A Comparative Study on Machine Learning Methods for EEG-Based Human Emotion Recognition
Abstract
1. Introduction
- Presentation of efficient techniques for analyzing and visualizing EEG datasets.
- A detailed methodology for utilizing key EEG features such as power spectral density (PSD) and differential entropy (DE).
- An effective approach for integrating feature selection, feature extraction, and classification algorithms.
- A comparative analysis of shallow and deep networks for EEG-based emotion classification.
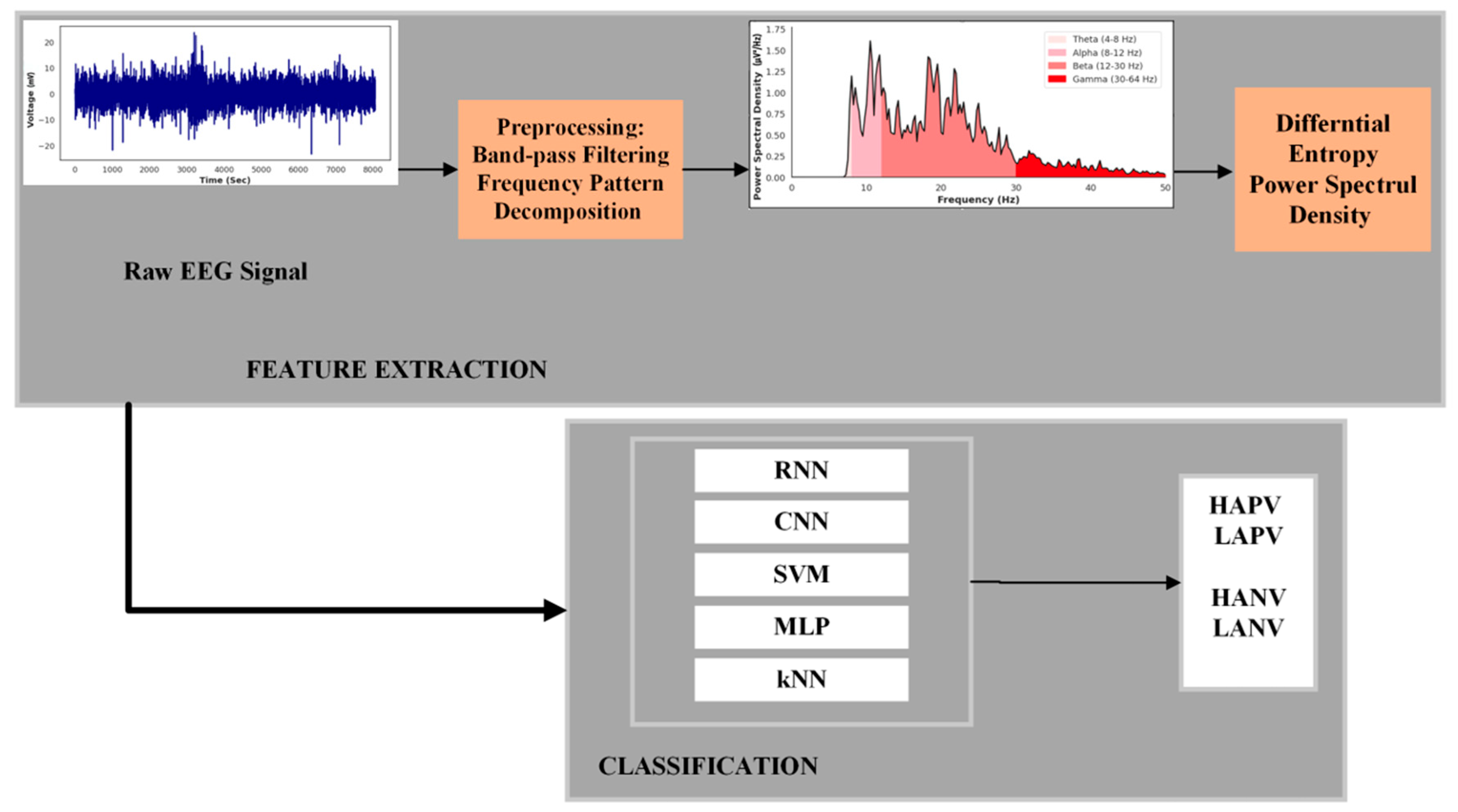
2. Materials and Methods
2.1. Emotional EEG Dataset
2.2. Preprocessing
2.3. Frequency Pattern Decomposition and Feature Extraction
2.3.1. Differential Entropy
2.3.2. Power Spectral Density
2.4. Classification
3. Results
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, X.; Zhang, Y.; Tiwari, P.; Song, D.; Hu, B.; Yang, M.; Zhao, Z.; Kumar, N.; Marttinen, P. EEG Based Emotion Recognition: A Tutorial and Review. ACM Comput. Surv. 2022, 55, 79. [Google Scholar] [CrossRef]
- Alam, M.S.; Jalil, S.Z.A.; Upreti, K. Analyzing recognition of EEG based human attention and emotion using Machine learning. Mater. Today Proc. 2022, 56, 3349–3354. [Google Scholar] [CrossRef]
- Yu, C.; Wang, M. Survey of emotion recognition methods using EEG information. Cogn. Robot. 2022, 2, 132–146. [Google Scholar] [CrossRef]
- Li, W.; Long, Y.; Yan, Y.; Xiao, K.; Wang, Z.; Zheng, D.; Leal-Junior, A.; Kumar, S.; Ortega, B.; Marques, C.; et al. Wearable photonic smart wristband for cardiorespiratory function assessment and biometric identification. Opto-Electron. Adv. 2025, 8, 240254. [Google Scholar] [CrossRef]
- Huang, N.; Zhou, M.; Bian, D.; Mehta, P.; Shah, M.; Rajput, K.S.; Selvaraj, N. Novel Continuous Respiratory Rate Monitoring Using an Armband Wearable Sensor. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS, Guadalajara, Mexico, 1–5 November 2021; Institute of Electrical and Electronics Engineers Inc.: New York, NY, USA, 2021; pp. 7470–7475. [Google Scholar] [CrossRef]
- Mai, N.D.; Lee, B.G.; Chung, W.Y. Affective computing on machine learning-based emotion recognition using a self-made eeg device. Sensors 2021, 21, 5135. [Google Scholar] [CrossRef]
- Nasir, A.N.K.; Ahmad, M.A.; Najib, M.S.; Wahab, Y.A.; Othman, N.A.; Ghani, N.M.A.; Irawan, A.; Khatun, S.; Ismail, R.M.T.R.; Saari, M.M.; et al. ECCE 2019; Springer: Singapore, 2019. [Google Scholar]
- Zhang, Y.; Chen, J.; Tan, J.H.; Chen, Y.; Chen, Y.; Li, D. An Investigation of Deep Learning Models for EEG-Based Emotion Recognition. Front. Neurosci. 2020, 14, 622759. [Google Scholar] [CrossRef]
- Yanagimoto, M. Recognition of Persisting Emotional Valence from EEG Using Convolutional Neural Networks. In Proceedings of the 2016 IEEE 9th International Workshop on Computational Intelligence and Applications (IWCIA), Hiroshima, Japan, 5 November 2016; pp. 27–32. [Google Scholar] [CrossRef]
- Padhmashree, V.; Bhattacharyya, A. Knowledge-Based Systems Human emotion recognition based on time–Frequency analysis of multivariate EEG signal. Knowl. Based Syst. 2022, 238, 107867. [Google Scholar] [CrossRef]
- Jaratrotkamjorn, A. Bimodal Emotion Recognition using Deep Belief Network. In Proceedings of the 2019 23rd International Computer Science and Engineering Conference (ICSEC), Phuket, Thailand, 30 October–1 November 2019; pp. 103–109. [Google Scholar]
- Mahmoud, A.; Amin, K.; Al Rahhal, M.M.; Elkilani, W.S.; Mekhalfi, M.L.; Ibrahim, M. A CNN Approach for Emotion Recognition via EEG. Symmetry 2023, 15, 1822. [Google Scholar] [CrossRef]
- Wen, T.; Zhang, Z. Deep Convolution Neural Network and Autoencoders-Based Unsupervised Feature Learning of EEG Signals. IEEE Access 2018, 6, 25399–25410. [Google Scholar] [CrossRef]
- Song, T.; Zheng, W.; Song, P.; Cui, Z. EEG Emotion Recognition Using Dynamical Graph Convolutional Neural Networks. IEEE Trans. Affect. Comput. 2020, 11, 532–541. [Google Scholar] [CrossRef]
- Ma, J.; Lu, B. Emotion Recognition using Multimodal Residual LSTM Network. In Proceedings of the 27th Acm International Conference on Multimedia, Nice, France, 21–25 October 2019; pp. 176–183. [Google Scholar] [CrossRef]
- Chakladar, D.D.; Dey, S.; Roy, P.P.; Dogra, D.P. EEG-based mental workload estimation using deep BLSTM-LSTM network and evolutionary algorithm. Biomed. Signal Process. Control 2020, 60, 101989. [Google Scholar] [CrossRef]
- Khateeb, M.; Anwar, S.M.; Alnowami, M. Multi-Domain Feature Fusion for Emotion Classification Using DEAP Dataset. IEEE Access 2021, 9, 12134–12142. [Google Scholar] [CrossRef]
- Chaudhary, R.; Jaswal, R.A. A Review of Emotion Recognition Based on EEG using DEAP Dataset. Int. J. Sci. Res. Sci. Eng. Technol. 2021, 8, 298–303. [Google Scholar] [CrossRef]
- Koelstra, S.; Muhl, C.; Soleymani, M.; Lee, J.S.; Yazdani, A.; Ebrahimi, T.; Pun, T.; Nijholt, A.; Patras, I. DEAP: A Database for Emotion Analysis using Physiological Signals. IEEE Trans. Affect. Comput. 2011, 3, 18–31. [Google Scholar] [CrossRef]
- Jafari, M.; Shoeibi, A.; Khodatars, M.; Bagherzadeh, S.; Shalbaf, A.; García, D.L.; Gorriz, J.M.; Acharya, U.R. Emotion recognition in EEG signals using deep learning methods: A review. Comput. Biol. Med. 2023, 165, 107450. [Google Scholar] [CrossRef]
- Moon, S.E.; Chen, C.J.; Hsieh, C.J.; Wang, J.L.; Lee, J.S. Emotional EEG classification using connectivity features and convolutional neural networks. Neural Netw. 2020, 132, 96–107. [Google Scholar] [CrossRef]
- Same, M.H.; Gandubert, G.; Gleeton, G.; Ivanov, P.; Landry, R. Simplified welch algorithm for spectrum monitoring. Appl. Sci. 2021, 11, 86. [Google Scholar] [CrossRef]
- Cho, R.; Puli, S.; Hwang, J. Machine Learning Techniques for Distinguishing Hand Gestures from Forearm Muscle Activity; International Society for Occupational Ergonomics and Safety: Freising, Germany, 2023. [Google Scholar] [CrossRef]
- Pfurtscheller, G.; Da Silva, F.H.L. Event-related EEG/MEG synchronization and desynchronization: Basic principles. Clin. Neurophysiol. 1999, 110, 1842–1857. [Google Scholar] [CrossRef]
- Thurnhofer-Hemsi, K.; López-Rubio, E.; Molina-Cabello, M.A.; Najarian, K. Radial Basis Function Kernel Optimization for Support Vector Machine Classifiers. July 2020. Available online: http://arxiv.org/abs/2007.08233 (accessed on 1 July 2025).
- Xiong, L.; Yao, Y. Study on an adaptive thermal comfort model with K-nearest-neighbors (KNN) algorithm. Build. Environ. 2021, 202, 108026. [Google Scholar] [CrossRef]
- Hancock, P.A.; Al-juaid, A. brain sciences Neural Decoding of EEG Signals with Machine Learning: A Systematic Review. Brain Sci. 2021, 11, 1525. [Google Scholar]
- Sherstinsky, A. Fundamentals of Recurrent Neural Network (RNN) and Long Short-Term Memory (LSTM) network. Phys. D 2020, 404, 132306. [Google Scholar] [CrossRef]
- Rini, D.P.; Sari, W.K. Optimizing Hyperparameters of CNN and DNN for Emotion Classification Based on EEG Signals. Int. J. Inf. Commun. Technol. (IJoICT) 2024, 10, 1–12. [Google Scholar] [CrossRef]
- Mao, S.; Sejdic, E. A Review of Recurrent Neural Network-Based Methods in Computational Physiology. IEEE Trans. Neural Netw. Learn. Syst. 2023, 34, 6983–7003. [Google Scholar] [CrossRef]
- Aliyu, I.; Mahmood, M.; Lim, G. LSTM Hyperparameter Optimization for an EEG-Based Efficient Emotion Classification in BCI. J. Korea Inst. Electron. Commun. Sci. 2019, 14, 1171–1180. [Google Scholar] [CrossRef]



| Total | |||||
|---|---|---|---|---|---|
| Subjects | EEG Channels | Videos | Sampling rate | Label | Scale range |
| 32 | 32 | 40 | 128 Hz | Valence and arousal | Continuous scale of 1–9 |
| The format of EEG data for one subject (preprocessed version) | |||||
| Array | Dimension | Content | |||
| Labels | 2 × 40 | Label (valence, arousal) × trial/video | |||
| Data | 32 × 8064 × 40 | Channels × data × trial/video | |||
| Class | DE (CNN) | PSD (CNN) | DE (RNN-LSTM) | PSD (RNN-LSTM) |
|---|---|---|---|---|
| HAPV | 128 | 64 | 256 | 128 |
| LAPV | 128 | 64 | 256 | 128 |
| HANV | 128 | 64 | 256 | 128 |
| LANV | 128 | 64 | 256 | 128 |
| Algorithm | Parameter Name | Value |
|---|---|---|
| MLP | Dimension of hidden layers | 128, 64, 32, 16, 8 |
| Activation function | Tanh | |
| Optimizer | Adam | |
| Learning rate | 0.01 | |
| Maximum iteration | 500 | |
| Epoch number | 25 | |
| kNN | Number of neighbors | 3 ≤ d ≤ 60 |
| Distance metric | Euclidean | |
| Epoch number | 25 | |
| SVM | Kernel | RBF |
| Scale factor (γ) | 0.05, 2 | |
| Epoch number | 25 | |
| CNN | Number of layers | 8 |
| Learning rate | 0.001 | |
| Pooling type | Max pooling | |
| Activation function | ReLU | |
| Padding | Same | |
| Optimizer | Adam | |
| RNN | Number of layers | 3 |
| Type of layers | LSTM | |
| Activation functions | ReLU, Softmax | |
| Learning rate | 0.001 | |
| Optimizer | Adam |
| Layer | Type | Size | Kernel Size | Stride |
|---|---|---|---|---|
| Input | Input | 128 × 384 × 32 | - | - |
| Convolution1 | Conv2D | 128 × 384 × 64 | 3 × 3 | 1 × 1 |
| Activation | Leaky ReLU | 128 × 384 × 64 | - | - |
| Spatial Dropout | Dropout | 128 × 384 × 64 | - | - |
| Convolution2 | Conv2D | 128 × 384 × 128 | 3 × 3 | 1 × 1 |
| Batch Normalization | BatchNorm | 128 × 384 × 128 | - | - |
| Activation | Leaky ReLU | 128 × 384 × 128 | - | - |
| Max Pooling 1 | MaxPooling2D | 64 × 192 × 128 | 2 × 2 | 2 × 2 |
| Convolution3 | Conv2D | 64 × 192 × 256 | 3 × 3 | 1 × 1 |
| Activation | Leaky ReLU | 64 × 192 × 256 | - | - |
| Spatial Dropout | Dropout | 64 × 192 × 256 | - | - |
| Convolution4 | Conv2D | 64 × 192 × 256 | 3 × 3 | 1 × 1 |
| Activation | Leaky ReLU | 64 × 192 × 256 | - | - |
| Spatial Dropout | Dropout | 64 × 192 × 256 | - | - |
| Max Pooling 2 | MaxPooling2D | 32 × 96 × 256 | 2 × 2 | 2 × 2 |
| Flatten | Flatten | 786,432 | - | - |
| Fully Connected | Dense | 1024 | - |
| Model | Class | R% | P% | F1-Score | Total Accuracy | Training Time (s) |
|---|---|---|---|---|---|---|
| MLP | HAPV | 68.47 | 0.68 | 0.66 | 0.669 | |
| LAPV | 65.4 | 0.62 | 0.62 | |||
| HANV | 58.1 | 0.59 | 0.58 | 24.30 | ||
| LANV | 61.23 | 0.61 | 0.61 | |||
| kNN | HAPV | 70.5 | 0.72 | 0.72 | 0.722 | |
| LAPV | 69.12 | 0.75 | 0.72 | 5.53 | ||
| HANV | 69.5 | 0.69 | 0.69 | |||
| LANV | 69.12 | 0.75 | 0.72 | |||
| SVM | HAPV | 72.31 | 0.75 | 0.73 | 0.740 | |
| LAPV | 70.1 | 0.7 | 0.7 | 38.99 | ||
| HANV | 71.31 | 0.76 | 0.73 | |||
| LANV | 70.82 | 0.73 | 0.72 | |||
| CNN | HAPV | 91.32 | 0.94 | 0.93 | 0.921 | |
| LAPV | 90.14 | 0.88 | 0.89 | |||
| HANV | 89.47 | 0.87 | 0.91 | 304.65 | ||
| LANV | 88.63 | 0.86 | 0.89 | |||
| RNN-LSTM | HAPV | 94.28 | 0.91 | 0.94 | 0.933 | |
| LAPV | 90.14 | 0.91 | 0.91 | 477.90 | ||
| HANV | 89.47 | 0.9 | 0.9 | |||
| LANV | 88.63 | 0.88 | 0.92 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Davarzani, S.; Masihi, S.; Panahi, M.; Olalekan Yusuf, A.; Atashbar, M. A Comparative Study on Machine Learning Methods for EEG-Based Human Emotion Recognition. Electronics 2025, 14, 2744. https://doi.org/10.3390/electronics14142744
Davarzani S, Masihi S, Panahi M, Olalekan Yusuf A, Atashbar M. A Comparative Study on Machine Learning Methods for EEG-Based Human Emotion Recognition. Electronics. 2025; 14(14):2744. https://doi.org/10.3390/electronics14142744
Chicago/Turabian StyleDavarzani, Shokoufeh, Simin Masihi, Masoud Panahi, Abdulrahman Olalekan Yusuf, and Massood Atashbar. 2025. "A Comparative Study on Machine Learning Methods for EEG-Based Human Emotion Recognition" Electronics 14, no. 14: 2744. https://doi.org/10.3390/electronics14142744
APA StyleDavarzani, S., Masihi, S., Panahi, M., Olalekan Yusuf, A., & Atashbar, M. (2025). A Comparative Study on Machine Learning Methods for EEG-Based Human Emotion Recognition. Electronics, 14(14), 2744. https://doi.org/10.3390/electronics14142744







