Empowering Kiwifruit Cultivation with AI: Leaf Disease Recognition Using AgriVision-Kiwi Open Dataset
Abstract
1. Introduction
- A novel open image dataset for kiwifruit disease recognition and kiwifruit leaf detection is introduced. With the contribution of two different annotations, the proposed dataset can be used for a variety of applications, beyond disease recognition.
- The dataset contains leaf images for the detection of Nematodes disease, which have not been previously available, as far as the authors’ knowledge.
- The identification of Nematodes disease in kiwifruit plants is achieved using leaf images for the first time. By utilizing non-invasive techniques, Nematodes disease has been studied through laboratory images, e.g., hyperspectral or microscopic images of roots, and not directly on the kiwifruit leaves. The proposed approach aims to investigate whether there are early visual symptoms of Nematodes on the leaves, as well as to identify, among several DL models, the one that could better detect it.
- The proposed dataset is evaluated by using six deep classification models of varying architectures.
2. Related Works
3. Materials and Methods
3.1. Dataset Presentation
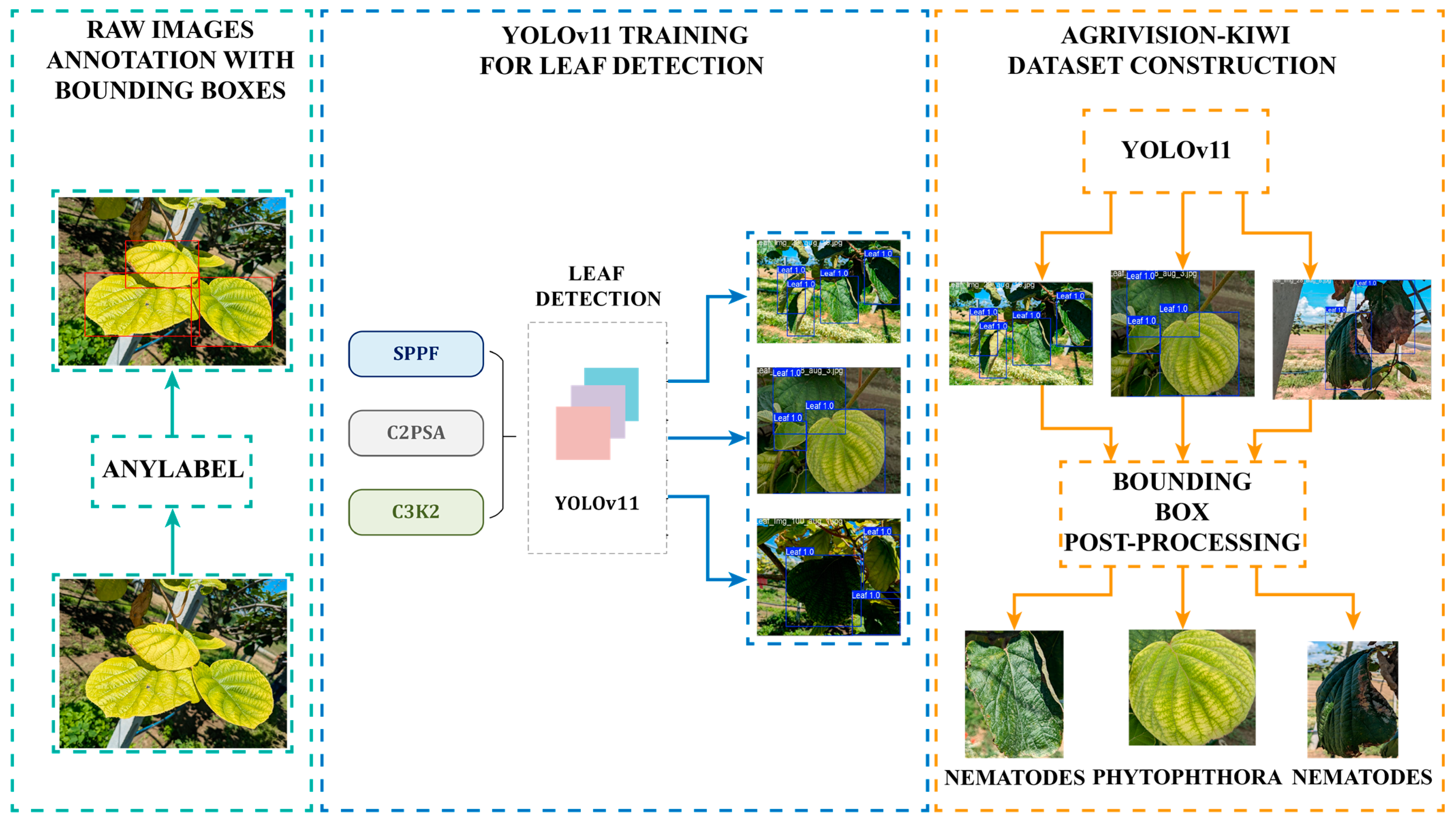
3.2. Leaf Detection and Disease Recognition Using YOLOv11
3.3. Leaf Detection
- The kiwi leaf must appear fully within the ROI. Kiwifruit leaves have a characteristic shape that can be affected by certain diseases, like Nematodes. This rule ensures that the selected samples are appropriate for distinguishing the diseases.
- The kiwifruit leaf must be large enough to provide sufficient information about the disease. The term “large enough” is defined based on the contour area of the largest leaf in the image.
3.4. Disease Recognition
- Alexnet was proposed by Alex Krizhevsky, et al. in 2012 [31]. It is one of the most influential DL models in the CV domain. It was the first deep CNN model that demonstrated the power of DL. This model’s performance could provide a strong verification that the targeted classes are distinguishable from the image samples.
- Densenet-121 was proposed by Gao Huang et al. in 2017 [32] and it is known for its dense connectivity pattern, where each layer is directly connected to every other subsequent layer. This design helps in feature reuse, alleviating the vanishing gradient problem and reducing the number of parameters in comparison to other similar deep models. This model achieved top results in the ImageNet competition.
- Efficientnet-B3 was proposed by Mingxing Tan and Quoc Le in 2019 [33] and it is one of the Efficientnet family models. Its novelty is a compound scaling method that helps the model to be considered state-of-the-art with an outstanding balance between performance and efficiency. This model focuses on the trade-off between efficiency and performance, making it a good candidate for edge device integration, i.e. for smartphones.
- MobileNet-V3 was proposed by Andrew Howard et al. in 2019 [34]. Its strong characteristic is its trade-off between performance and efficiency for resource-constrained devices, achieving real-time performance. This model is designed for edge devices like smartphones, which makes it suitable for the proposed application.
- Resnet-50 was proposed by Kaiming He et al. in 2016 [35] and is a part of the groundbreaking Resnet family models. Its novelty is in the introduced residual connections to tackle the gradient vanishing problem. This model was selected since it is considered the ground base of DL models for CV applications. Additionally, it is one of the most reusable models in the CV domain due to its robustness and versatility.
- VGG-16 was proposed by Karen Simonyan and Andrew Zisserman in 2015 [36]. Its simplified architecture significantly improves its performance compared to earlier models like AlexNet. It is considered a benchmark architecture for image classification and is widely adopted in industry and research.
3.5. Experimental Setup and Evaluation Metrics
4. Results
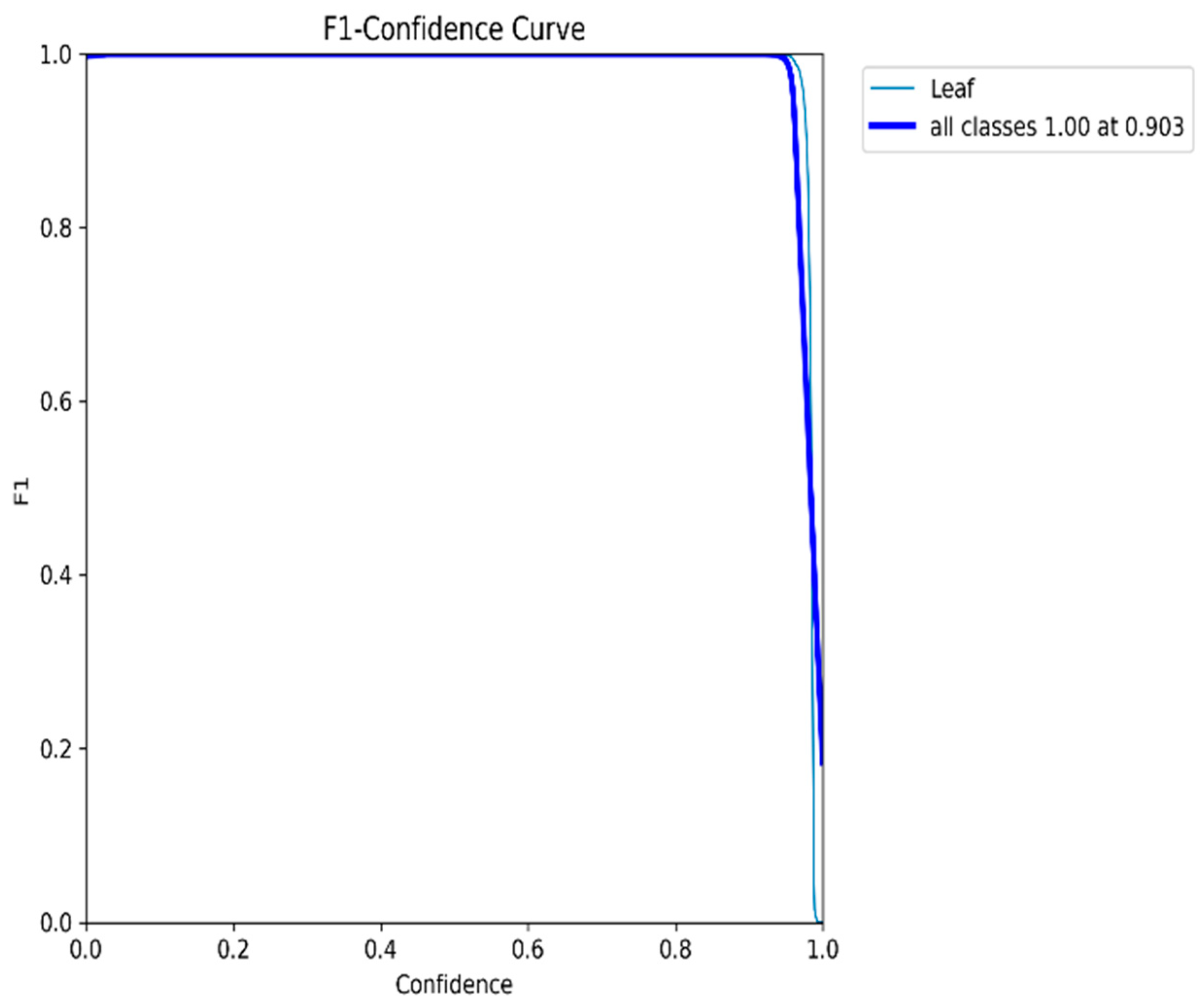
4.1. Leaf Detection Results
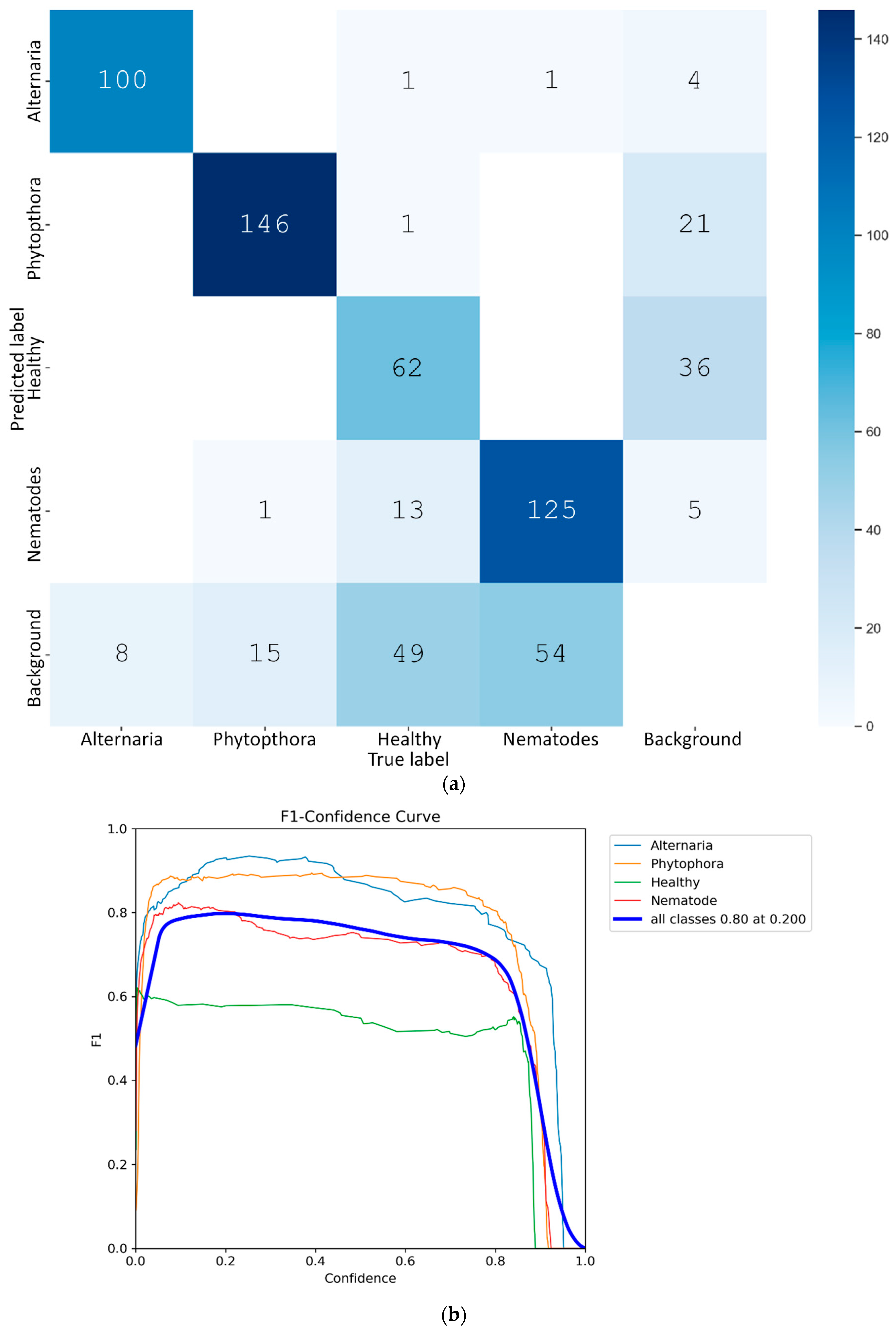
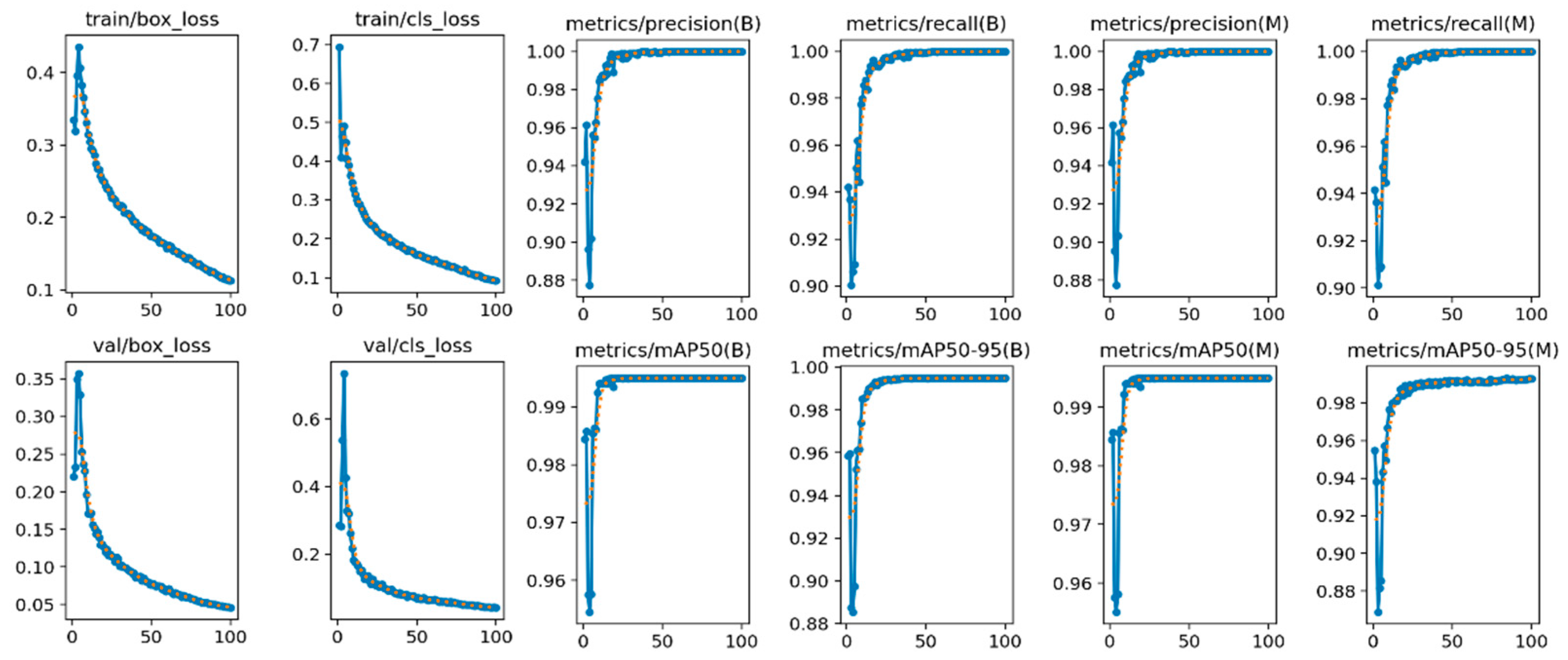
- The box loss (first column subfigures of Figure 6) indicates the performance of the model localizing the kiwi leaves with strict bounding boxes that contain the most significant features. In our case, the box loss during training continually decreases throughout the epochs, reaching 0.053 at the final epoch.
- The cls loss (second column subfigures of Figure 6) indicates the performance of the model in classifying the detected bounding boxes into target classes, of which, in this specific application, there are two: leaf and no leaf. In our case, the cls loss during training also decreases throughout the epochs, reaching 0.060 at the final epoch.
- Regarding the performance metrics of precision and recall, the reported results are high, reaching 0.995 and 0.990, respectively, at the final epoch (3–6 column subfigures of Figure 6). The symbol (B) in the subfigures (columns 3 and 4) of Figure 6 refers to the best performance by the model, while (M) (columns 5 and 6) represents the average macro, which calculates the average performance across all classes.
4.2. Leaf Disease Recognition Results
5. Discussion
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ciacci, C.; Russo, I.; Bucci, C.; Iovino, P.; Pellegrini, L.; Giangrieco, I.; Tamburrini, M.; Ciardiello, M.A. The Kiwi Fruit Peptide Kissper Displays Anti-Inflammatory and Anti-Oxidant Effects in in-Vitro and Ex-Vivo Human Intestinal Models. Clin. Exp. Immunol. 2014, 175, 476–484. [Google Scholar] [CrossRef]
- Richardson, D.P.; Ansell, J.; Drummond, L.N. The Nutritional and Health Attributes of Kiwifruit: A Review. Eur. J. Nutr. 2018, 57, 2659–2676. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Tang, J.; Zou, Y.; Sun, X.; Lan, J.; Wang, W.; Xu, P.; Wu, X.; Ma, R.; Wang, Q.; et al. Whole Genome Sequence of Alternaria Alternata, the Causal Agent of Black Spot of Kiwifruit. Front. Microbiol. 2021, 12, 713462. [Google Scholar] [CrossRef] [PubMed]
- Bi, X.; Hieno, A.; Otsubo, K.; Kageyama, K.; Liu, G.; Li, M. A Multiplex PCR Assay for Three Pathogenic Phytophthora Species Related to Kiwifruit Diseases in China. J. Gen. Plant Pathol. 2019, 85, 12–22. [Google Scholar] [CrossRef]
- Banihashemian, S.N.; Jamali, S.; Golmohammadi, M.; Ghasemnezhad, M. Isolation and Identification of Endophytic Bacteria Associated with Kiwifruit and Their Biocontrol Potential against Meloidogyne Incognita. Egypt. J. Biol. Pest Control 2022, 32, 111. [Google Scholar] [CrossRef]
- Hosny, K.M.; El-Hady, W.M.; Samy, F.M.; Vrochidou, E.; Papakostas, G.A. Multi-Class Classification of Plant Leaf Diseases Using Feature Fusion of Deep Convolutional Neural Network and Local Binary Pattern. IEEE Access 2023, 11, 62307–62317. [Google Scholar] [CrossRef]
- Yao, J.; Wang, Y.; Xiang, Y.; Yang, J.; Zhu, Y.; Li, X.; Li, S.; Zhang, J.; Gong, G. Two-Stage Detection Algorithm for Kiwifruit Leaf Diseases Based on Deep Learning. Plants 2022, 11, 768. [Google Scholar] [CrossRef]
- Liu, J.; Wang, X. Plant Diseases and Pests Detection Based on Deep Learning: A Review. Plant Methods 2021, 17, 22. [Google Scholar] [CrossRef]
- Kalampokas, T.; Vrochidou, E.; Tzampazaki, M.; Papakostas, G.A. Unraveling the Potential of AI Towards Digital and Green Transformation in Kiwifruit Farming. In Proceedings of the 2024 International Conference on Software, Telecommunications and Computer Networks (SoftCOM), Split, Croatia, 26 September 2024; pp. 1–6. [Google Scholar]
- El-Borai, F.E.; Duncan, L.W. Nematode Parasites of Subtropical and Tropical Fruit Tree Crops. In Plant Parasitic Nematodes in Subtropical and Tropical Agriculture; CABI Publishing: Wallingford, UK, 2005; pp. 467–492. [Google Scholar]
- Tao, Y.; Xu, C.; Yuan, C.; Wang, H.; Lin, B.; Zhuo, K.; Liao, J. Meloidogyne Aberrans Sp. Nov. (Nematoda: Meloidogynidae), a New Root-Knot Nematode Parasitizing Kiwifruit in China. PLoS ONE 2017, 12, e0182627. [Google Scholar] [CrossRef]
- AYDINLI, G.; MENNAN, S. Samsun İli (Türkiye) Kivi Bahçelerindeki Kök-Ur Nematodlarının Yaygınlığı ve Meyve Verimine Etkileri. Turk. J. Entomol. 2022, 46, 187–197. [Google Scholar] [CrossRef]
- Gonçalves, A.R.; Conceição, I.L.; Carvalho, R.P.; Costa, S.R. Meloidogyne Hapla Dominates Plant-parasitic Nematode Communities Associated with Kiwifruit Orchards in Portugal. Plant Pathol. 2025, 74, 171–179. [Google Scholar] [CrossRef]
- Khanam, R.; Hussain, M. YOLOv11: An Overview of the Key Architectural Enhancements. arXiv 2024, arXiv:2410.17725. [Google Scholar]
- Banerjee, D.; Kukreja, V.; Hariharan, S.; Sharma, V. Fast and Accurate Multi-Classification of Kiwi Fruit Disease in Leaves Using Deep Learning Approach. In Proceedings of the 2023 International Conference on Innovative Data Communication Technologies and Application (ICIDCA), Uttarakhand, India, 14 March 2023; pp. 131–137. [Google Scholar]
- Bansal, A.; Sharma, R.; Sharma, V.; Jain, A.K.; Kukreja, V. An Automated Approach for Accurate Detection and Classification of Kiwi Powdery Mildew Disease. In Proceedings of the 2023 IEEE 8th International Conference for Convergence in Technology (I2CT), Lonavla, India, 7 April 2023; pp. 1–4. [Google Scholar]
- Mir, T.A.; Gupta, S.; Aggarwal, P.; Pokhariya, H.S.; Banerjee, D.; Lande, A. Multiclass Kiwi Leaf Disease Detection Using CNN-SVM Fusion. In Proceedings of the 2024 IEEE International Conference on Information Technology, Electronics and Intelligent Communication Systems (ICITEICS), Bangalore, India, 28 June 2024; pp. 1–6. [Google Scholar]
- Liu, B.; Ding, Z.; Zhang, Y.; He, D.; He, J. Kiwifruit Leaf Disease Identification Using Improved Deep Convolutional Neural Networks. In Proceedings of the 2020 IEEE 44th Annual Computers, Software, and Applications Conference (COMPSAC), Madrid, Spain, 13–17 July 2020; pp. 1267–1272. [Google Scholar]
- Banerjee, D.; Kukreja, V.; Chamoli, S.; Thapliyal, S. Optimizing Kiwi Fruit Plant Leaves Disease Recognition: A CNN-Random Forest Integration. In Proceedings of the 2023 3rd International Conference on Smart Generation Computing, Communication and Networking (SMART GENCON), Bangalore, India, 29–31 December 2023; pp. 1–6. [Google Scholar]
- Li, X.; Chen, X.; Yang, J.; Li, S. Transformer Helps Identify Kiwifruit Diseases in Complex Natural Environments. Comput. Electron. Agric. 2022, 200, 107258. [Google Scholar] [CrossRef]
- Wu, J.; Zheng, H.; Zhao, B.; Li, Y.; Yan, B.; Liang, R.; Wang, W.; Zhou, S.; Lin, G.; Fu, Y.; et al. AI Challenger: A Large-Scale Dataset for Going Deeper in Image Understanding. arXiv 2017, arXiv:1711.06475. [Google Scholar] [CrossRef]
- Hughes, D.P.; Salathe, M. An Open Access Repository of Images on Plant Health to Enable the Development of Mobile Disease Diagnostics. arXiv 2015, arXiv:1511.08060. [Google Scholar]
- Saleem, M.H.; Potgieter, J.; Arif, K.M. A Performance-Optimized Deep Learning-Based Plant Disease Detection Approach for Horticultural Crops of New Zealand. IEEE Access 2022, 10, 89798–89822. [Google Scholar] [CrossRef]
- Senanu Ametefe, D.; Seroja Sarnin, S.; Mohd Ali, D.; Caliskan, A.; Tatar Caliskan, I.; Adozuka Aliu, A.; John, D. Enhancing Leaf Disease Detection Accuracy through Synergistic Integration of Deep Transfer Learning and Multimodal Techniques. Inf. Process. Agric. 2024, in press. [CrossRef]
- Wu, K.; Jia, Z.; Duan, Q. The Detection of Kiwifruit Sunscald Using Spectral Reflectance Data Combined with Machine Learning and CNNs. Agronomy 2023, 13, 2137. [Google Scholar] [CrossRef]
- Sekhukhune, M.K.; Shokoohi, E.; Maila, M.Y.; Mashela, P.W. Free-Living Nematodes Associated with Kiwifruit and Effect of Soil Chemical Properties on Their Diversity. Biologia 2022, 77, 435–444. [Google Scholar] [CrossRef]
- Kumar, S.; Rawat, S.; Joshi, V.; Kundu, A.; Somvanshi, V.S. Northern Root-Knot Nematode Meloidogyne Hapla Chitwood, 1949 Infecting Kiwi Fruit (Actinidia Chinensis) in Bageshwar District of Uttarakhand State. Indian Phytopathol. 2023, 76, 665–667. [Google Scholar] [CrossRef]
- Kang, H.; Je, H.; Choi, I. Occurrence and Distribution of Root-Knot Nematodes in Kiwifruit Orchard. Res. Plant Dis. 2023, 29, 45–51. [Google Scholar] [CrossRef]
- Neural Research Lab. AnyLabeling. Available online: https://anylabeling.nrl.ai/ (accessed on 27 January 2024).
- Liu, Z.; Wei, L.; Song, T. Optimized YOLOv11 Model for Lung Nodule Detection. Biomed. Signal Process. Control 2025, 107, 107830. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. arXiv 2016, arXiv:1608.06993. [Google Scholar]
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the 36th International Conference on Machine Learning, ICML 2019, Long Beach, CA, USA, 9–15 June 2019. [Google Scholar]
- Howard, A.; Sandler, M.; Chen, B.; Wang, W.; Chen, L.-C.; Tan, M.; Chu, G.; Vasudevan, V.; Zhu, Y.; Pang, R.; et al. Searching for MobileNetV3. In Proceedings of the 2019 IEEE/CVF International Conference on Computer Vision (ICCV), Paris, France, 6 October 2019; pp. 1314–1324. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015—Conference Track Proceedings, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015—Conference Track Proceedings, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]

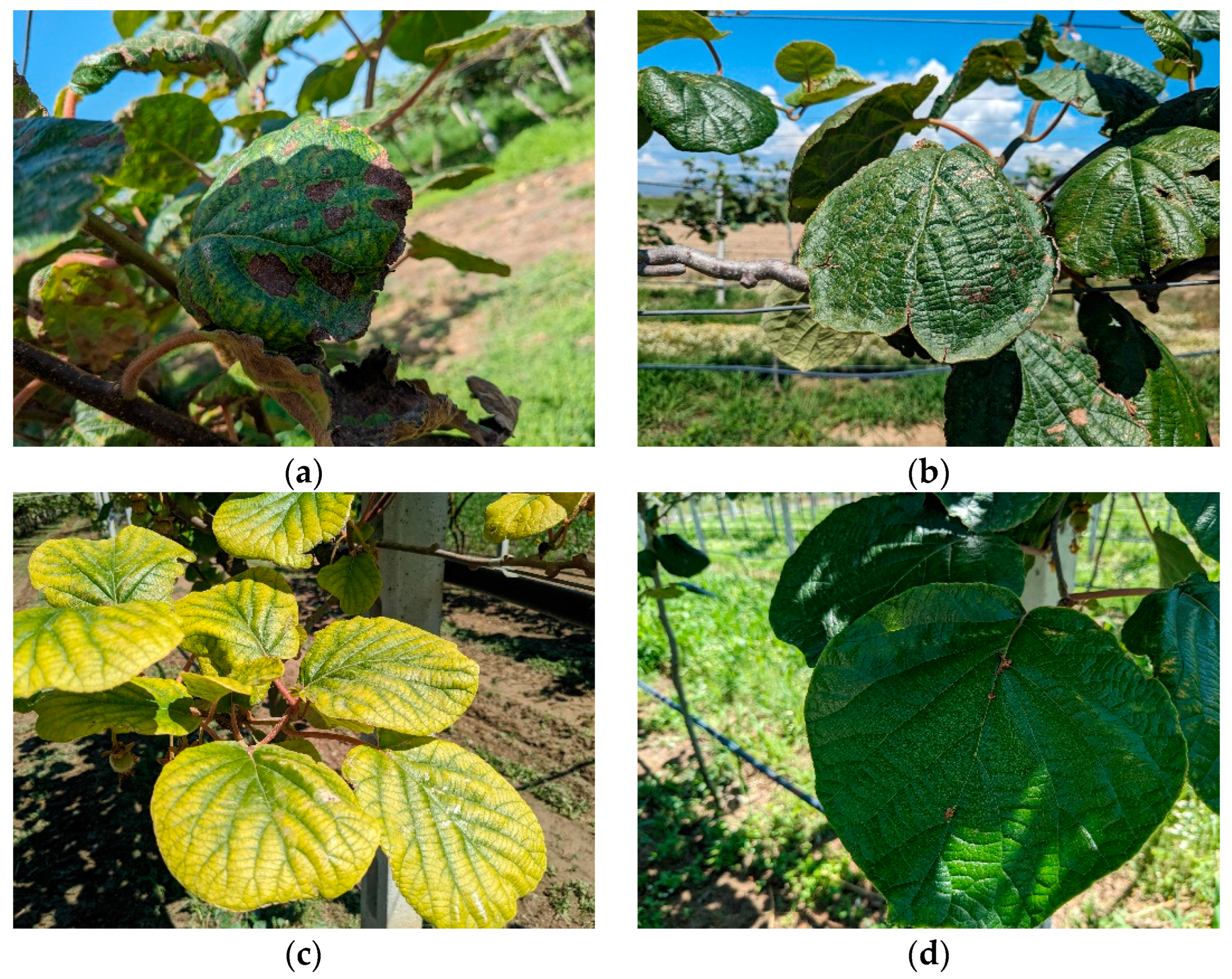
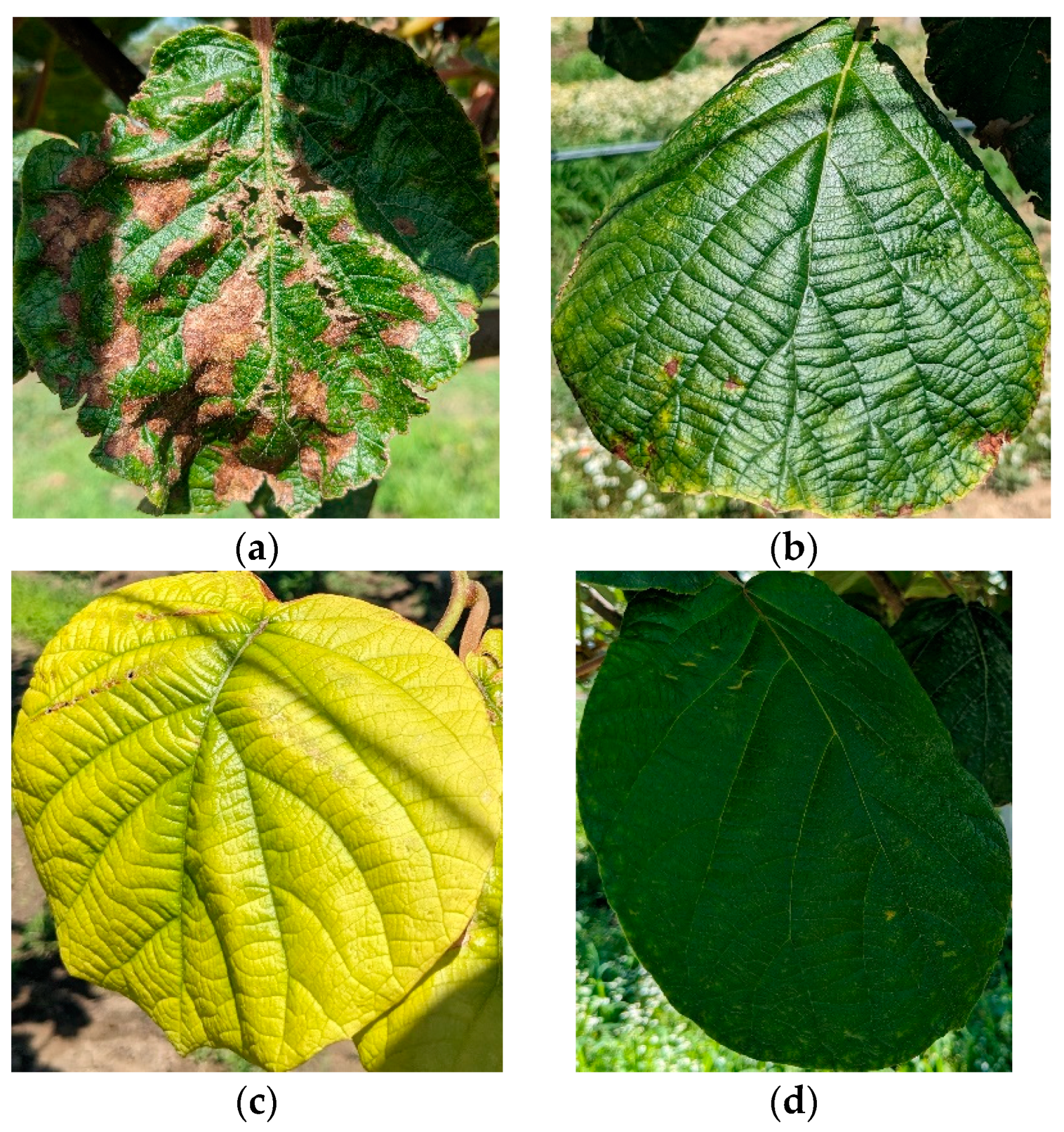





| Characteristics | Related Works | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| [7] | [15] | [16] | [17] | [18] | [19] | [20] | [23] | [24] | [25] | |||
| Disease | Brown Spot | ✓ | ✓ | - | - | ✓ | - | ✓ | - | ✓ | - | |
| Bacterial Canker | ✓ | ✓ | - | - | - | - | - | ✓ | - | - | ||
| Anthracnose | - | ✓ | - | - | ✓ | - | ✓ | - | ✓ | - | ||
| Mosaic | - | ✓ | - | - | ✓ | - | ✓ | - | ✓ | - | ||
| Powdery Mildew | - | - | ✓ | - | - | - | - | - | - | - | ||
| Rust | - | - | - | ✓ | - | - | - | - | - | - | ||
| Botrytis | - | - | - | ✓ | - | ✓ | - | - | - | - | ||
| Phomopsis | - | - | - | ✓ | - | - | - | - | - | - | ||
| PSA | - | - | - | ✓ | - | - | - | - | - | - | ||
| Armillaria Root Rot | - | - | - | - | - | ✓ | - | - | - | - | ||
| Bacterial Bligh | - | - | - | - | - | ✓ | - | - | - | - | ||
| Bleeding Canker | - | - | - | - | - | ✓ | - | - | - | - | ||
| Phytophthora Root | - | - | - | - | - | ✓ | - | - | - | - | ||
| Water Staining | - | - | - | - | - | ✓ | - | - | - | - | ||
| Juice Blotch | - | - | - | - | - | ✓ | - | - | - | - | ||
| Sooty Mold of Fruit | - | - | - | - | - | ✓ | - | - | - | - | ||
| Collar Rot | - | - | - | - | - | ✓ | - | - | - | - | ||
| Crown Rot | - | - | - | - | - | ✓ | - | - | - | - | ||
| Black Spot | - | - | - | - | - | - | ✓ | - | - | - | ||
| Yellow Leaf | - | - | - | - | - | - | ✓ | - | - | - | ||
| Ulcer | - | - | - | - | - | - | ✓ | ✓ | - | - | ||
| Sunscald | - | - | - | - | - | - | - | - | - | ✓ | ||
| Task | Leaf detection | Model | YOLOX | - | - | - | - | - | - | RFCN | - | - |
| Results | <na> | - | - | - | - | - | - | 0.938 prec. | - | - | ||
| Disease recognition | Model | DeepLabV3+ | CNN + SVM | CNN + LSTM | CNN + SVM | CNN | CNN + RF | ConvViT-B | RFCN | Densenet-201 | MS-CNN | |
| Results | 0.966 acc. | 0.8348 acc. | 0.9570 prec. | 0.9568 acc. | 0.9854 acc. | 0.8564 acc. | 0.9986 acc. | 0.9380 prec. | 0.9989 acc | 0.99 acc. | ||
| Data | Public dataset | No | No | No | No | No | No | No | No | No | No | |
| Task | Raw Dataset | Augmented Raw Dataset | Targets |
|---|---|---|---|
| Leaf Object Detection | 152 | 5832 | 2 Classes (Leaf, Background) |
| Task | Raw Dataset | AgriVision-Kiwi Dataset | Augmented AgriVision-Kiwi Dataset | Targets |
|---|---|---|---|---|
| Leaf Disease Recognition | 57 (Phytophthora), 24 (Healthy), 43 (Nematodes), 28 (Alternaria) | 108 (Phytophthora), 51 (Healthy), 70 (Nematodes), 33 (Alternaria) | 1680 (Phytophthora), 1588 (Healthy), 1588 (Nematodes), 1591 (Alternaria) | 4 Classes (Phytophthora, Healthy, Nematodes, Alternaria) |
| Training Loss | Validation Loss | Accuracy | Precision | Recall |
|---|---|---|---|---|
| 0.1123 | 0.134 | 0.67 | 0.83 | 0.86 |
| Model Name | Year | Layers | Layers |
|---|---|---|---|
| AlexNet | 2012 | 8 | First deep CNN to win ImageNet |
| DenseNet-121 | 2017 | 121 | Efficient depth with fewer parameters |
| EfficientNet-B3 | 2019 | 32 | Best trade-off between accuracy and efficiency |
| MobileNet-V3 | 2019 | 16 | Suitable architecture for mobile and edge devices |
| ResNet-50 | 2015 | 50 | Enabled very deep networks, first for dealing vanishing gradient problem |
| VGG-16 | 2014 | 16 | Simplicity of design and improved accuracy by increasing depth |
| Task | Library—Framework | Details |
|---|---|---|
| Image Processing | OpenCV, Pillow | Data handling, image pre-processing |
| Image Augmentation | Imgaug | Image augmentation |
| Leaves Object Detection | Ultralytics | YOLOv11 training and inference |
| Leaves Disease Recognition | Pytorch | Deep learning models training and inference for image classification |
| Metric | YOLOv11 |
|---|---|
| Train-box-loss | 0.053 |
| Train-cls-loss | 0.060 |
| Val-box-loss | 0.043 |
| Val-cls-loss | 0.041 |
| Precision(B) | 0.99 |
| Recall(B) | 0.99 |
| Precision(M) | 0.99 |
| Recall(M) | 0.99 |
| F1 score | 0.99 |
| Confidence score | 0.99 |
| Model | AlexNet | DenseNet-121 | EfficientNet-B3 | MobileNet-V3 | ResNet-50 | VGG-16 |
|---|---|---|---|---|---|---|
| Fold 1 | 0.069 | 0.063 | 0.088 | 0.075 | 0.064 | 0.073 |
| Fold 2 | 0.072 | 0.065 | 0.086 | 0.070 | 0.062 | 0.060 |
| Fold 3 | 0.078 | 0.060 | 0.087 | 0.069 | 0.063 | 0.058 |
| Fold 4 | 0.072 | 0.063 | 0.086 | 0.076 | 0.064 | 0.075 |
| Fold 5 | 0.072 | 0.065 | 0.086 | 0.071 | 0.064 | 0.075 |
| Fold 6 | 0.074 | 0.061 | 0.087 | 0.066 | 0.062 | 0.055 |
| Fold 7 | 0.070 | 0.064 | 0.085 | 0.074 | 0.061 | 0.070 |
| Fold 8 | 0.072 | 0.068 | 0.088 | 0.070 | 0.064 | 0.081 |
| Fold 9 | 0.072 | 0.069 | 0.083 | 0.070 | 0.061 | 0.070 |
| Fold 10 | 0.075 | 0.064 | 0.085 | 0.069 | 0.062 | 0.071 |
| Mean | 0.073 | 0.064 | 0.086 | 0.071 | 0.063 | 0.069 |
| Model | AlexNet | DenseNet-121 | EfficientNet-B3 | MobileNet-V3 | ResNet-50 | VGG-16 |
|---|---|---|---|---|---|---|
| Fold 1 | 0.989 | 0.996 | 0.981 | 0.982 | 0.989 | 0.984 |
| Fold 2 | 0.989 | 0.995 | 0.987 | 0.987 | 0.992 | 0.990 |
| Fold 3 | 0.990 | 0.989 | 0.990 | 0.984 | 0.986 | 0.984 |
| Fold 4 | 0.982 | 0.995 | 0.976 | 0.989 | 0.993 | 0.995 |
| Fold 5 | 0.982 | 0.993 | 0.981 | 0.981 | 0.981 | 0.986 |
| Fold 6 | 0.993 | 0.995 | 0.982 | 0.993 | 0.987 | 0.986 |
| Fold 7 | 0.984 | 0.992 | 0.981 | 0.981 | 0.990 | 0.987 |
| Fold 8 | 0.982 | 0.990 | 0.984 | 0.987 | 0.987 | 0.978 |
| Fold 9 | 0.990 | 0.993 | 0.986 | 0.989 | 0.990 | 0.982 |
| Fold 10 | 0.979 | 0.995 | 0.979 | 0.992 | 0.989 | 0.973 |
| Mean | 0.986 | 0.993 | 0.983 | 0.986 | 0.989 | 0.984 |
| Model | AlexNet | DenseNet-121 | EfficientNet-B3 | MobileNet-V3 | ResNet-50 | VGG-16 |
|---|---|---|---|---|---|---|
| Fold 1 | 0.986 | 0.988 | 0.988 | 0.989 | 0.987 | 0.986 |
| Fold 2 | 0.986 | 0.987 | 0.988 | 0.989 | 0.987 | 0.987 |
| Fold 3 | 0.986 | 0.987 | 0.987 | 0.985 | 0.987 | 0.987 |
| Fold 4 | 0.986 | 0.987 | 0.987 | 0.985 | 0.987 | 0.987 |
| Fold 5 | 0.986 | 0.987 | 0.987 | 0.985 | 0.987 | 0.987 |
| Fold 6 | 0.986 | 0.987 | 0.987 | 0.985 | 0.987 | 0.987 |
| Fold 7 | 0.986 | 0.987 | 0.987 | 0.985 | 0.987 | 0.987 |
| Fold 8 | 0.986 | 0.987 | 0.986 | 0.986 | 0.987 | 0.987 |
| Fold 9 | 0.986 | 0.988 | 0.985 | 0.985 | 0.987 | 0.987 |
| Fold 10 | 0.986 | 0.988 | 0.986 | 0.986 | 0.987 | 0.987 |
| Mean | 0.986 | 0.987 | 0.987 | 0.986 | 0.987 | 0.987 |
| Model | AlexNet | DenseNet-121 | EfficientNet-B3 | MobileNet-V3 | ResNet-50 | VGG-16 |
|---|---|---|---|---|---|---|
| Fold 1 | 0.986 | 0.988 | 0.988 | 0.984 | 0.988 | 0.987 |
| Fold 2 | 0.986 | 0.987 | 0.987 | 0.984 | 0.987 | 0.987 |
| Fold 3 | 0.986 | 0.987 | 0.987 | 0.985 | 0.987 | 0.987 |
| Fold 4 | 0.986 | 0.987 | 0.987 | 0.985 | 0.987 | 0.987 |
| Fold 5 | 0.986 | 0.987 | 0.987 | 0.985 | 0.987 | 0.987 |
| Fold 6 | 0.986 | 0.987 | 0.987 | 0.985 | 0.987 | 0.987 |
| Fold 7 | 0.986 | 0.987 | 0.987 | 0.986 | 0.987 | 0.987 |
| Fold 8 | 0.986 | 0.987 | 0.987 | 0.985 | 0.987 | 0.987 |
| Fold 9 | 0.986 | 0.987 | 0.987 | 0.985 | 0.987 | 0.987 |
| Fold 10 | 0.986 | 0.988 | 0.987 | 0.985 | 0.987 | 0.987 |
| Mean | 0.986 | 0.987 | 0.987 | 0.985 | 0.987 | 0.987 |
| Model | AlexNet | DenseNet-121 | EfficientNet-B3 | MobileNet-V3 | ResNet-50 | VGG-16 |
|---|---|---|---|---|---|---|
| Time | 192 ms | 676 ms | 602 ms | 496 ms | 600 ms | 579 ms |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalampokas, T.; Vrochidou, E.; Mavridou, E.; Iliadis, L.; Voglitsis, D.; Michalopoulou, M.; Broufas, G.; Papakostas, G.A. Empowering Kiwifruit Cultivation with AI: Leaf Disease Recognition Using AgriVision-Kiwi Open Dataset. Electronics 2025, 14, 1705. https://doi.org/10.3390/electronics14091705
Kalampokas T, Vrochidou E, Mavridou E, Iliadis L, Voglitsis D, Michalopoulou M, Broufas G, Papakostas GA. Empowering Kiwifruit Cultivation with AI: Leaf Disease Recognition Using AgriVision-Kiwi Open Dataset. Electronics. 2025; 14(9):1705. https://doi.org/10.3390/electronics14091705
Chicago/Turabian StyleKalampokas, Theofanis, Eleni Vrochidou, Efthimia Mavridou, Lazaros Iliadis, Dionisis Voglitsis, Maria Michalopoulou, George Broufas, and George A. Papakostas. 2025. "Empowering Kiwifruit Cultivation with AI: Leaf Disease Recognition Using AgriVision-Kiwi Open Dataset" Electronics 14, no. 9: 1705. https://doi.org/10.3390/electronics14091705
APA StyleKalampokas, T., Vrochidou, E., Mavridou, E., Iliadis, L., Voglitsis, D., Michalopoulou, M., Broufas, G., & Papakostas, G. A. (2025). Empowering Kiwifruit Cultivation with AI: Leaf Disease Recognition Using AgriVision-Kiwi Open Dataset. Electronics, 14(9), 1705. https://doi.org/10.3390/electronics14091705












