1. Introduction
The epidermal surface in digital epidermal microscopic (DEM) images displays a netlike pattern, characterized by an interwoven network of skin furrows and ridges [
1]. Quantitative analysis of DEM images is crucial for investigating the extent of skin aging. This is because skin furrows and ridges undergo gradual modifications linked to both intrinsic and extrinsic aging processes [
2]. These analyses calculate structural features for furrows, such as length, width, and angle. They also calculate ridge features, including number, perimeter, and area. To obtain these features, DEM images must first be segmented to separate ridges from furrows [
3]. To extract these features, DEM images must be segmented to accurately separate ridges from furrows. In practice, reliable DEM segmentation has two key applications. In clinical dermatology, it supports the objective assessment of skin aging, enabling the early detection of age-related disorders and evaluation of treatment efficacy. In cosmetics, it provides quantitative indicators for monitoring anti-aging product effectiveness and developing personalized skincare strategies.
However, human skin microstructure exhibits significant geometric heterogeneity. As a result, furrows and ridges in DEM images present highly complex topological features. Additionally, uneven illumination and low contrast problems often occur during the imaging process. Together, these factors challenge the development of automatic and robust segmentation methods for DEM images.
Traditional image segmentation techniques, such as region growing and clustering segmentation [
4,
5], have been utilized for skin images. However, they often perform poorly when facing complex skin patterns, as well as issues like lighting variation and low contrast [
6]. In recent years, Convolutional Neural Networks (CNNs) and Transformer-based architectures have shown great potential for medical image segmentation tasks, including skin lesion detection and dermoscopic image analysis [
7,
8]. These methods enable models to automatically extract features through end-to-end learning and overcome the limitations of traditional methods to some extent. However, existing studies have mainly focused on the segmentation of skin lesions [
9,
10], while less research has been conducted on the automatic segmentation of furrows and ridges under the microscope.
Recent advances in deep learning have further promoted the development of medical image segmentation, including the wide application of models such as full convolutional networks FCN-8s [
11], UNet [
7], SegNet [
12], DeepLabV3+ [
13], and so on. UNet [
7] and its variants show good performance in medical image segmentation tasks due to their unique skip connection structure. Typical improvements to the UNet model include ResUNet [
14], which introduces residual connections; AttentionUNet [
15], which incorporates attention mechanisms; and NestedUNet [
16], which achieves multi-scale feature aggregation. In addition, Transformer-based networks (e.g., TransUNet [
17]) have further enhanced the capability of deep learning in medical image segmentation. However, the applicability of these existing methods requires further validation and optimization due to the specific challenges of DEM image segmentation, including ambiguity, individual variability, and complex texture. Furthermore, the scarcity of publicly available DEM datasets limits the development and evaluation of automatic segmentation methods.
This study first constructs a new DEM dataset to explore deep learning methods capable of accurately and automatically segmenting complex DEM images while meeting diverse performance requirements. This dataset contains microscopic skin images that are finely labeled and can provide a reliable benchmark for the training and evaluation of deep learning models. Moreover, this study thoroughly and systematically investigates a variety of state-of-the-art deep learning models, including FCN-8s [
11], SegNet [
12], UNet [
7], DeepLabV3+ [
13], ResUNet [
14], NestedUNet [
16], AttentionUNet [
15], and TransUNet [
17], to compare and analyze their effectiveness in the DEM image segmentation task. This study provides a comprehensive evaluation of the segmentation effectiveness of these models. Furthermore, it analyzes their performance in terms of computational efficiency, robustness, and generalization ability. This study finds better and more flexible methods for segmenting skin furrows and ridges in DEM images, thereby laying the foundation for subsequent applications, such as skin aging analysis based on the relevant features of skin furrows and ridges.
2. Materials and Methods
This section outlines the experimental framework used to evaluate deep learning models for DEM image segmentation. It begins with a detailed description of data collection, including image acquisition from healthy volunteers and subsequent preprocessing steps such as cropping, labeling, and augmentation. The aim is to build a robust dataset for training and testing. Following this, the section introduces eight segmentation models—ranging from classical CNNs to hybrid architectures—and explains their structural features. Finally, it presents the training setup and evaluation metrics used to assess both segmentation accuracy and computational efficiency.
2.1. Data Collection and Processing
DEM images were captured by a specialized skin imaging device developed by Boseview Technology Company [
18] (Guangzhou, China). This advanced instrument integrates flexible plug-and-play technology and connects conveniently to a computer via a standard USB interface. The system includes an ergonomic handheld probe, designed to minimize operator fatigue during prolonged use. The probe allows for flexible positioning and stable contact with the skin surface, ensuring high-quality image acquisition. This design provides researchers with convenient and reliable access to detailed skin microstructural information.
For this study, DEM images were obtained from a cohort of 46 healthy volunteers (age range: 20–50 years) residing in Yichang, China. The majority of participants reported indoor occupations, which helped to reduce the influence of long-term ultraviolet radiation and environmental exposure on skin morphology. The forearm, specifically the dorsal and ventral regions between the elbow and wrist, was selected as the sampling site. These anatomical sites are commonly used in dermatological research because they offer relatively uniform skin thickness, easy accessibility, and minimal interference from dense hair growth.
In total, 46 raw DEM images were acquired, corresponding to one image from each participant. The collected dataset provides a valuable resource for analyzing the microtopography and biophysical properties of normal skin.
Figure 1 presents four representative raw DEM images, illustrating the variations in texture, fine lines, and microstructures among different individuals.
To mitigate the adverse impacts of uneven illumination and provide more effective training samples, we preprocessed each raw 704 × 576-pixel DEM image. From each raw image, we cropped four to seven local images at a resolution of 256 × 256 pixels (see regions ②–⑤ in
Figure 2). This process resulted in a total of 261 local DEM images. Subsequently, an experienced researcher used Adobe Photoshop 2023 to accurately label the skin furrows and ridges in the 261 local DEM images, generating a corresponding set of labeled images. Additionally, a larger test image (greater than 256 × 256 but smaller than the original 704 × 576) was cropped from each raw DEM image (see region ① in
Figure 2), yielding a total of 46 test images.
To reduce the risk of overfitting during training models and to improve the robustness of models [
19], we implemented data augmentation on 261 local DEM images using Python 3.9. The data augmentation operations include horizontal flipping, rotation, geometric transformations, brightness and contrast adjustment, and gamma correction. The details of the augmentation operations are as follows (see
Figure 3): flip horizontally with 50% probability; rotate with 50% probability by randomly selecting one of the four angles, namely, 0°, 90°, 180°, and 270°; perform a combination of translation, scaling, and rotation with 50% probability; adjust the brightness and contrast of the image with 50% probability; and change the gamma value of the image with 50% probability. All of the above operations use random sampling to ensure a high degree of diversity in the results at each data augmentation. This random application ensures that multiple augmentations of the same source image produce unique outputs, thereby effectively increasing the diversity of the training data.
After data augmentation for the 261 local DEM images, we generated five augmented versions of each image, resulting in a total of 1566 local DEM images (256 × 256 pixels) and the corresponding 1566 labeled images. These images are randomly divided into a training set and a validation set in the ratio of 8:2. The test set consists of 46 images, each larger than the 256 × 256 pixel images used for training. This setup implies that the models learn from local patches during training but must make predictions on larger images during testing, a process that better evaluates their generalization ability [
20].
2.2. Deep Learning Models for Segmentation
2.2.1. Model Descriptions
The architectural design of the eight deep learning models selected for this study largely follows the classic encoder–decoder paradigm. This structure employs a contracting path (the encoder) to capture the contextual semantic information of an image and a symmetric expanding path (the decoder) to restore spatial resolution, enabling precise pixel-level localization. The primary distinctions among these models lie in their feature extraction modules, decoder upsampling strategies, and the methods used to fuse features from different levels. The most distinctive core design of each model is briefly introduced below.
- (i)
FCN-8s
The Fully Convolutional Network (FCN) is a pioneering work in the field of semantic segmentation [
11]. It replaces the final fully-connected layers of a traditional CNN with convolutional layers, enabling end-to-end, pixel-level prediction for inputs of arbitrary size. The FCN-8s version used in this study merges feature maps from three different depths (with strides of 8, 16, and 32), which more effectively combines deep semantic information with shallow details compared to earlier versions, thus improving segmentation fineness.
- (ii)
SegNet
The core innovation of SegNet lies in its efficient decoding mechanism [
12]. During max-pooling in the encoder, it stores the positional indices of the maximum values. In the decoder, it uses these indices to perform non-parametric upsampling (unpooling), which helps restore object boundary details with minimal additional computational cost.
- (iii)
UNet
UNet’s signature designs are its U-shaped symmetric architecture and “skip connections” [
7]. By directly concatenating high-resolution feature maps from each stage of the encoder to the corresponding stage in the decoder, it significantly mitigates information loss during downsampling. This allows the model to leverage both deep semantic information and shallow texture details simultaneously, achieving great success in medical image segmentation.
- (iv)
ResUNet
ResUNet integrates the core ideas of UNet with Residual Networks (ResNet) [
14]. It introduces residual connections within each convolutional block of the UNet architecture, allowing information to “skip” across layers. This effectively addresses the vanishing gradient problem in deep network training, making it possible to build deeper networks to learn more complex feature representations.
- (v)
NestedUNet
NestedUNet redesigns the decoder path of UNet by introducing dense, nested skip connections [
16]. This design enables the decoder at each node to fuse features from multiple scales, achieving deep feature aggregation and flexible network pruning. Consequently, it delivers excellent performance across segmentation tasks of varying complexity.
- (vi)
DeepLabV3+
The distinguishing feature of DeepLabV3+ is its powerful capability for capturing multi-scale contextual information [
13]. It employs an Atrous Spatial Pyramid Pooling (ASPP) module with atrous convolutions in the encoder to obtain a larger receptive field without reducing spatial resolution [
21,
22]. It also adopts an encoder–decoder structure to better recover object boundary information.
- (vii)
TransUNet
TransUNet was among the first models to successfully integrate the Transformer architecture into medical image segmentation. It innovatively embeds a Transformer module at the bottom of the UNet encoder (the bottleneck layer), using its self-attention mechanism to capture global long-range dependencies in the image, thereby compensating for the limitations of traditional CNNs in global context modeling [
17].
- (viii)
AttentionUNet
AttentionUNet incorporates Attention Gates into the skip connections of the UNet architecture [
15]. Guided by high-level semantic information from the decoder, these gates automatically learn to focus on feature regions most relevant to the current segmentation task while suppressing feature transmission from irrelevant areas, such as the background. This allows the model to more intelligently “attend” to important details [
23,
24].
2.2.2. Model Setting and Training
The eight network models in this study were all implemented with Python 3.9 in the PyCharm 2025.1 development environment, using the PyTorch 2.0.0+cu118 deep learning framework. All experiments are conducted on a unified hardware platform equipped with the Windows 10 operating system, an NVIDIA GeForce RTX 3060 Laptop GPU (12 GB graphics memory), an Intel Core i9-11980HK CPU (2.60 GHz), and 32 GB RAM.
To ensure the fairness and comparability of the experiments, after data preprocessing, experimental environment construction, and model building, the training process for each dataset is set to 200 epochs, and the same training parameters are uniformly adopted. The learning rate is set to 0.001, a value that has been proven in most image segmentation tasks to balance convergence speed and stability, thus serving as a common default setting [
25].
In the model training phase, we set the batch sizes to 2, 4, 8, and 16 to evaluate their impact on model performance. The choice of batch size is primarily constrained by model complexity and GPU memory capacity. While ensuring the memory limits were not exceeded, values following the 2n pattern are adjusted. Under the same number of training rounds and hyperparameters, the performance of each set of settings is evaluated on the validation dataset. The batch size that achieves the best performance on the validation dataset is selected as the optimal one for the model, and accordingly, final evaluation and model comparison analysis are performed on the test dataset with that batch size.
To ensure reproducibility and minimize human intervention, we used PyTorch’s default settings for all hyperparameters not explicitly mentioned, such as optimizer parameters and weight initialization methods.
Table 1 shows the specific parameters used in the training phase of each model, including image input size, batch size, learning rate, and number of training rounds.
2.3. Quantitative Metrics for Model Evaluation
A set of mainstream performance metrics is used to provide a comprehensive assessment of the model’s performance. The metrics used to evaluate segmentation accuracy include the Dice Similarity Coefficient (DSC), Intersection over Union (IoU), Recall, and Precision [
26,
27].
DSC is defined as follows:
TP is defined as the number of pixels correctly predicted as furrows and confirmed by the ground truth. FP is defined as the number of pixels predicted as furrows but actually belonging to the background in the ground truth. FN is defined as the number of pixels predicted as background but actually corresponding to furrows in the ground truth. TN is defined as the number of pixels predicted as background and confirmed by the ground truth.
IoU is defined as follows:
Recall is defined as follows:
Precision is defined as follows:
In addition, three metrics were employed to quantify the processing efficiency of the models under study [
28]: the number of parameters (Params), floating-point operations (FLOPs), and inference time. Params measure memory resource usage and are reported in millions (M). FLOPs indicate computational complexity and are reported in GFLOPs, calculated with the thop toolkit by estimating the number of operations required for a single forward pass with a 256 × 256 input. Inference time, including average CPU and GPU times, reflects segmentation speed and is reported in milliseconds (ms).
3. Results
This section presents a comprehensive evaluation of the eight deep learning models based on segmentation accuracy and computational efficiency. Quantitative metrics such as DSC, IoU, Recall, and Precision are used to measure segmentation performance, while Params, FLOPs, and inference time assess efficiency. A normalized comparison is also provided to rank the models holistically.
3.1. Segmentation Accuracy of Each Model
This study evaluates the segmentation accuracy of eight deep learning models in the DEM image segmentation task, using DSC, IoU, Recall, and Precision for comparative analysis.
Table 2 presents the specific segmentation accuracy values (mean ± standard deviation across test samples) of the eight models. Due to computational constraints, all results are based on a single training run. To further assess the robustness of the findings, we computed the evaluation metrics for each test image individually and then reported their mean and standard deviation. The relatively small per-sample fluctuations indicate stable performance on the test set.
As shown in
Table 2, AttentionUNet achieved the highest segmentation accuracy (DSC: 0.8696, IoU: 0.7703, Recall: 0.8819, Precision: 0.8587), demonstrating strong ability in capturing complex furrow boundaries. FCN-8s (0.8690) and NestedUNet (0.8642) also perform competitively: FCN-8s offers comparable accuracy with faster inference, while NestedUNet benefits from dense skip connections and deep supervision for robust training. The baseline UNet (0.8641) remains solid, whereas ResUNet (0.8548) and DeepLabV3+ (0.8470) are slightly weaker, reflecting the limitations of deeper or more complex designs under limited data. In contrast, SegNet (0.8272) and TransUNet (0.8281) show the lowest accuracy, due to information loss in unpooling and the underutilization of Transformer modules in small, localized datasets. Overall, AttentionUNet excels in accuracy, FCN-8s offers the best balance of accuracy and efficiency, while SegNet and TransUNet illustrate the risks of oversimplified or overly complex architectures in small-sample scenarios.
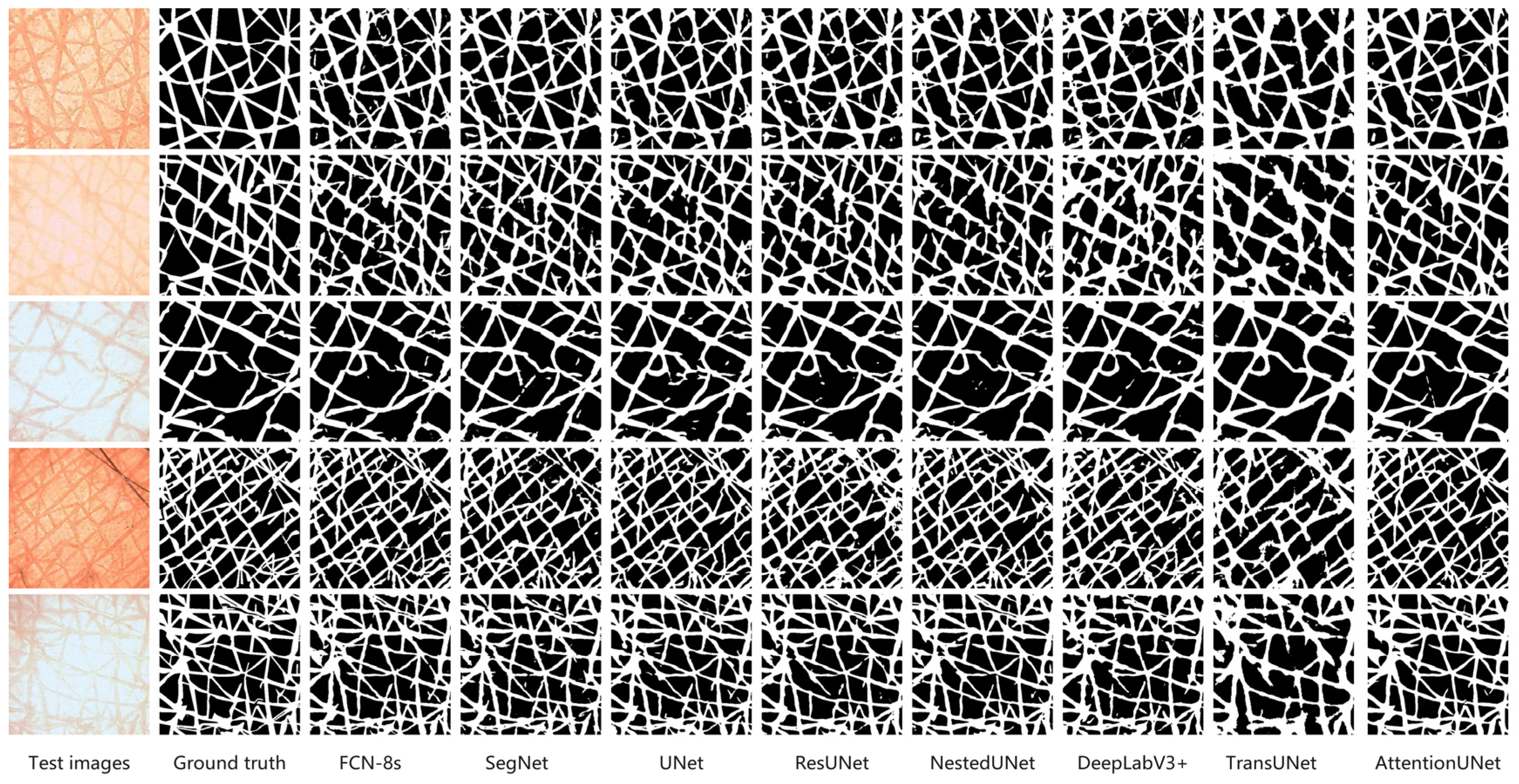
Figure 4 shows segmentation results of the eight network models on five representative DEM images. We evaluated the performance of each network model by comparing its segmentation results against the ground truth images. AttentionUNet and FCN-8s maintained clear furrow boundaries. Both models accurately captured fine structures. UNet and NestedUNet also exhibit good segmentation capabilities and demonstrate strong resistance to interference from complex backgrounds. During segmentation, DeepLabV3+ and SegNet tend to produce boundary blurring, particularly within high-complexity regions, making discontinuous results more likely. TransUNet maintains high accuracy under certain conditions; however, its performance becomes less stable when dealing with complex textures, leading to substantial uncertainty along the boundaries.
In summary, AttentionUNet and FCN-8s demonstrate relatively higher segmentation accuracy for the DEM image segmentation, while UNet and NestedUNet also exhibit good robustness. DeepLabV3+ and SegNet, however, exhibit a tendency for boundary blurring in specific scenarios, and TransUNet still has space for further optimization in segmenting complex skin textures.
3.2. Segmentation Efficiency of Each Model
This study also evaluates the efficiency performance of eight deep learning network models in the DEM image segmentation task. Comparative analysis is conducted using the Params, FLOPs, average CPU inference time (ms), and average GPU inference time (ms).
Table 3 shows that different models exhibit significant efficiency differences in processing the DEM image segmentation task.
FCN-8s is a lightweight model with only 18.64 M parameters and 25.50 G FLOPs. It achieves the fastest inference speed among all of the tested models (CPU: 12.62 ms; GPU: 37.36 ms), making it highly suitable for scenarios that require computational efficiency. The computational complexity of SegNet is slightly higher than that of FCN-8s, but its overall efficiency remains at a relatively high level (CPU: 23.92 ms, GPU: 54.00 ms), achieving a certain balance between speed and model capability. NestedUNet achieves good inference speed (CPU: 29.11 ms, GPU: 80.18 ms) with a minimal number of parameters (9.16 M) and moderate FLOPs (34.90 G). Despite having fewer parameters (13.04 M), ResUNet exhibits a higher FLOPs count (80.98 G), reflecting a more intricate computational structure. While it performs well on CPU (18.67 ms), its longer average GPU inference time (98.86 ms) suggests it benefits more from CPU computation optimization.
In contrast to the previously discussed models, both UNet and TransUNet suffer from longer inference times. UNet involves 31.04 M parameters and 54.74 G FLOPs, leading to an average GPU inference time as high as 130.08 ms. TransUNet has the largest parameter count (67.08 M) and the longest GPU inference time (207.27 ms). AttentionUNet and DeepLabV3+ exhibit good segmentation accuracy but have higher computational overheads, with average GPU inference times of 97.97 ms and 94.26 ms, respectively.
Overall, FCN-8s demonstrated the highest efficiency in both CPU and GPU processing. UNet demonstrates lower efficiency for both CPU and GPU processing. Although TransUNet shows moderate CPU processing efficiency, it has the lowest efficiency for GPU processing. SegNet and NestedUNet achieve a good balance between CPU and GPU processing efficiency. AttentionUNet, while exhibiting higher CPU processing efficiency, shows lower efficiency for GPU processing.
3.3. Comprehensive Performance Evaluation of Each Model
To comprehensively evaluate the segmentation performance of the eight network models, it is essential to consider both the segmentation accuracy and efficiency metrics. However, because the various quantitative metrics have different units and numerical ranges, they cannot be directly compared. To remove the effect of these differing units, each metric must undergo scaling to ensure all values fall within the [0, 1] range.
Regarding positive metrics like DSC, IoU, Recall, and Precision (higher values denote superior performance), Max Scaling [
29] is applied. The normalization equation is presented below, as follows:
where
denotes the raw metric value,
denotes the maximum value of that metric across all models.
For negative metrics like Params, FLOPs, CPU runtime, and GPU runtime (where lower values signify superior performance), reciprocal normalization [
30] is used. The formula is presented below as follows:
where
denotes the minimum value of that metric across all models.
After the above normalization processing, all indicators adhere to the following principle: the higher the indicator value, the better the model’s performance in that metric. The normalized quantitative evaluation results are shown in
Table 4.
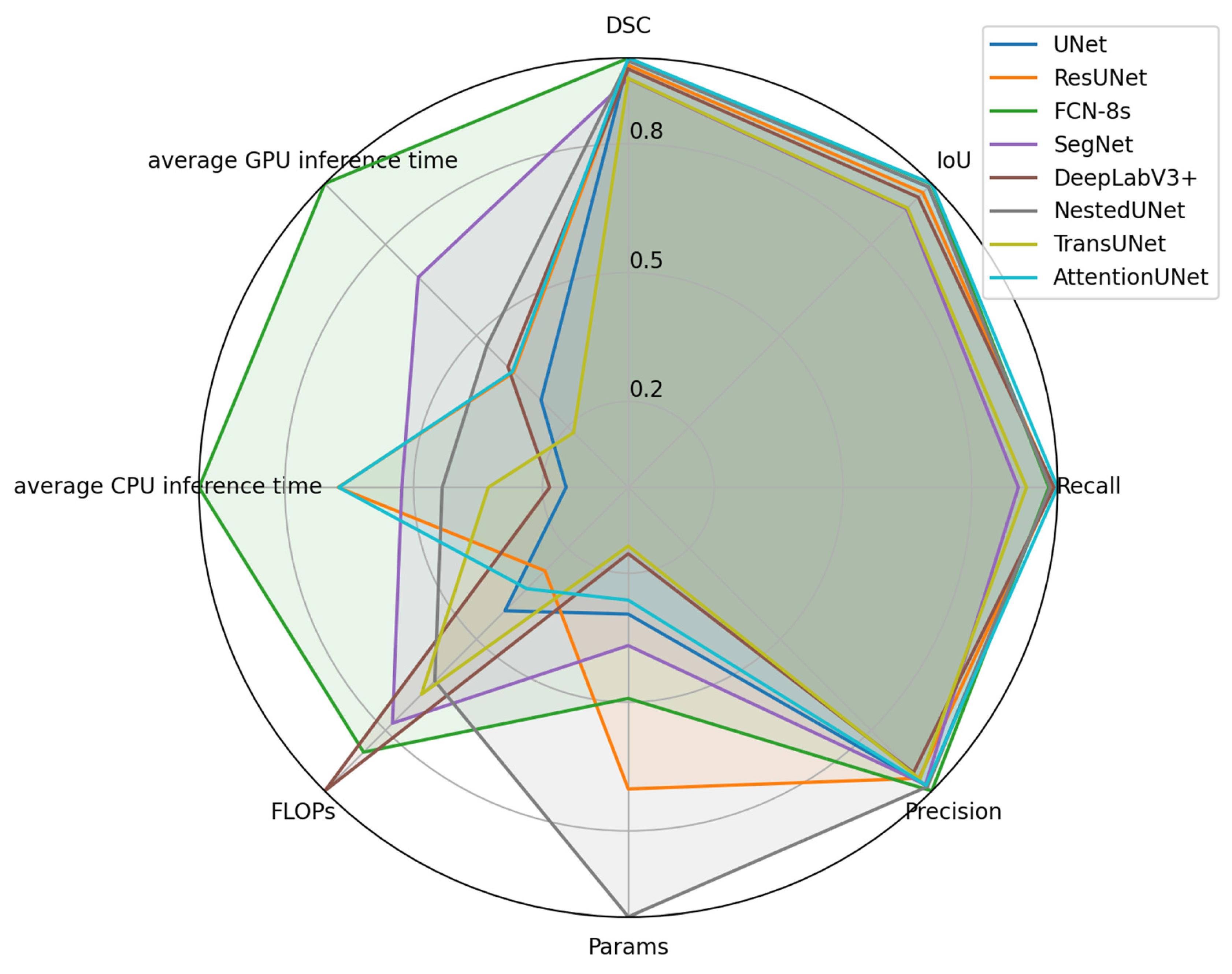
Normalized metrics are visualized in a radar chart (see
Figure 5). Since higher normalized values correspond to superior model performance, values nearer to the outer edge of the chart indicate better performance. Based on the results of the area calculations from the radar charts (see
Table 5), the overall model performance ranking from highest to lowest is as follows: FCN-8s, NestedUNet, SegNet, ResUNet, AttentionUNet, DeepLabV3+, UNet, and TransUNet.
FCN-8s shows the largest area on the radar chart, suggesting the best overall performance in segmentation accuracy and efficiency, whereas TransUNet has the smallest area, indicating the worst overall performance. Traditional Convolutional Neural Network architectures like FCN-8s and SegNet are more fundamental compared to newer architectures, but they still maintain stable performance across multiple evaluation metrics. In particular, FCN-8s achieves the best overall performance in this task. In contrast, TransUNet, which integrates attention mechanisms and Transformer structures, fails to demonstrate the expected advantages. This might be influenced by the data characteristics or training strategies.
These results indicate that model performance depends not only on the complexity of the network architecture but also closely relates to the task type and data characteristics. In the DEM image segmentation task, traditional network architectures still possess significant competitiveness, whereas the advantages of newer architectures require further validation and optimization within specific contexts.
4. Discussion
Focusing on automatic segmentation for DEM images, this study systematically assesses the performance of eight representative deep learning models based on eight metrics that cover segmentation accuracy and computational efficiency.
Regarding segmentation accuracy, AttentionUNet demonstrates relatively better performance across multiple evaluation metrics (DSC: 0.8696, IoU: 0.7703, Recall: 0.8819), effectively highlighting its relative advantage in handling complex boundary structures. AttentionUNet extends the traditional UNet architecture by integrating attention gates. These gates dynamically inhibit irrelevant regions during feature transfer, strengthening the model’s focus on significant areas like epidermal groove edges. This mechanism enhances both spatial perception and feature selection accuracy. As a result, it is especially effective for DEM images characterized by indistinct boundaries and intricate textures. Additionally, the attention module addresses the traditional UNet’s weakness in differentiating complex tissue structures through the introduction of non-linear correlation modeling.
NestedUNet significantly improves the fusion of multi-scale semantic and spatial features. It achieves this by introducing dense, nested skip connections, which are further enhanced by a deep supervision mechanism. Its unique decoding path adopts a multi-level aggregation strategy. This strategy enhances semantic consistency while preserving spatial details. As a result, it effectively bridges the information gap between shallow features and deep semantic alignment that exists in the original UNet structure. Additionally, the integration of deep supervision enables loss calculation at all sub-network levels during training, which improves convergence speed and segmentation robustness.
ResUNet, on the other hand, incorporates residual blocks into UNet. This integration enables efficient gradient transfer through constant mappings, thereby stabilizing the network’s deeper layers during training. This structure enhances the nonlinear expression ability during the feature extraction phase. It effectively addresses the common issues of gradient disappearance and feature degradation in deeper networks while preserving the texture details in lower layers. ResUNet demonstrates good performance in both training stability and accuracy.
Overall, classic models based on the encoder–decoder architecture, along with their improved variants, continue to demonstrate strong adaptability and scalability when addressing the challenges posed by complex local structures and dense textures in DEM images. These model designs generally emphasize multi-scale feature fusion (e.g., NestedUNet’s cascaded structure), deep semantic extraction (e.g., ResUNet’s residual blocks), and local boundary enhancement (e.g., AttentionUNet’s attention mechanism), providing a solid structural foundation for segmenting complex medical images.
However, greater architectural complexity does not necessarily translate into better performance. TransUNet integrates a vision transformer module into the UNet encoder. This design theoretically enhances global context modeling and long-range dependency capture. However, in this study, its segmentation performance (DSC: 0.8281) was significantly lower than that of several CNN-based models. Several factors may explain this result. First, the texture features in DEM images are primarily localized. Consequently, the Transformer’s global attention mechanism struggles to precisely capture small, irregular edge details. Second, the Transformer module’s high parameter complexity and data dependency hinder its structural benefits when data volume is limited, potentially causing reduced generalization and overfitting risks. This suggests that Transformer-based modules are better suited to large datasets with pronounced global structures. However, for tasks involving dense local textures, their performance benefits remain to be validated and optimized.
In terms of computational efficiency, FCN-8s demonstrates a relative advantage. This model employs a relatively shallow convolutional architecture and a fully convolutional up-sampling strategy, resulting in a concise structure. With only 18.64 M parameters and 25.50 G FLOPs, it achieves an average GPU inference time of 37.36 ms, which is the lowest among all the tested models. The skip connection mechanism in FCN-8s preserves high-resolution features and facilitates semantic information flow, effectively controlling computational resource consumption. This model features a short fusion path and a straightforward up-sampling process, making it suitable for resource-constrained or computationally efficient application scenarios. However, its down-sampling stage relies on traditional max-pooling, which can compromise the recovery of high-frequency details.
Although SegNet also uses an encoder–decoder structure, its reliance on max unpooling with indices during the decoding phase leads to significant information loss. This deficiency results in blurred segmentation boundaries and the poor recovery of high-frequency details. Additionally, while this structure reduces storage overhead, it has limitations in complex texture reconstruction tasks.
Notably, although AttentionUNet demonstrates superior accuracy, its complex attention computation and deep feature fusion structure result in relatively slow inference speed. This makes it more suitable for tasks requiring high segmentation accuracy and tolerable delay.
Based on a normalized multi-metric comprehensive analysis using radar charts, FCN-8s demonstrates stable performance in both accuracy and efficiency. It achieves the largest radar chart area (2.3856), indicating the best overall performance. In contrast, NestedUNet and SegNet manage to maintain a certain level of segmentation accuracy while also achieving structural light-weighting and edge-awareness, placing them among the top performers as well. However, TransUNet, despite incorporating structurally complex Transformer modules, fails to demonstrate the expected advantages in the DEM image segmentation task. This result further underscores the tension between architectural complexity and task suitability. It suggests that adapting a model’s structure to a specific task is more effective than simply increasing its complexity.
Nevertheless, several limitations of this study should be acknowledged. First, the dataset is relatively small (46 volunteers; 261 original images) and relies heavily on offline augmentation, which may restrict the generalization ability of the models. This constraint may particularly disadvantage data-intensive architectures like TransUNet and DeepLabV3+. Consequently, their suboptimal performance in our experiments is likely driven by data scarcity rather than inherent architectural limitations. Second, the training was conducted on 256 × 256 cropped images, while testing used larger images, creating a domain gap that may bias performance evaluation. Moreover, because hyperparameters were not systematically tuned for each model, the robustness of our comparative conclusions may be limited.
5. Conclusions
The performance of eight representative deep learning models was systematically evaluated for DEM image segmentation. AttentionUNet achieved the best segmentation accuracy, particularly in capturing complex boundary structures, while FCN-8s demonstrated the highest computational efficiency. In contrast, TransUNet showed the lowest performance in both accuracy and efficiency.
Considering both segmentation accuracy and efficiency, FCN-8s provides a good trade-off and can serve as a baseline model for DEM segmentation tasks. Different models may suit different scenarios: lightweight models (e.g., FCN-8s) are preferable for real-time clinical applications; high-accuracy models (e.g., AttentionUNet) are suitable for research requiring precise recognition; and balanced models (e.g., NestedUNet) show promise for small-scale or collaborative studies.
The dataset has notable limitations regarding demographic diversity. It was collected from healthy volunteers in a single region, featuring relatively concentrated age ranges and skin types. This limitation may affect the generalization ability of the models across populations with different ages, genders, skin tones, and lifestyles. Therefore, future research should focus on collecting larger and more diverse datasets to further validate the comprehensive performance of the eight network models. In addition, future work will emphasize enhancing model robustness and generalization, including lightweight optimization for AttentionUNet, accuracy improvement for FCN-8s, and the exploration of larger, more diverse datasets to support broader applicability.