An Explainable Deep Learning Framework with Adaptive Feature Selection for Smart Lemon Disease Classification in Agriculture
Abstract
1. Introduction
1.1. Research Motivation
1.2. Feature Selection Using Nature-Inspired Algorithms
1.3. Importance and Contributions
- Development of the adaptive PSO-LemonNetX model, which integrates DL with adaptive PSO-based feature selection, grid search hyperparameter tuning, and lightweight architectural components for effective and efficient classification of lemon diseases.
- Integration of XAI LIME to enhance the transparency of the model’s predictions, providing actionable insights for agricultural stakeholders and researchers.
1.4. Paper Organization
2. Related Work
2.1. ML Appraoches
2.2. DL and TL Approaches
| Study | Method/Model | Dataset | Classes | Advantages | Limitations |
|---|---|---|---|---|---|
| Sahu et al. (2022) [19] | K-means segmentation + GLCM features + SVM | Lemon leaf dataset | Diseased vs. healthy | Simple ML pipeline, low computational cost | Requires manual segmentation, less scalable to large datasets |
| Hanh et al. (2023) [2] | Yolov4 + image processing with conveyor belt | Rotating lemon dataset | Quality categories | Enables real-time inspection, covers entire fruit surface | Limited generalization to unseen environments, high setup cost |
| Hernandez et al. (2021) [12] | 8 CNN models (color vs. grayscale) | 913 Mexican lemons | Healthy vs. unhealthy | Compared multiple CNNs, achieved high separation accuracy | Small dataset, limited class diversity |
| Sharma et al. (2022) [11] | CNN + LSTM (CLTN hybrid) | 3000 lemon citrus canker images | Four disease levels | Captures spatial and temporal features, effective for severity levels | Computationally intensive, requires large annotated dataset |
| Pourdarbani et al. (2023) [20] | Hyperspectral imaging + 3D-CNN (ResNet, DenseNet, etc.) | Bruised lemon fruits | Bruised vs. normal | Early detection of bruising, considers spatial-frequency info. | Needs hyperspectral imaging, expensive and less practical in field |
| Kaushik et al. (2025) [21] | DenseNet121 with preprocessing and augmentation | 2076 lemon images | Good vs. bad quality | Robust preprocessing pipeline, high classification reliability | High model complexity, limited interpretability |
| Pramanik et al. (2021) [22] | TL models (ResNet50, DenseNet201, Xception, ResNet152V2) | Field-level lemon leaves | Multiple diseases | Low-cost TL-based training, strong generalization | Limited explainability, performance depends on pretrained model |
| He et al. (2021) [23] | TL (VGG16) + visual feature extraction | Lemon fruit dataset | Green vs. mold defect | Combined TL with handcrafted features, improved accuracy over ML | Dataset-specific preprocessing, limited scalability |
2.3. Addressing Gaps in Existing Lemon Disease Classification Methods
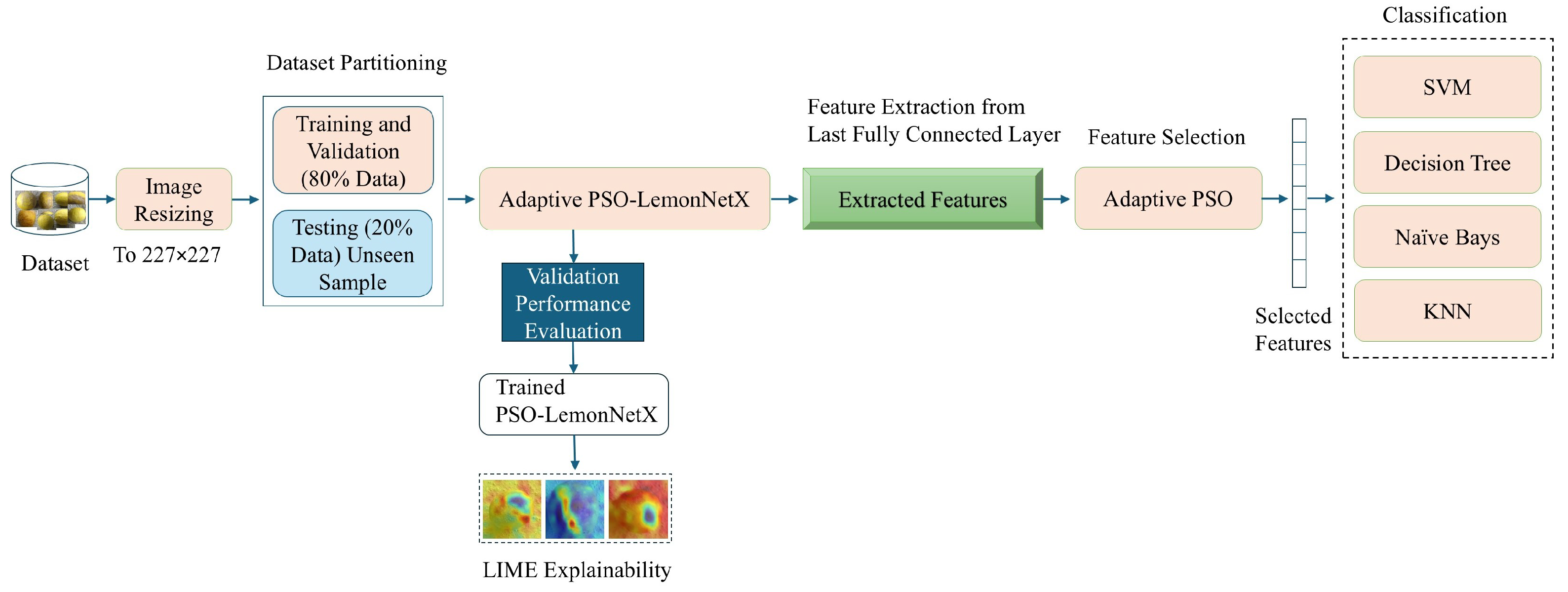
3. Methodology
3.1. Image Resizing
3.2. Dataset Partitioning
3.3. Adaptive PSO-LemonNetX Architecture Details
Motivation Behind Adaptive PSO-LemonNetX Architecture
3.4. Particle Swarm Optimization
Advantages of Using PSO for Feature Selection
- PSO efficiently explores the search space while convergently approaching potential solutions by combining global exploration and local exploitation through particle interaction.
- It supports parallel computation, enabling scalability to high-dimensional data.
- It does not require gradient information, making it suitable for noisy or complex fitness landscapes.
3.5. Classification Methods
3.6. Hyperparameters Optimization
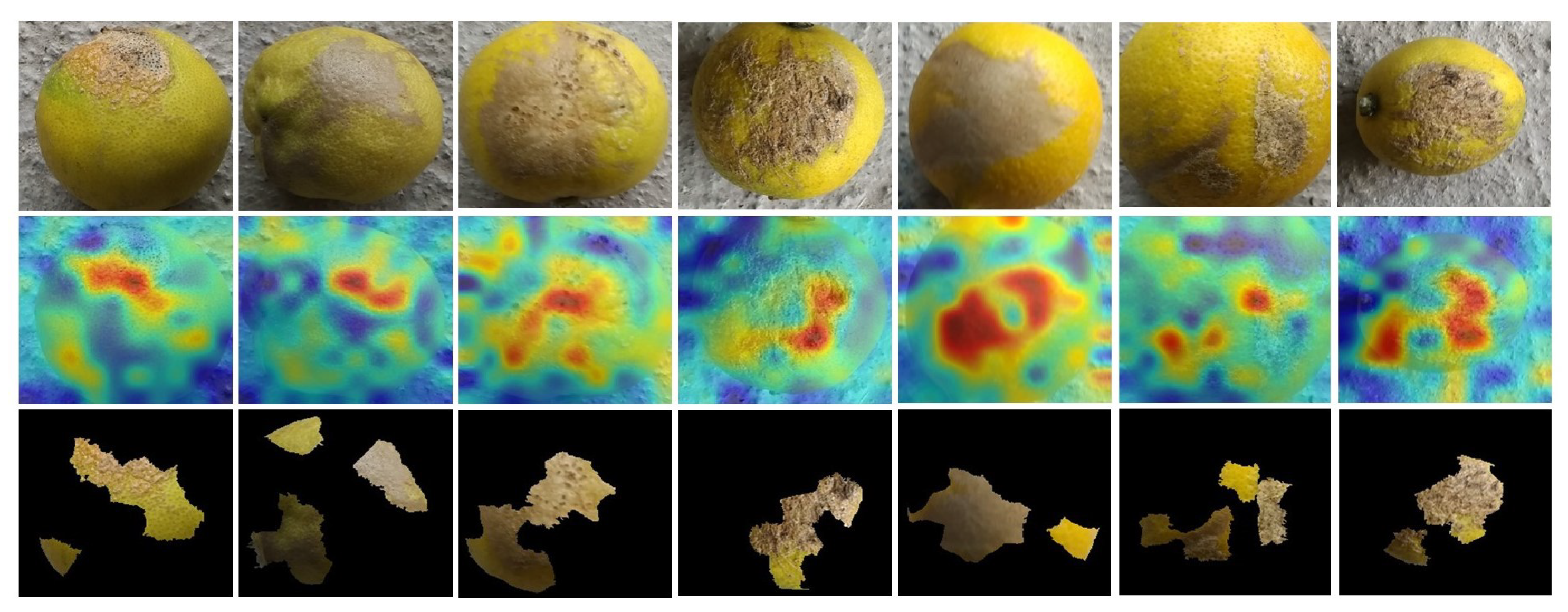
3.7. Explainability Using LIME
4. Experimental Setup
4.1. Dataset
4.2. Experimental Design
4.3. Baseline Classifiers and Parameters
4.4. Evaluation Metrics
5. Results and Discussion
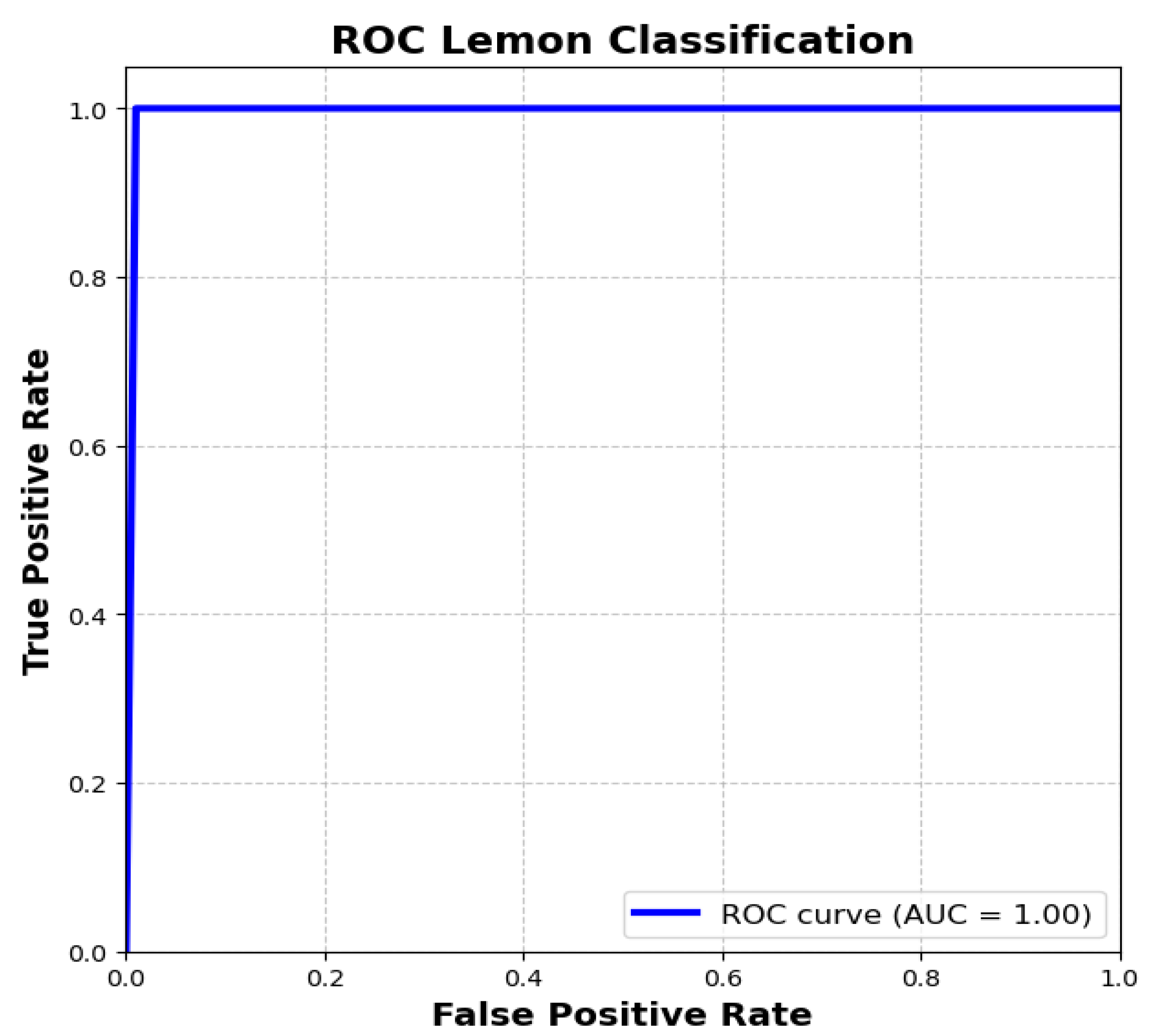
5.1. First Phase: Lemon Quality Classification Using Adaptive PSO-LemonNetX Without PSO Feature Selection
Testing on Unseen Samples
5.2. Second Phase: PSO Feature Selection and Classification Using ML Classifiers
5.2.1. Comparison of Different ML Classifiers with and Without Adaptive PSO Feature Selection
5.2.2. Comparison with Other Feature Selection Approaches
5.2.3. Comparative Analysis with Established CNN Feature Extractors
5.3. Third Phase: Explainability of Adaptive PSO-LemonNetX Decisions: The Significance of LIME
5.4. Comparison with State-of-the-Art Methods
5.5. Edge-Optimized Model Efficiency and Suitability for IoT-Driven Smart Agriculture Applications
5.6. Limitations
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Jahanbakhshi, A.; Momeny, M.; Mahmoudi, M.; Zhang, Y.D. Classification of sour lemons based on apparent defects using stochastic pooling mechanism in deep convolutional neural networks. Sci. Hortic. 2020, 263, 109133. [Google Scholar] [CrossRef]
- Hanh, L.D.; Bao, D.N.T. Autonomous lemon grading system by using machine learning and traditional image processing. Int. J. Interact. Des. Manuf. (IJIDeM) 2023, 17, 445–452. [Google Scholar] [CrossRef]
- Ullah, N.; Khan, J.A.; Almakdi, S.; Alshehri, M.S.; Al Qathrady, M.; Aldakheel, E.A.; Khafaga, D.S. A lightweight deep learning-based model for tomato leaf disease classification. Comput. Mater. Contin. 2023, 77, 3969–3992. [Google Scholar] [CrossRef]
- Xu, L.; Lv, J. Recognition method for apple fruit based on SUSAN and PCNN. Multimed. Tools Appl. 2018, 77, 7205–7219. [Google Scholar] [CrossRef]
- Nasiri, A.; Taheri-Garavand, A.; Zhang, Y.D. Image-based deep learning automated sorting of date fruit. Postharvest Biol. Technol. 2019, 153, 133–141. [Google Scholar] [CrossRef]
- Sunoj, S.; Subhashree, S.N.; Dharani, S.; Igathinathane, C.; Franco, J.; Mallinger, R.; Prasifka, J.; Archer, D. Sunflower floral dimension measurements using digital image processing. Comput. Electron. Agric. 2018, 151, 403–415. [Google Scholar] [CrossRef]
- Ullah, N.; Khan, J.A.; Al Qathrady, M.; El-Sappagh, S.; Ali, F. An effective approach for plant leaf diseases classification based on a novel DeepPlantNet deep learning model. Front. Plant Sci. 2023, 14, 1212747. [Google Scholar] [CrossRef] [PubMed]
- Mousavi, S.M.H.; MiriNezhad, S.Y.; Mosleh, M.S.; Dezfoulian, M.H. A PSO fuzzy-expert system: As an assistant for specifying the acceptance by NOET measures, at PH.D level. In In Proceedings of the 2017 Artificial Intelligence and Signal Processing Conference (AISP), Shiraz, Iran, 25–27 October 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 11–18. [Google Scholar]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why should i trust you?” Explaining the predictions of any classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 1135–1144. [Google Scholar]
- Maraveas, C.; Asteris, P.G.; Arvanitis, K.G.; Bartzanas, T.; Loukatos, D. Application of bio and nature-inspired algorithms in agricultural engineering. Arch. Comput. Methods Eng. 2023, 30, 1979–2012. [Google Scholar] [CrossRef]
- Sharma, R.; Kukreja, V. Amalgamated convolutional long term network (CLTN) model for Lemon Citrus Canker Disease Multi-classification. In Proceedings of the 2022 International Conference on Decision Aid Sciences and Applications (DASA), Chiangrai, Thailand, 23–25 March 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 326–329. [Google Scholar]
- Hernández, A.; Ornelas-Rodríguez, F.J.; Hurtado-Ramos, J.B.; González-Barbosa, J.J. Accuracy Comparison Between Deep Learning Models for Mexican Lemon Classification. In Proceedings of the Telematics and Computing: 10th International Congress, WITCOM 2021, Virtual Event, 8–12 November 2021; Proceedings 10. Springer: Berlin/Heidelberg, Germany, 2021; pp. 62–73. [Google Scholar]
- Singh, H.; Rani, R.; Mahajan, S. Detection and classification of citrus leaf disease using hybrid features. In Soft Computing: Theories and Applications: Proceedings of SoCTA 2018; Springer: Singapore, 2020; pp. 737–745. [Google Scholar]
- Ullah, N.; Javed, A. Deep Features Comparative Analysis for COVID-19 Detection from the Chest Radiograph Images. In Proceedings of the 2021 International Conference on Frontiers of Information Technology (FIT), Islamabad, Pakistan, 13–14 December 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 258–263. [Google Scholar]
- Maheshwar; Kumar, G. Breast cancer detection using decision tree, naïve bayes, KNN and SVM classifiers: A comparative study. In Proceedings of the 2019 International Conference on Smart Systems and Inventive Technology (ICSSIT); Tirunelveli, India, 27–29 November 2019, IEEE: Piscataway, NJ, USA, 2019; pp. 683–686. [Google Scholar]
- Ali Khan, S.; Hussain, A.; Basit, A.; Akram, S. Kruskal-Wallis-Based Computationally Efficient Feature Selection for Face Recognition. Sci. World J. 2014, 2014, 672630. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Alshouiliy, K.; Roy, A.; AlGhamdi, A.; Agrawal, D.P. Chi-squared based feature selection for stroke prediction using AzureML. In Proceedings of the 2020 Intermountain Engineering, Technology and Computing (IETC), Orem, UT, USA, 2–3 October 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1–6. [Google Scholar]
- Spolaôr, N.; Cherman, E.A.; Monard, M.C.; Lee, H.D. ReliefF for multi-label feature selection. In Proceedings of the 2013 Brazilian Conference on Intelligent Systems; IEEE: Piscataway, NJ, USA, 2013; pp. 6–11. [Google Scholar]
- Sahu, S.R.; Jena, S.; Panda, S. A Machine Learning Approach for Classification of Lemon Leaf Diseases. In Proceedings of the International Conference on Computing, Communication and Learning, Warangal, India, 29–31 August 2022; Springer: Berlin/Heidelberg, Germany, 2022; pp. 254–265. [Google Scholar]
- Pourdarbani, R.; Sabzi, S.; Dehghankar, M.; Rohban, M.H.; Arribas, J.I. Examination of lemon bruising using different CNN-based classifiers and local spectral-spatial hyperspectral imaging. Algorithms 2023, 16, 113. [Google Scholar] [CrossRef]
- Kaushik, P.; Khurana, S. Automated Lemon Quality Detection using DenseNet121: A Deep Learning Approach. In Proceedings of the 2025 International Conference on Electronics and Renewable Systems (ICEARS), Tuticorin, India, 11–13 February 2025; IEEE: Piscataway, NJ, USA, 2025; pp. 1907–1911. [Google Scholar]
- Pramanik, A.; Khan, A.Z.; Biswas, A.A.; Rahman, M. Lemon leaf disease classification using cnn-based architectures with transfer learning. In Proceedings of the 2021 12th International Conference on Computing Communication and Networking Technologies (ICCCNT), Kharagpur, India, 6–8 July 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 1–6. [Google Scholar]
- He, Y.; Zhu, T.; Wang, M.; Lu, H. On lemon defect recognition with visual feature extraction and transfers learning. J. Data Anal. Inf. Process. 2021, 9, 233–248. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Ullah, N.; Khan, J.A.; De Falco, I.; Sannino, G. Bridging clinical gaps: Multi-dataset integration for reliable multi-class lung disease classification with DeepCRINet and occlusion sensitivity. In Proceedings of the 2024 IEEE Symposium on Computers and Communications (ISCC), Paris, France, 26–29 June 2024; IEEE: Piscataway, NJ, USA, 2024; pp. 1–6. [Google Scholar]
- Lemon Quality Dataset. 2023. Available online: https://www.kaggle.com/datasets/yusufemir/lemon-quality-dataset (accessed on 6 January 2024).
- Corder, G.W.; Foreman, D.I. Nonparametric Statistics for Non-Statisticians; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2011. [Google Scholar]
- Liu, H.; Setiono, R. Chi2: Feature selection and discretization of numeric attributes. In Proceedings of the 7th IEEE International Conference on Tools with Artificial Intelligence, Herndon, VA, USA, 5–8 November 1995; IEEE: Piscataway, NJ, USA, 1995; pp. 388–391. [Google Scholar]
- Robnik-Šikonja, M.; Kononenko, I. Theoretical and empirical analysis of ReliefF and RReliefF. Mach. Learn. 2003, 53, 23–69. [Google Scholar] [CrossRef]
- Sawyer, S.F. Analysis of variance: The fundamental concepts. J. Man. Manip. Ther. 2009, 17, 27E–38E. [Google Scholar] [CrossRef]
- Ding, C.; Peng, H. Minimum redundancy feature selection from microarray gene expression data. J. Bioinform. Comput. Biol. 2005, 3, 185–205. [Google Scholar] [CrossRef] [PubMed]
- Yılmaz, E.K.; Adem, K.; Kılıçarslan, S.; Aydın, H.A. Classification of lemon quality using hybrid model based on Stacked AutoEncoder and convolutional neural network. Eur. Food Res. Technol. 2023, 249, 1655–1667. [Google Scholar] [CrossRef]
- Kaur, A.; Govindrao, P.S.; Mahajan, S. Lemon Perfection: Leveraging VGG16 Deep Learning for Superior Quality Assessment. In Proceedings of the 2024 Second International Conference Computational and Characterization Techniques in Engineering & Sciences (IC3TES), Lucknow, India, 15–16 November 2024; IEEE: Piscataway, NJ, USA, 2024; pp. 1–5. [Google Scholar]
- Bird, J.J.; Barnes, C.M.; Manso, L.J.; Ekárt, A.; Faria, D.R. Fruit quality and defect image classification with conditional GAN data augmentation. Sci. Hortic. 2022, 293, 110684. [Google Scholar] [CrossRef]
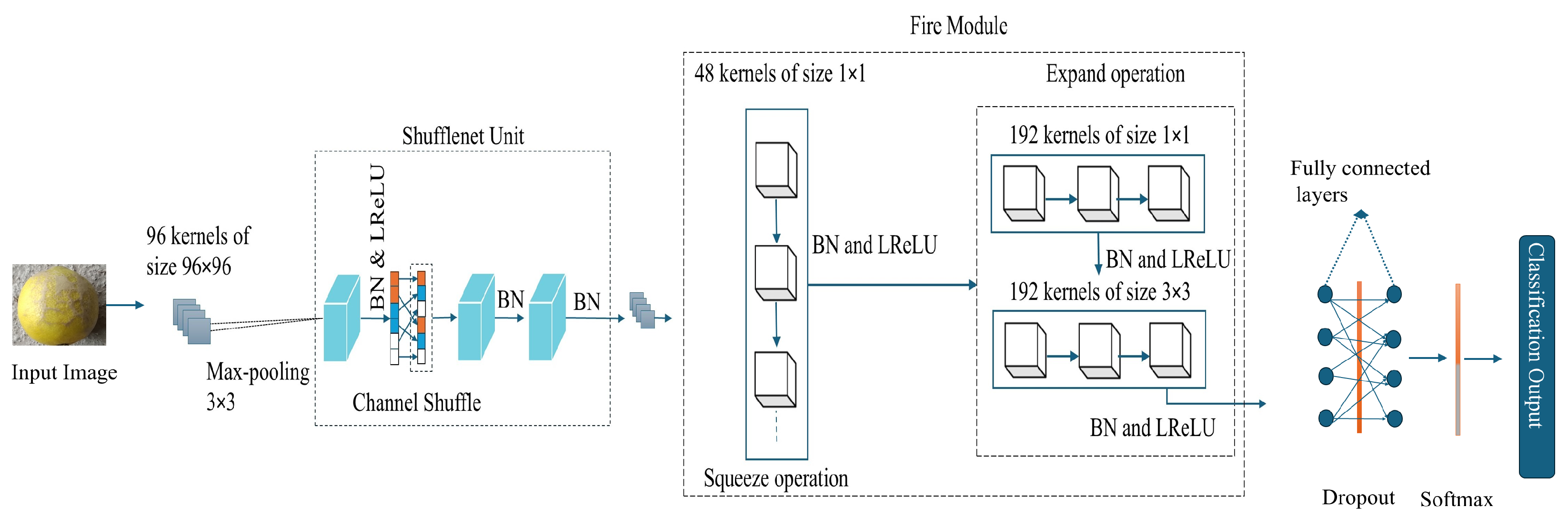



| S. No | Operation | Layers | No of Filters | Filter Size | Padding | Stride |
|---|---|---|---|---|---|---|
| 1 | Input | Input Layer | ||||
| 2 | Convolution | Convolution (LReLU, CCN) | 96 | 11 × 11 | 4 × 4 | |
| 3 | Pooling | Max pooling | 3 × 3 | 2 × 2 | ||
| 4 | Shufflenet unit | Group convolution (LReLU, BN) | 34 | 1 × 1 | ||
| Channel shuffling layer | ||||||
| Group convolution (LReLU, BN) | 1 | 3 × 3 | 1 × 1 | |||
| Group convolution (LReLU, BN) | 34 | 3 × 3 | ||||
| 5 | Pooling | Max pooling | 3 × 3 | 2 × 2 | ||
| 6 | Fire-module | Convolution (BN, LReLU) | 48 | 1 × 1 | ||
| Convolution (BN, LReLU) | 192 | 1 × 1 | ||||
| Convolution (BN, LReLU) | 192 | 3 × 3 | 1 × 1 | |||
| 7 | Pooling | Max-Pooling | 3 × 3 | 2 × 2 | ||
| 8 | Flatten Layer | |||||
| 9 | SelfAttention Layer | |||||
| 10 | FC + LReLU + BN + Dropout | |||||
| 11 | FC + Softmax + Classification | |||||
| Parameters | Values |
|---|---|
| Training Epochs | 20 |
| Optimization algorithm | SGDM |
| K-fold | 5 |
| Dropout | 0.5 |
| Activation function | LReLU |
| Learning rate | 0.001 |
| Iterations per epoch | 10 |
| Validation frequency | 30 |
| Classifier | Parameter Settings |
|---|---|
| KNN | MATLAB fitcknn, default parameters (Euclidean distance, ) |
| SVM | MATLAB fitcsvm, default parameters (linear kernel) |
| Decision Tree | MATLAB fitctree, default parameters (binary splits, Gini index) |
| Naive Bayes | MATLAB fitcnb, default parameters (Gaussian distribution) |
| Metrics | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|
| Average | 98.5 | 98.3 | 98.4 | 98.3 |
| Median | 98.4 | 98.0 | 98.5 | 98.2 |
| Maximum | 99.1 | 99.0 | 99.0 | 99.0 |
| Minimum | 98.2 | 98.0 | 98.0 | 98.0 |
| Standard deviation | 0.3 | 0.4 | 0.4 | 0.4 |
| Variance | 0.1 | 0.2 | 0.1 | 0.1 |
| Metrics | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|
| Average | 97.1 | 97.0 | 97.0 | 97.0 |
| Median | 97.1 | 97.0 | 97.0 | 97.0 |
| Maximum | 97.3 | 97.5 | 97.0 | 97.2 |
| Minimum | 96.3 | 96.5 | 96.0 | 96.2 |
| Standard deviation | 0.4 | 0.4 | 0.4 | 0.4 |
| Variance | 0.1 | 0.2 | 0.2 | 0.1 |
| Predicted Good | Predicted Bad | |
|---|---|---|
| Actual Good (225) | 219 (TP) | 6 (FN) |
| Actual Bad (190) | 6 (FP) | 184 (TN) |
| Parameter | Value |
|---|---|
| Number of particles (N) | 10 |
| Maximum iterations (max_Iter) | 100 |
| Cognitive factor () | 2 |
| Social factor () | 2 |
| Inertia weight (w) | 1 |
| Objective function | Maximize classifier performance |
| Generic solution structure | Binary feature vector |
| Execution time | 37 s |
| Metrics | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|
| PSO features and KNN | 99.1 | 99.0 | 99.0 | 99.0 |
| PSO features and SVM | 99.7 | 99.5 | 99.5 | 99.5 |
| PSO features and DT | 99.4 | 99.5 | 99.5 | 99.5 |
| PSO features and Naive Bayes | 100.0 | 100.0 | 100.0 | 100.0 |
| Metrics | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|
| PSO features and KNN | 95.2 | 95.0 | 95.5 | 95.2 |
| PSO features and SVM | 98.8 | 98.5 | 99.0 | 98.7 |
| PSO features and DT | 97.6 | 97.5 | 98.0 | 97.7 |
| PSO features and Naive Bayes | 98.8 | 98.5 | 99.0 | 98.7 |
| Features + ML Classifier | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) |
|---|---|---|---|---|
| All features + KNN | 95.18 | 94.5 | 96 | 95.25 |
| PSO features + KNN | 96.39 | 96.5 | 96.5 | 96.5 |
| All features + SVM | 96.39 | 96.0 | 97 | 96.5 |
| PSO features + SVM | 98.80 | 98.5 | 99 | 98.75 |
| All features + DT | 96.57 | 95.5 | 96 | 96.75 |
| PSO features + DT | 97.59 | 97.5 | 98 | 97.75 |
| All features + Naive Bayes | 96.39 | 96.0 | 97 | 96.5 |
| PSO features + Naive Bayes | 98.80 | 98.5 | 99 | 98.75 |
| Metrics | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|
| Kruskal–Wallis | 96.9 | 96.5 | 97.0 | 96.7 |
| () | 96.1 | 96.0 | 96.5 | 96.2 |
| ReliefF | 96.4 | 96.0 | 96.5 | 96.2 |
| ANOVA | 96.4 | 96.0 | 96.5 | 96.2 |
| MRMR | 96.7 | 96.5 | 96.5 | 96.5 |
| Adaptive PSO | 98.8 | 98.5 | 99.0 | 98.75 |
| Model + Adaptive PSO + Classifier | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) |
|---|---|---|---|---|
| VGG16 + PSO (NB) | 95.2 | 94.9 | 94.6 | 94.7 |
| VGG16 + PSO (SVM) | 95.8 | 95.5 | 95.2 | 95.3 |
| ResNet50 + PSO (NB) | 97.0 | 96.7 | 96.6 | 96.6 |
| ResNet50 + PSO (SVM) | 97.4 | 97.1 | 97.0 | 97.0 |
| DenseNet121 + PSO (NB) | 97.8 | 97.5 | 97.3 | 97.4 |
| DenseNet121 + PSO (SVM) | 98.0 | 97.8 | 97.6 | 97.7 |
| EfficientNet-B0 + PSO (NB) | 98.2 | 97.9 | 97.7 | 97.8 |
| EfficientNet-B0 + PSO (SVM) | 98.4 | 98.1 | 97.9 | 98.0 |
| PSO-LemonNetX + PSO (NB) | 98.8 | 98.5 | 99.0 | 98.7 |
| PSO-LemonNetX + PSO (SVM) | 98.8 | 98.5 | 99.0 | 98.7 |
| Ref. | Method | Dataset | Accuracy (%) | Explainability |
|---|---|---|---|---|
| [32] (2023) | SAE–CNN Hybrid: Combined GLCM, Color Space, Morphological features; SAE for dimensionality reduction; hybrid CNN with ML classifiers (SVC, Ridge, Subspace Discriminant) | Lemon quality dataset | 98.96 | None |
| [33] (2024) | VGG16: Deep CNN architecture trained on lemon images; three-class classification (good, poor, background) | Lemon quality dataset | 97.0 | None |
| [34] (2022) | GAN–VGG16: Transfer learning with VGG16; appended 4096-neuron FC layer; Conditional GAN for data augmentation; model pruning for efficiency | Public Lemon Dataset (2690 images) | 88.75 | Grad-CAM |
| [21] (2025) | DenseNet121: Preprocessing with resizing, normalization, CLAHE, augmentation; binary classification (good vs bad lemons) | Kaggle Lemon Dataset (2076 images) | 96.0 | None |
| This Work | Adaptive PSO-LemonNetX (Proposed): PSO-based adaptive feature selection, lightweight CNN, edge-friendly design | Lemon quality dataset | 100 | LIME |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ullah, N.; Ruocco, M.; Della Cioppa, A.; De Falco, I.; Sannino, G. An Explainable Deep Learning Framework with Adaptive Feature Selection for Smart Lemon Disease Classification in Agriculture. Electronics 2025, 14, 3928. https://doi.org/10.3390/electronics14193928
Ullah N, Ruocco M, Della Cioppa A, De Falco I, Sannino G. An Explainable Deep Learning Framework with Adaptive Feature Selection for Smart Lemon Disease Classification in Agriculture. Electronics. 2025; 14(19):3928. https://doi.org/10.3390/electronics14193928
Chicago/Turabian StyleUllah, Naeem, Michelina Ruocco, Antonio Della Cioppa, Ivanoe De Falco, and Giovanna Sannino. 2025. "An Explainable Deep Learning Framework with Adaptive Feature Selection for Smart Lemon Disease Classification in Agriculture" Electronics 14, no. 19: 3928. https://doi.org/10.3390/electronics14193928
APA StyleUllah, N., Ruocco, M., Della Cioppa, A., De Falco, I., & Sannino, G. (2025). An Explainable Deep Learning Framework with Adaptive Feature Selection for Smart Lemon Disease Classification in Agriculture. Electronics, 14(19), 3928. https://doi.org/10.3390/electronics14193928










