automatic correction of artifacts was not an aim of the present study; artifacts were selected and rejected manually. Meanwhile, different approaches have been proposed to remove and correct for ECG artifacts [
26,
27,
28,
29].
In the future, medical professionals should visually verify whether the corrected uECG can be used for purposes of diagnosis. Additional features of interest include: the PQ length, QRS width, ST length, and the P and T length. These features are not readily available and require more sophisticated computing or manual extraction by a medical professional. Although segmentation algorithms for ECG exist, they are reported to fail for, e.g., the capacitive ECG [
30].
A short calibration procedure is necessary for the correction using the models. A recording procedure with the uECG is proposed as follows:
- (1)
Simultaneous recording of the uECG and the reference ECG for 30 s.
- (2)
Automated data processing:
- (a)
Alignment of the uECG and the reference ECG channel data.
- (b)
Performance of system identification to obtain models.
- (c)
Choice of best model for uECG correction.
- (3)
Removal of the reference ECG.
- (4)
Long-term uECG recording using the obtained model for correction.
The short calibration phase (steps 1–3) is added to the time needed to apply a standard ECG. In step 4, either a real-time correction can be implemented in the ECG software, or an offline correction is applied afterwards. The need for calibration for each subject or even each acquisition is known in clinical practice, i.e., in EEG acquisition. Artifacts from eye movements are removed by different methods and most require calibration before each EEG recording [
31]. Instead of calibration, more general models might be found, when a larger study group is available. However, individualized filters have the advantage that they correct more precisely for the given ECG setup, as demonstrated in the present leave-one-out cross-validation.