Quality Assurance of the Whole Slide Image Evaluation in Digital Pathology: State of the Art and Development Results
Abstract
1. Introduction
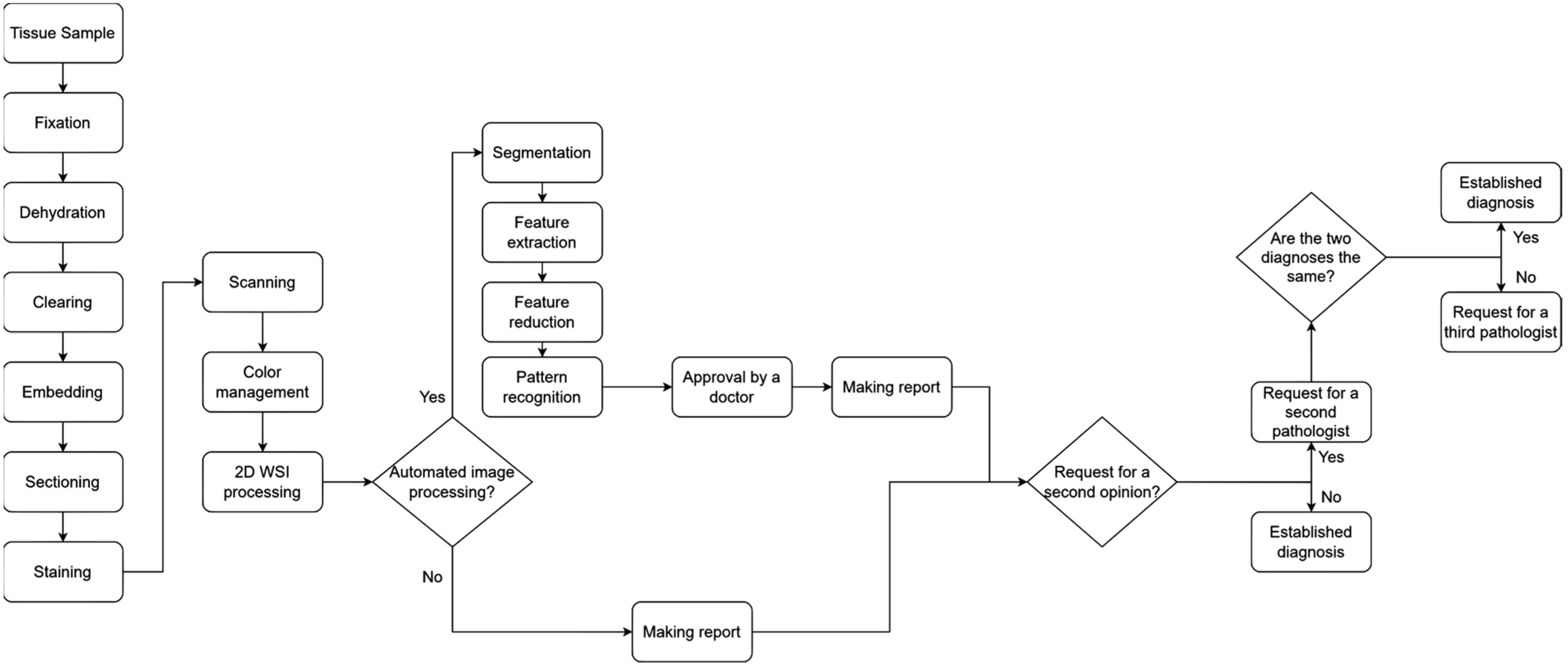
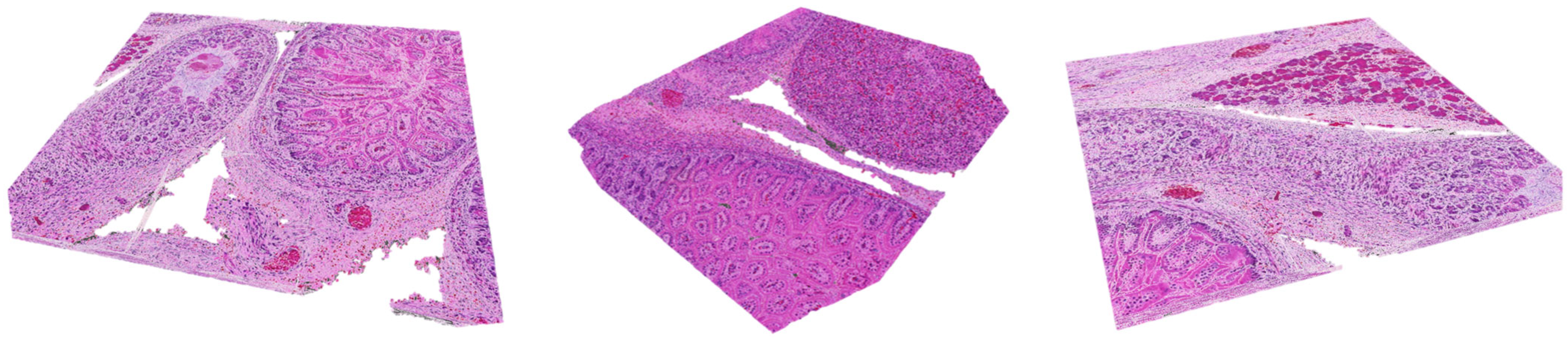
1.1. Whole Slide Images and Quality Control in Digital Pathology
- Fixation: In this step, various chemicals are used to fix the tissue structure in its natural shape. The practical purpose of fixation is to prevent or stop the degenerative processes that start when a tissue is deprived of blood supply. The most-used fixative is 10% neutral buffered formalin.
- Dehydration: In this step, ethanol is added to the tissue sample. The water removed with ethanol causes the sample to harden, which supports the examination of the tissue samples with light microscopes.
- Clearing: In this step, organic solvents are added to the sample to help remove ethanol and allow the wax to infiltrate the sample. One such solvent is xylene.
- Embedding: In this step, the tissue sample is infiltrated with paraffin wax. This results in a paraffin block which, after hardening, allows thin slices (layers) to be cut from it.
- Sectioning: In this step, the hardened sample is cut into layers using a microtome. The most commonly used layer thickness is 4–5 μm.
- Staining: Most cells are transparent and therefore would not show up on examination. To avoid this, some staining material should be used to highlight features that are relevant to the current study. The most commonly used stain is hematoxylin and eosin (H&E).
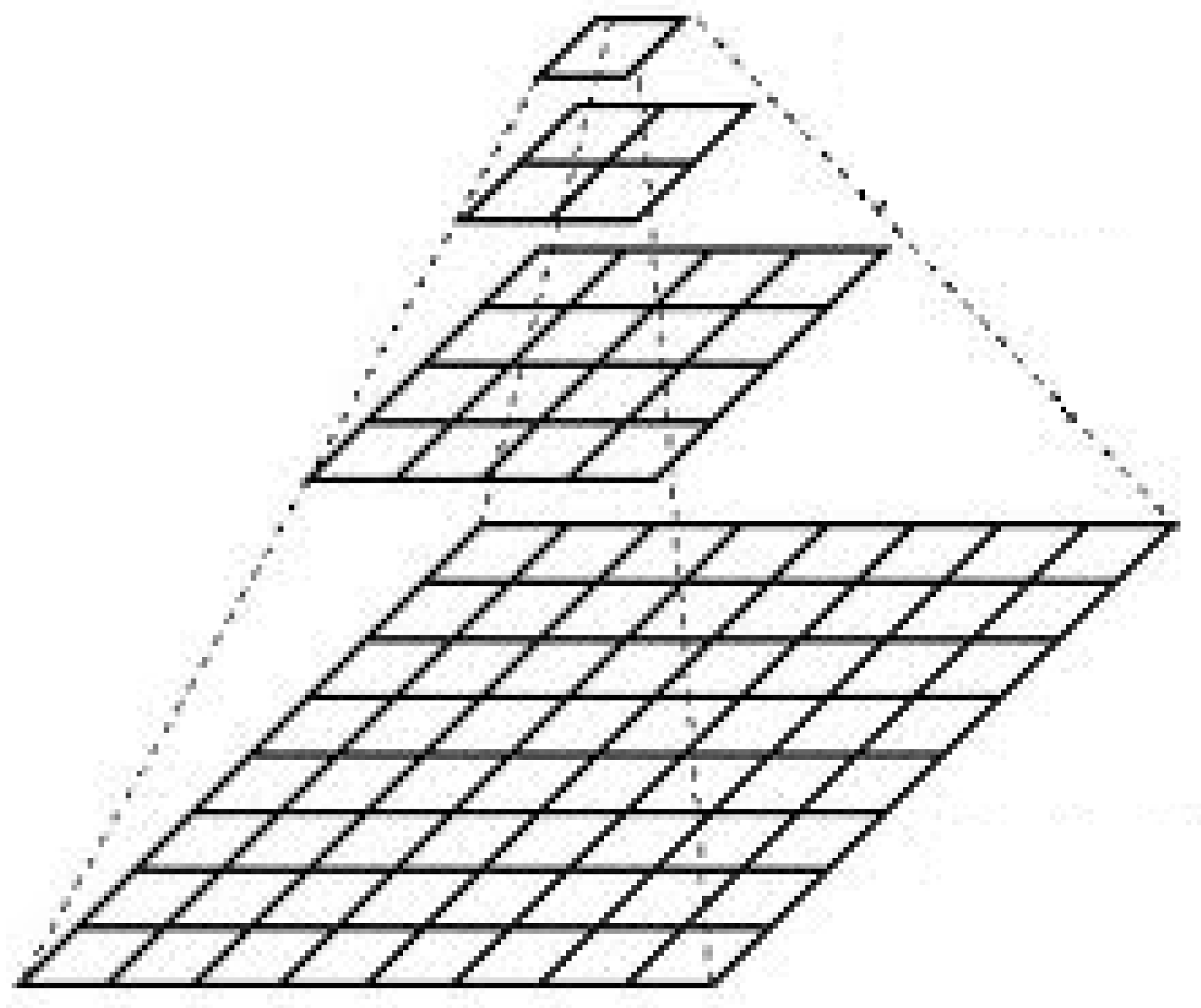
- Base image: The highest-resolution image, which contains the most detail.
- Pyramidal levels: The original image is broken down into lower-resolution versions at different levels, with each level containing an image of progressively reduced size and resolution.
- Processing software: The image pyramid is often used in software that allows navigation between different levels, so the user can quickly switch between high-resolution and low-resolution images.
- The complexity of establishing a diagnosis;
- The need to analyze huge amounts of complex clinical data;
- The great progress in computer science.
- Developing and controlling workflows from the pre-analytical until post-analytical phases;
- Managing resources;
- Performing assessments and continuous improvement to ensure consistent quality results.
1.2. Sensing Methods in Medicine
- Automatically calculate how much of each image has been examined by the professional;
- Determine how much time has been spent on the different areas of the sample;
- Determine which areas may have been missed during processing.
1.3. Consistency and Reproducibility of Medical Diagnoses
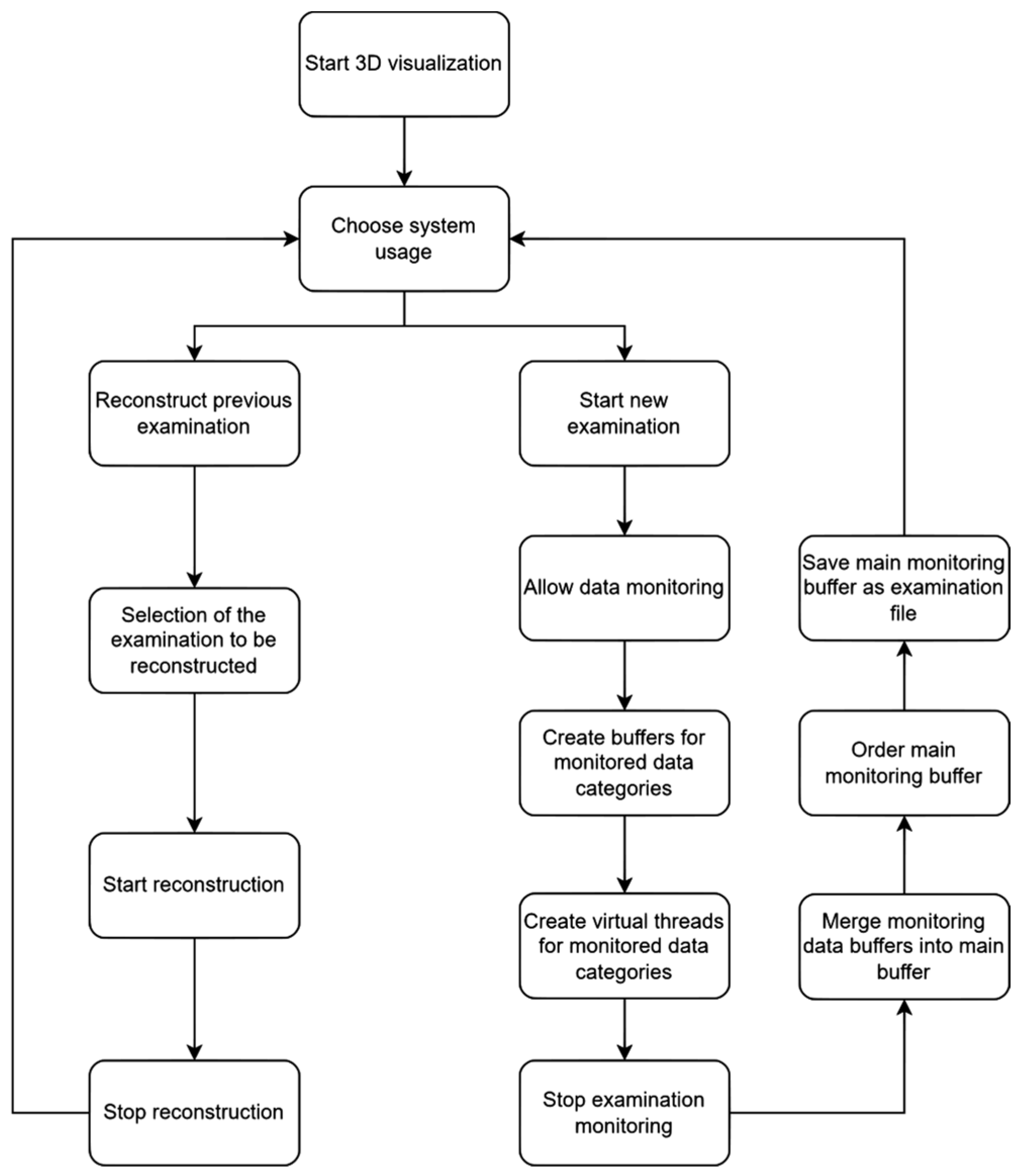
2. Materials and Methods
3. Results
- Monitoring and reproduction of 3D diagnosis;
- Millisecond data recording in 3D virtual reality environment;
- Introduction and definition of primary and adjacent zooming areas;
- Determination of sample evaluation by eye-tracking.
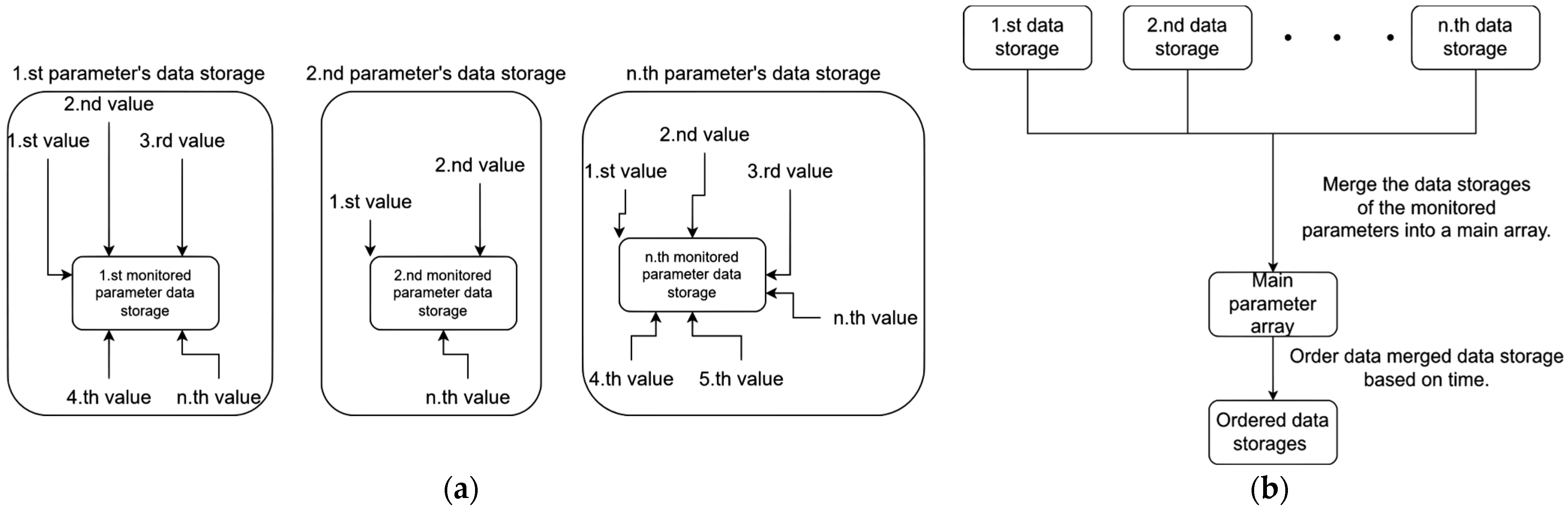
3.1. Parameter Recording During 3D Medical Sample Evaluation
- User movement in 3D space;
- User rotation in 3D space;
- The user’s left hand’s movement in 3D;
- The user’s left hand’s rotation in 3D;
- The user’s right hand’s movement in 3D;
- The user’s right hand’s rotation in 3D;
- The movement of the medical sample in 3D;
- The rotation of the medical sample in 3D;
- The graphical user interface usage in the software;
- The medical sample loading.
3.2. Testing the Parameter Monitoring
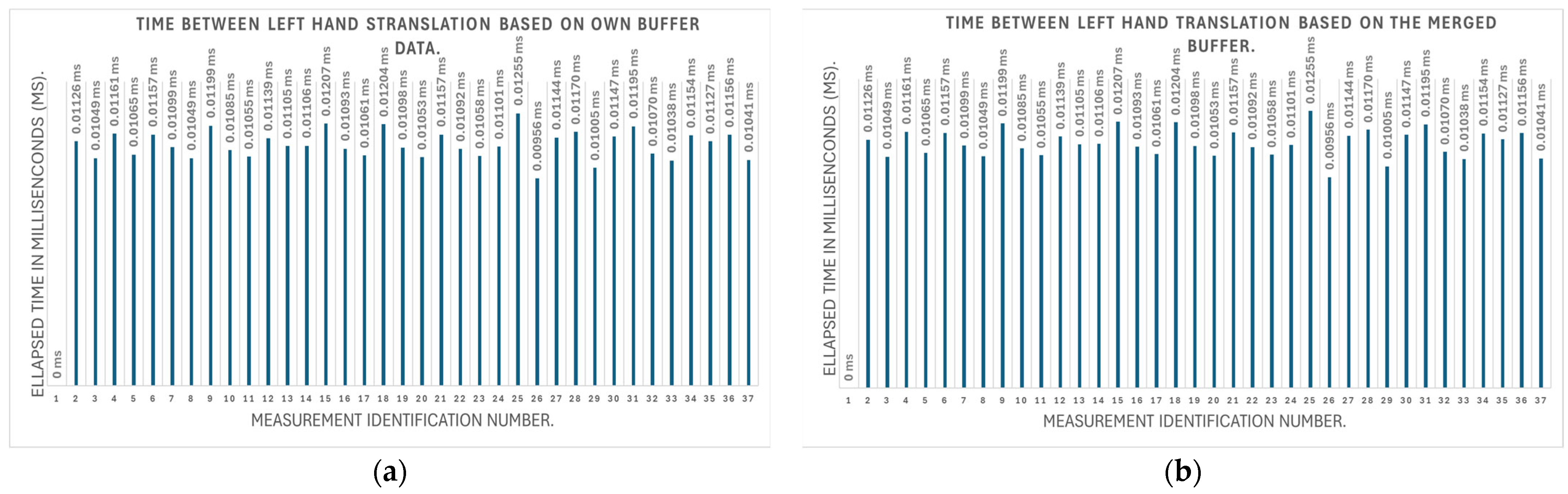
3.2.1. Testing the Accuracy of Parameter Monitoring
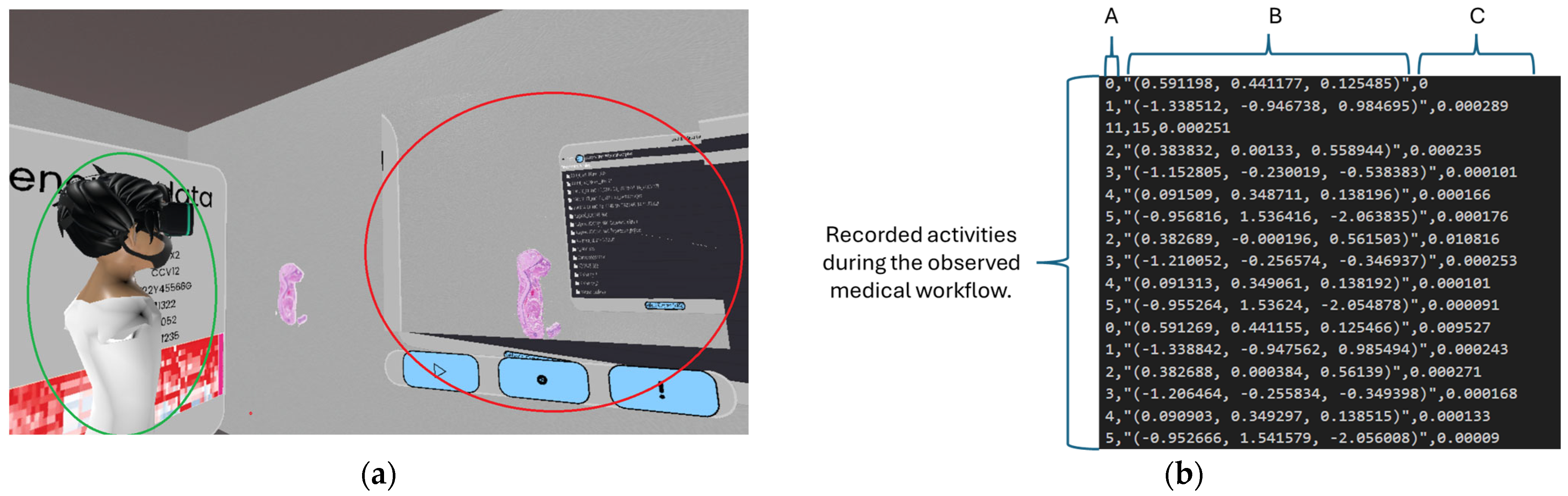
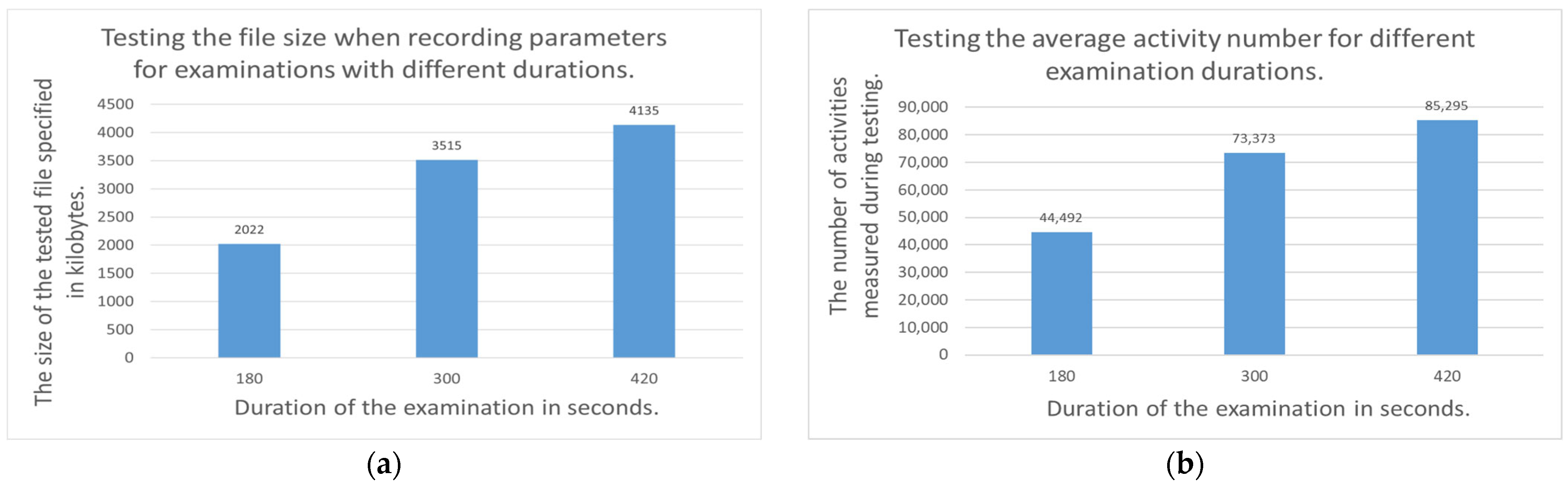
3.2.2. Examination of the Recording File Generated During Parameter Monitoring
3.3. The Definition of Primary and Adjacent Zooming Areas in 3D Medical Image Evaluation
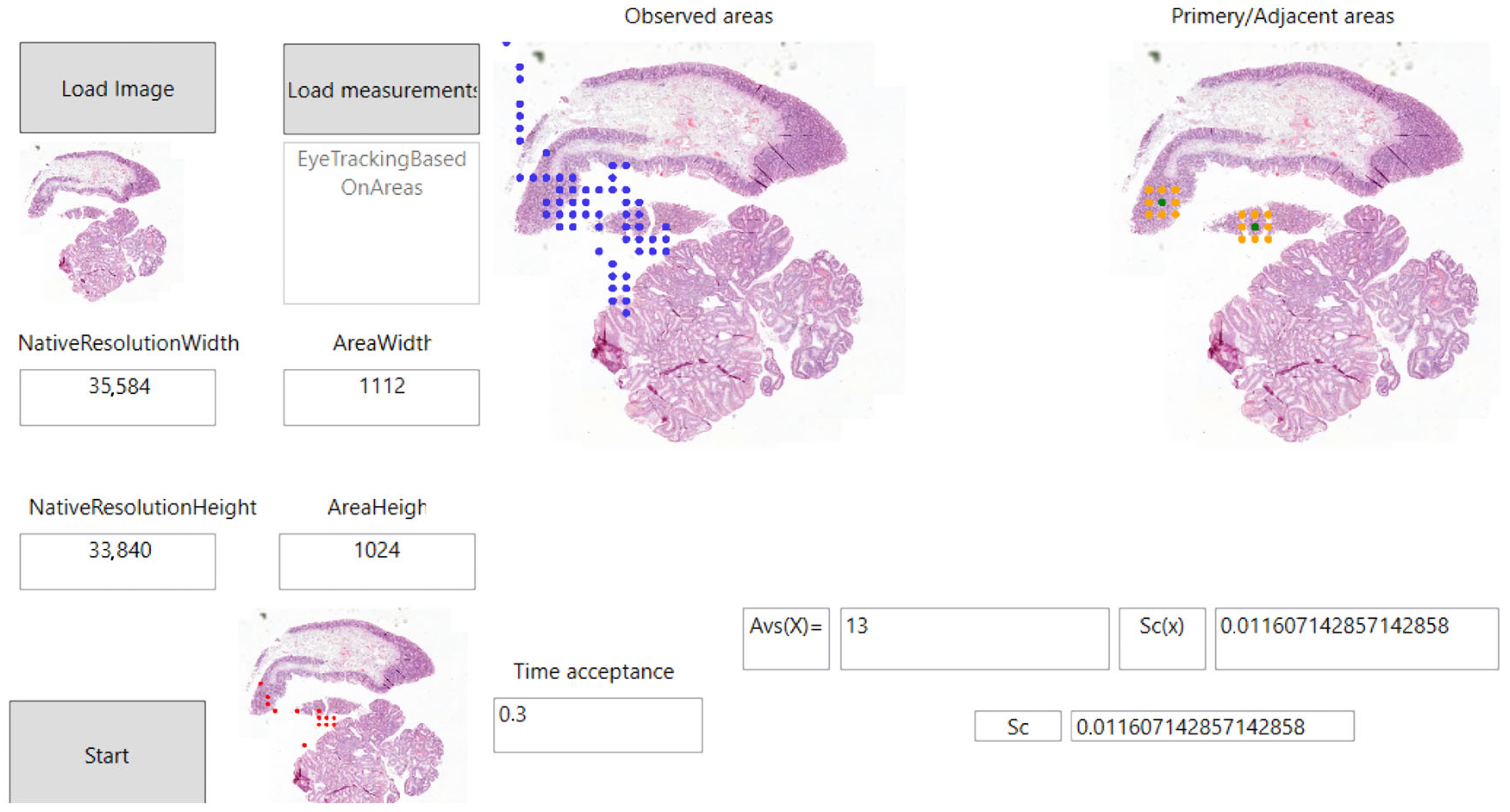
- Primary zooming area: We consider areas on which the user has applied a magnification function as the primary area in our research. The area around the selected area can be examined in 3D by the user in the depth of the full serial section. Primary zooming areas can be seen in Figure 9 with a green color.
- Adjacent zooming area: In our research, we consider as adjacent zooming areas those areas that have been displayed in 3D by the user, but for which the user did not zoom in directly, but on one of their adjacent areas. This is shown in orange in Figure 9.
- : Matrix containing the coordinates of the primary and adjacent areas;
- : The total number of samples in the total pathological serial section;
- : The index of the currently selected sample in the series section.
3.4. Using Eye-Tracking for the Quality Assurance of Medical Image Processing
- Vive Focus 3 VR device;
- Vive Focus 3 Eye tracker extension.
- : Evaluation status of areas currently being examined;
- : Time spent on the area being examined;
- : The limit value at which the area currently being examined is considered to have been evaluated.
- : The evaluation status of a given digitized pathology sample;
- : The total number of areas in a given digitized pathology sample.
- : The total number of samples in the total pathological serial section;
- : The number of areas in a given sample of a digitized pathology serial section.
3.5. Testing the Eye-Tracking Solution and the Primary and Adjacent Areas
- Total number of areas viewed by the user;
- How many times did the user look inside the defined testing area;
- Total time spent by the user reviewing the defined test area;
- Total time spent examining parts of the tissue outside the defined testing area.
4. Discussion
- Full sessions can be shared:Sharing full sessions can make it easier to obtain second/third opinions. The user can share the entire recorded 3D session with a colleague, so that his/her colleague will see exactly what he/she saw in 3D. This solution avoids misunderstandings when diagnosing image data.
- Verifiability of the medical image data examination process:Since the entire 3D session is recorded, we can reconstruct past 3D evaluations using different algorithms. This allows us to examine which parts of the sample were examined by the physician and for how long. We can also check whether the whole area of the sample was examined when making the diagnosis, and whether any parts were excluded.
- Improving medical education materials:Three-dimensional VR sessions captured with our solution could be suitable for use in medical education. It is sufficient for the instructor/researcher to record a complete session only once, which can then be shared with students. The students can then replay the 3D diagnostic session recorded by the instructor as many times as they wish.
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Angel Arul Jothi, J.; Mary Anita Rajam, V. A survey on automated cancer diagnosis from histopathology images. Artif. Intell. Rev. 2017, 48, 31–81. [Google Scholar] [CrossRef]
- Pena, G.; Andrade-Filho, J. How Does a Pathologist Make a Diagnosis? Arch. Pathol. Lab. Med. 2009, 133, 124–132. [Google Scholar] [CrossRef]
- Saalfeld, S.; Saalfeld, P.; Berg, P.; Merten, N.; Preim, B. How to Evaluate Medical Visualizations on the Example of 3D Aneurysm Surfaces. In Proceedings of the Eurographics Workshop on Visual Computing for Biology and Medicine, Bergen, Norway, 7–9 September 2016. [Google Scholar]
- Smith, B.; Hermsen, M.; Lesser, E.; Ravichandar, D.; Kremers, W. Developing image analysis pipelines of whole-slide images: Pre- and post-processing. J. Clin. Trans. Sci. 2021, 5, e38. [Google Scholar] [CrossRef] [PubMed]
- Sorell, T.; Li, R. Digital Pathology Scanners and Contextual Integrity. Digit. Soc. 2023, 2, 56. [Google Scholar] [CrossRef]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef]
- Sobirov, I.; Saeed, N.; Yaqub, M. Super Images—A New 2D Perspective on 3D Medical Imaging Analysis. arXiv 2023, arXiv:2205.02847. [Google Scholar]
- Wang, S.; Feng, W.; Guo, W. A Survey on 3D Medical Image Visualization. Adv. Mater. Res. 2012, 546–547, 416–419. [Google Scholar] [CrossRef]
- Gao, Y.; Chen, X.; Yang, Q.; Lasso, A.; Kolesov, I.; Pieper, S.; Kikinis, R.; Tannenbaum, A.; Zhu, L. An effective and open source interactive 3D medical image segmentation solution. Sci. Rep. 2024, 14, 29878. [Google Scholar] [CrossRef]
- Falk, M.; Ynnerman, A.; Treanor, D.; Lundström, C. Interactive Visualization of 3D Histopathology in Native Resolution. IEEE Trans. Vis. Comput. Graph. 2018, 25, 1008–1017. [Google Scholar] [CrossRef]
- Umirzakova, S.; Ahmad, S.; Khan, L.U.; Whangbo, T. Medical image super-resolution for smart healthcare applications: A comprehensive survey. Inf. Fusion 2024, 103, 102075. [Google Scholar] [CrossRef]
- Marques Godinho, T.; Lebre, R.; Silva, L.B.; Costa, C. An efficient architecture to support digital pathology in standard medical imaging repositories. J. Biomed. Inform. 2017, 71, 190–197. [Google Scholar] [CrossRef]
- Ruusuvuori, P.; Valkonen, M.; Kartasalo, K.; Valkonen, M.; Visakorpi, T.; Nykter, M.; Latonen, L. Spatial analysis of histology in 3D: Quantification and visualization of organ and tumor level tissue environment. Heliyon 2022, 8, e08762. [Google Scholar] [CrossRef] [PubMed]
- Adelson, E.H.; Anderson, C.H.; Bergen, J.R.; Burt, P.J.; Ogden, J.M. Pyramid methods in image processing. RCA Eng. 1984, 29, 33–41. [Google Scholar]
- Ashman, K.; Zhuge, H.; Shanley, E.; Fox, S.; Halat, S.; Sholl, A.; Summa, B.; Brown, J.Q. Whole slide image data utilization informed by digital diagnosis patterns. J. Pathol. Inf. 2022, 13, 100113. [Google Scholar] [CrossRef]
- Arimura, H.; Magome, T.; Yamashita, Y.; Yamamoto, D. Computer-Aided Diagnosis Systems for Brain Diseases in Magnetic Resonance Images. Algorithms 2009, 2, 925. [Google Scholar] [CrossRef]
- Chan, H.-P.; Doi, K.; Galhotra, S.; Vyborny, C.J.; MacMahon, H.; Jokich, P.M. Image feature analysis and computer-aided diagnosis in digital radiography. I. Automated detection of microcalcifications in mammography. Med. Phys. 1987, 14, 538–548. [Google Scholar] [CrossRef]
- Giger, M.L.; Doi, K.; MacMahon, H. Image feature analysis and computer-aided diagnosis in digital radiography. 3. Automated detection of nodules in peripheral lung fields. Med. Phys. 1988, 15, 158–166. [Google Scholar] [CrossRef]
- Yanase, J.; Triantaphyllou, E. A Systematic Survey of Computer-Aided Diagnosis in Medicine: Past and Present Developments. Expert Syst. Appl. 2019, 138, 112821. [Google Scholar] [CrossRef]
- Williams, B.; Knowles, C.; Treanor, D. Maintaining quality diagnosis with digital pathology: A practical guide to ISO 15189 accreditation. J. Clin. Pathol. 2019, 72, 663–668. [Google Scholar] [CrossRef]
- Mcauliffe, M.; Lalonde, F.; McGarry, D.P.; Gandler, W.; Csaky, K.; Trus, B. Medical Image Processing, Analysis & Visualization in Clinical Research. In Proceedings of the 14th IEEE Symposium on Computer-Based Medical Systems, CBMS 2001, Bethesda, MD, USA, 26–27 July 2001; Volume 14, p. 386. [Google Scholar] [CrossRef]
- Chong, Y.; Bae, J.; Kang, D.W.; Kim, G.; Han, H. Development of quality assurance program for digital pathology by the Korean Society of Pathologists. J. Pathol. Transl. Med. 2022, 56, 370–382. [Google Scholar] [CrossRef]
- Weng, Z.; Seper, A.; Pryalukhin, A.; Mairinger, F.; Wickenhauser, C.; Bauer, M.; Glamann, L.; Bläker, H.; Lingscheidt, T.; Hulla, W.; et al. GrandQC: A comprehensive solution to quality control problem in digital pathology. Nat. Commun. 2024, 15, 10685. [Google Scholar] [CrossRef]
- Aeffner, F.; Zarella, M.D.; Buchbinder, N.; Bui, M.M.; Goodman, M.R.; Hartman, D.J.; Lujan, G.M.; Molani, M.A.; Parwani, A.V.; Lillard, K.; et al. Introduction to Digital Image Analysis in Whole-slide Imaging: A White Paper from the Digital Pathology Association. J. Pathol. Inform. 2019, 10, 9. [Google Scholar] [CrossRef] [PubMed]
- Xi, C.; Cao, D. Quality management in anatomic pathology: The past, present, and future. iLABMED 2023, 1, 75–81. [Google Scholar] [CrossRef]
- Wright, A.I.; Dunn, C.M.; Hale, M.; Hutchins, G.G.A.; Treanor, D.E. The Effect of Quality Control on Accuracy of Digital Pathology Image Analysis. IEEE J. Biomed. Health Inf. 2021, 25, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Brixtel, R.; Bougleux, S.; Lézoray, O.; Caillot, Y.; Lemoine, B.; Fontaine, M.; Nebati, D.; Renouf, A. Whole Slide Image Quality in Digital Pathology: Review and Perspectives. IEEE Access 2022, 10, 131005–131035. [Google Scholar] [CrossRef]
- World Health Organization. Laboratory Quality Management System: Handbook; World Health Organization: Geneva, Switzerland, 2011; p. 247. [Google Scholar]
- Beard, D.V.; Johnston, R.E.; Toki, O.; Wilcox, C. A study of radiologists viewing multiple computed tomography examinations using an eyetracking device. J. Digit. Imag. 1990, 3, 230–237. [Google Scholar] [CrossRef]
- Suwa, K.; Furukawa, A.; Matsumoto, T.; Yosue, T. Analyzing the eye movement of dentists during their reading of CT images. Odontology 2001, 89, 54–61. [Google Scholar] [CrossRef]
- Kundel, H.L.; Nodine, C.F.; Krupinski, E.A.; Mello-Thoms, C. Using gaze-tracking data and mixture distribution analysis to support a holistic model for the detection of cancers on mammograms. Acad. Radiol. 2008, 15, 881–886. [Google Scholar] [CrossRef]
- Holmqvist, K.; Nyström, M.; Andersson, R.; Dewhurst, R.; Jarodzka, H.; Van de Weijer, J. Eye Tracking: A Comprehensive Guide to Methods and Measures; Oxford University Press: Oxford, UK, 2011; Available online: https://global.oup.com/academic/product/eye-tracking-9780199697083?cc=nl&lang=en& (accessed on 18 March 2025).
- Gong, H.; Hsieh, S.S.; Holmes, D.R., III; Cook, D.A.; Inoue, A.; Bartlett, D.J.; Baffour, F.; Takahashi, H.; Leng, S.; Yu, L.; et al. An interactive eye-tracking system for measuring radiologists’ visual fixations in volumetric CT images: Implementation and initial eye-tracking accuracy validation. Med. Phys. 2021, 48, 6710–6723. [Google Scholar] [CrossRef]
- Leveque, L.; Bosmans, H.; Cockmartin, L.; Liu, H. State of the Art: Eye-Tracking Studies in Medical Imaging. IEEE Access 2018, 6, 37023–37034. [Google Scholar] [CrossRef]
- Wang, S.; Ouyang, X.; Liu, T.; Wang, Q.; Shen, D. Follow My Eye: Using Gaze to Supervise Computer-Aided Diagnosis. IEEE Trans. Med. Imaging 2022, 41, 1688–1698. [Google Scholar] [CrossRef] [PubMed]
- Sqalli, M.T.; Aslonov, B.; Gafurov, M.; Mukhammadiev, N.; Sqalli Houssaini, Y. Eye tracking technology in medical practice: A perspective on its diverse applications. Front. Med. Technol. 2023, 5, 1253001. [Google Scholar] [CrossRef]
- Lev, A.; Braw, Y.; Elbaum, T.; Wagner, M.; Rassovsky, Y. Eye tracking during a continuous performance test: Utility for assessing ADHD patients. J. Atten. Disord. 2020, 26, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.-H.A.; Ghazi, A.; Sridhar, A.; Stoyanov, D.; Slack, M.; Kelly, J.D.; Collins, J.W. Evolving robotic surgery training, improving patient safety, with the integration of novel technologies. World J. Urol. 2020, 39, 2883–2893. [Google Scholar] [CrossRef]
- Picton, T. The P300 Wave of the Human Event-Related Potential. J. Clin. Neurophysiol. Off. Publ. Am. Electroencephalogr. Soc. 1992, 9, 456–479. [Google Scholar] [CrossRef]
- Polich, J.; Herbst, K. P300 as a clinical assay: Rationale, evaluation, and findings. Int. J. Psychophysiol. Off. J. Int. Organ. Psychophysiol. 2000, 38, 3–19. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Sodani, A.K.; Chouksey, D.; Jain, R. Value of P300 as a Screening Tool of Cognitive Impairment in Epilepsy: A Prospective Study from India. Rom. J. Neurol. 2021, 20, 161–168. [Google Scholar] [CrossRef]
- Pan, J.-B.; Takeshita, T.; Morimoto, K. P300 as a measure of cognitive dysfunction from occupational and environmental insults. Env. Health Prev. Med. 1999, 4, 103–110. [Google Scholar] [CrossRef]
- Jackson, S.L.; Frederick, P.D.; Pepe, M.S.; Nelson, H.D.; Weaver, D.L.; Allison, K.H.; Carney, P.A.; Geller, B.M.; Tosteson, A.N.; Onega, T.; et al. Diagnostic Reproducibility: What Happens When the Same Pathologist Interprets the Same Breast Biopsy Specimen at Two Points in Time? Ann. Surg. Oncol. 2017, 24, 1234–1241. [Google Scholar] [CrossRef]
- Haggenmüller, S.; Wies, C.; Abels, J.; Winterstein, J.T.; Heinlein, L.; Nogueira Garcia, C.; Utikal, J.S.; Wohlfeil, S.A.; Meier, F.; Hobelsberger, S.; et al. Discordance, accuracy and reproducibility study of pathologists’ diagnosis of melanoma and melanocytic tumors. Nat. Commun. 2025, 16, 789. [Google Scholar] [CrossRef]
- The Site of the SideMatch Software. Available online: https://www.micro-dimensions.com/slidematch (accessed on 29 February 2024).
- Vincze, M.; Molnar, B.; Kozlovszky, M. 3D Visualization in Digital Medicine Using XR Technology. Future Internet 2023, 15, 284. [Google Scholar] [CrossRef]
- Biricz, B.; Jónás, V.; Vincze, M.; Benhamida, A.; Paulik, R.; Kozlovszky, M. User friendly virtual reality software development and testing. In Proceedings of the 2022 13th IEEE International Conference on Cognitive Infocommunications (CogInfoCom), Budapest, Hungary, 21–23 September 2022; pp. 000087–000092. [Google Scholar] [CrossRef]
- Monitoring the Examination of Digitised Pathology Samples in a 3D VR Environment (MTMT). Available online: https://m2.mtmt.hu/gui2/?mode=browse¶ms=publication;35167933 (accessed on 17 February 2025).
- Padmapriya, S.T.; Parthasarathy, S. Ethical Data Collection for Medical Image Analysis: A Structured Approach. ABR 2024, 16, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Manjunath, K.N.; Rajaram, C.; Hegde, G.; Kulkarni, A.; Kurady, R.; Manuel, K. A Systematic Approach of Data Collection and Analysis in Medical Imaging Research. Asian Pac. J. Cancer Prev. APJCP 2021, 22, 537–546. [Google Scholar] [CrossRef]
- Kiryati, N.; Landau, Y. Dataset Growth in Medical Image Analysis Research. J. Imaging 2021, 7, 155. [Google Scholar] [CrossRef]
- Egevad, L.; Cheville, J.; Evans, A.J.; Hörnblad, J.; Kench, J.G.; Kristiansen, G.; Leite, K.R.; Magi-Galluzzi, C.; Pan, C.C.; Samaratunga, H.; et al. Pathology Imagebase—A reference image database for standardization of pathology. Histopathology 2017, 71, 677–685. [Google Scholar] [CrossRef]



| Tester ID | Number of All Measurements | Number of Measurements Within the Testing Area | Time Spent Inside the Testing Area. (Second) | Time Spent Outside the Testing Area. (Second) | Measurements in the Test Area (%) |
|---|---|---|---|---|---|
| 1 | 238 | 49 | 123,131 | 17,003 | 20.58 |
| 2 | 124 | 42 | 118,558 | 12,211 | 33.87 |
| 3 | 126 | 25 | 19,919 | 11,411 | 19.84 |
| Findings from the Research Used in the Current Study. | Results of the Actual Study |
|---|---|
| In the article, the authors state that the analysis and evaluation of digitized pathology samples is typically performed manually by highly skilled physicians/researchers [51]. | In the article, the authors state that the analysis and evaluation of digitized pathology samples is typically performed manually by highly skilled physicians/researchers. We fully agree with this statement, which is why we have created a system capable of monitoring, saving, and reconstructing the evaluation of a skilled pathologist. |
| The article notes that there are currently few quality assurance programs for users in digital pathology [24]. | We agree with the findings of the research, but the number of such programs is growing. The aim of our research presented in this paper was also to design and build a system that can track and score the evaluation status of a given serial section. |
| In their paper, the authors present the design and creation of a reference image database in which pathologists can further train themselves [52]. | In our research, we designed and built a system similar to the one in the linked article. The presented system can capture and share complete 3D diagnostic sessions, allowing doctors/researchers to further train themselves by examining the diagnostic routine of a colleague. |
| In their paper, the authors mention that gaze-tracking is a subjective parameter and that there is a large variation between users. The authors suggest that a separate solution should be developed to increase the accuracy of gaze-tracking [35]. | We agree with the author’s statement. Consequently, our future development goals include the design and development of a system that eliminates variability in input data. |
| The authors present a solution that can find and categorize artifacts that may be generated during the preparation of tissue samples [23]. Detection and identification of these defects can improve the quality of diagnosis. | The authors’ solution and the solution presented in this paper can complement each other well, to provide the widest possible range of quality control for the evaluation of tissue samples. The authors’ work can detect and categorize artifacts generated during sample preparation, while the solution presented in this paper can capture the diagnoses performed on samples that are considered good. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vincze, M.; Molnár, B.; Kozlovszky, M. Quality Assurance of the Whole Slide Image Evaluation in Digital Pathology: State of the Art and Development Results. Electronics 2025, 14, 1943. https://doi.org/10.3390/electronics14101943
Vincze M, Molnár B, Kozlovszky M. Quality Assurance of the Whole Slide Image Evaluation in Digital Pathology: State of the Art and Development Results. Electronics. 2025; 14(10):1943. https://doi.org/10.3390/electronics14101943
Chicago/Turabian StyleVincze, Miklós, Béla Molnár, and Miklós Kozlovszky. 2025. "Quality Assurance of the Whole Slide Image Evaluation in Digital Pathology: State of the Art and Development Results" Electronics 14, no. 10: 1943. https://doi.org/10.3390/electronics14101943
APA StyleVincze, M., Molnár, B., & Kozlovszky, M. (2025). Quality Assurance of the Whole Slide Image Evaluation in Digital Pathology: State of the Art and Development Results. Electronics, 14(10), 1943. https://doi.org/10.3390/electronics14101943






