Abstract
Medical image segmentation is a critical task in image analysis and plays an essential role in computer-aided diagnosis. Despite the promising performance of hybrid models combining U-Net and transformer architectures, these approaches face challenges in extracting local features and optimizing attention mechanisms. To address these limitations, we propose the Depthwise Composite Transformer and Depthwise Polarized Attention Network (DC-TransDPANet), a novel framework designed for medical image segmentation. The proposed DC-TransDPANet introduces a Depthwise Composite Attention Module (DW-CAM), which integrates depthwise convolution, and a Composite Attention mechanism to enhance local feature extraction and fuse contextual information. Additionally, a Depthwise Polarized Attention (DPA) block is employed to improve global context representation while preserving high-resolution details, achieving a fine balance between local and global feature extraction. Extensive experiments on benchmark datasets demonstrate that DC-TransDPANet significantly outperforms existing methods in segmentation accuracy.
1. Introduction
Deep learning models, particularly Convolutional Neural Networks (CNNs) like U-Net [1], have achieved notable success in segmentation across various modalities, such as radiography [2], endoscopy [3], and computed tomography (CT) [4,5]. Despite their advancements, CNNs often suffer from limited receptive fields, making it challenging to capture long-range dependencies and global context, which are crucial for accurate segmentation in boundary-rich and small-structure-dense regions.
Transformers [6], initially developed for Natural Language Processing, have been successfully applied to various Computer Vision tasks. In particular, the Vision Transformer (ViT) [7] has demonstrated superior performance in image classification and segmentation by capturing long-range dependencies and global context, thereby addressing the locality limitation of CNNs. Medical segmentation models like TransUnet [8] and Swin-Unet [9] have achieved promising results in medical image segmentation by leveraging transformer-based architectures. To mitigate the feature resolution degradation introduced by transformers, TransUNet [8] integrates CNNs and transformers in a hybrid architecture. This enables the model to preserve high-resolution spatial information from CNNs while harnessing the global context encoded by transformers. Swin-Unet [9] builds upon the Swin Transformer, utilizing hierarchical feature representation and shifted window attention mechanisms to enhance computational efficiency and improve segmentation accuracy. However, these models still face significant challenges, including limited efficiency in capturing multi-scale and hierarchical features, which are vital for accurate medical image segmentation.
We propose a novel method, named DC-TransDPANet, to address the limitations of existing medical image segmentation models. The method consists of two main components: an encoder and a decoder. The encoder integrates CNN-based feature extraction with transformer-based global modeling, leveraging the local feature capturing capabilities of convolutional networks and the global context modeling power of transformers. The initial CNN layers capture local features. The Depthwise Composite Attention Module (DW-CAM) combines depthwise convolution [10] with a Composite Attention Module (CAM) to strengthen local and intermediate feature representation. The Depthwise Polarized Attention (DPA) module refines the feature representation further by incorporating depthwise convolution with Polarized Self-Attention, addressing the challenge of global context modeling while preserving high-resolution spatial details. The decoder progressively restores spatial details through a combination of upsampling, skip connections, and feature fusion, enabling precise segmentation with well-defined anatomical boundaries. This process ensures accurate reconstruction of high-resolution features from the input image, facilitating precise segmentation in medical imaging tasks.
We conducted extensive experiments on the Synapse, CVC-ClinicDB, and Kvasir-SEG datasets, where DC-TransDPANet outperformed existing state-of-the-art methods in segmentation accuracy. Additionally, ablation studies were performed to evaluate the impact of different kernel sizes in the DW-CAM module and various combinations of loss functions, further validating the effectiveness of the proposed framework. The key contributions include the following:
- We develop the DW-CAM, a novel transformer-based encoder that integrates depthwise convolution with CAM, effectively capturing local features and enhancing the segmentation of small and complex structures.
- We introduce the DPA module, which leverages depthwise convolution and Polarized Self-Attention to strengthen global feature representation and preserve high-resolution information. Extensive ablation studies demonstrate that our framework improves accuracy.
2. Ralated Works
2.1. CNN-Based Image Segmentation
Convolution-based segmentation models laid the foundation for modern approaches. FCNs (Fully Convolutional Networks) [11] pioneered end-to-end segmentation by replacing fully connected layers with convolutional layers, enabling pixel-wise predictions. Later, U-Net [1] introduced an encoder–decoder structure with symmetric skip connections, significantly improving segmentation performance, especially in biomedical imaging. Subsequent U-Net variants refined its architecture to address specific limitations. On this basis, U-Net++ [12] introduced nested skip connections, enabling more precise feature fusion across different scales, which proved beneficial for segmenting complex structures with fine-grained boundaries. In parallel, Res-UNet [13] incorporated residual connections, mitigating the vanishing gradient problem and enhancing training stability in deeper architectures. On the other hand, Dense-UNet [14] further optimized feature propagation by integrating dense blocks, reducing redundant parameters while maintaining high segmentation accuracy. Moving forward, R2U-Net [15] leveraged recurrent connections to enhance feature representation over sequential layers, capturing richer contextual information. Acc-UNet [16] revisited the classic U-Net architecture, integrating modern convolutional techniques to enhance feature extraction and segmentation accuracy. Additionally, TinyU-Net [17] optimized the U-Net architecture by incorporating cascaded multi-receptive fields, achieving high segmentation accuracy with a significantly reduced model size, ideal for resource-limited environments. Lastly, MAGRes-UNet [18] advanced medical image segmentation by incorporating four multi-attention gate (MAG) modules and residual blocks into a standard U-Net structure, while employed Mish and ReLU activation functions to enhance feature learning and segmentation accuracy.
Recently, various methods have been proposed to integrate attention mechanisms into U-Net to enhance segmentation performance by improving feature selection and contextual awareness. Examples such as Attention Res-UNet [19] have utilized soft attention [20], which effectively enhances the network’s ability to segment structures with subtle intensity variations. However, the reliance on soft attention can increase the sensitivity to noise, particularly in low-contrast regions, potentially impacting robustness. Similarly, SA-UNet [21] integrated spatial attention mechanisms to focus on specific locations within the image, achieving higher segmentation accuracy for regions with irregular or diffuse boundaries. Following this, SelfRegUNet [22] incorporated self-regularization mechanisms within the U-Net framework, improving generalization in diverse medical imaging datasets. EmCAD [23] combined multi-scale convolutional attention with efficient decoding strategies, optimizing both accuracy and computational cost. Subsequently, WMV-KM-L2 [24], a weighted multiview k-means algorithm, incorporated feature and view weights to improve the clustering of multiview data, outperformed existing methods in seven diverse benchmark datasets. Lastly, DCSSGA-UNet [25] leveraged DenseNet201 with channel spatial attention (CSA) and semantic guidance attention (SGA) mechanisms, improved biomedical image segmentation by addressing lesion variability and semantic gaps, and achieved high precision in medical datasets.
2.2. Transformer-Based Works in Medical Image Segmentation
Recently, transformer-based models have emerged as a promising alternative in medical image segmentation. The Vision Transformer (ViT) [7] introduced a novel approach to image classification, which paved the way for subsequent variants such as DeiT [26], Swin Transformer [27], and LeViT [28], each of which further advanced the application of transformer-based models for visual tasks.
The integration of transformers into U-Net architectures marked a significant breakthrough in medical image segmentation. TransUNet [8] was one of the first hybrid models to incorporate ViT with a U-Net backbone, demonstrating superior performance by combining CNN-based local feature extraction with transformer-based global context modeling. Following this, TransU-Net++ [29] further refined this approach by introducing attention-enhanced skip connections, improving multi-scale feature fusion across different resolutions. Another significant development was the replacement of convolutional operations with transformer blocks. Swin-Unet [9] leveraged Swin Transformer, which introduced a hierarchical attention mechanism, enabling effective multi-scale representation learning while maintaining computational efficiency. On this basis, DS-TransUNet [30] improved upon this by integrating the TIF (Token-based Interaction Fusion) module, enhancing the model’s ability to capture multi-scale contextual information more effectively. Additionally, to further optimize performance and efficiency, HiFormer [31] leveraged hierarchical transformer-based representations to model multi-scale features, enabling precise segmentation of medical images with varying anatomical scales. LVit [32] integrated language-guided vision transformers to leverage multimodal information, combining textual and visual cues to improve segmentation performance in medical imaging tasks. EmF-Former [33] introduced a transformer-based model optimized for memory efficiency, enabling effective segmentation of high-resolution medical images without excessive computational demands. Lastly, EFFResNet-ViT [34], a hybrid model, employed Grad-CAM visualizations to enhance interpretability and achieved outstanding performance on BT CE-MRI and retinal datasets.
However, balancing local and global feature extraction remains challenging. Convolutional operations can extract fine-grained local features but lack the capability to model long-range dependencies. In contrast, transformers provide robust global context.
Our DC-TransDPANet addresses these limitations with the DW-CAM module and the DPA module. DW-CAM leveraged depthwise convolutions and composite attention mechanisms to enhance local feature extraction while maintaining efficiency. Meanwhile, the DPA module combines depthwise convolutions with Polarized Self-Attention, refining global feature representation and preserving spatial consistency. These innovations enable DC-TransDPANet to achieve a balanced extraction of local and global features, significantly improving segmentation accuracy and efficiency in medical image analysis.
3. Methodology
The DC-TransDPANet framework proposed (Figure 1) consists of four key components: encoder, Depthwise Composite Attention Module (DW-CAM), Depthwise Polarized Attention (DPA) module, and decoder.
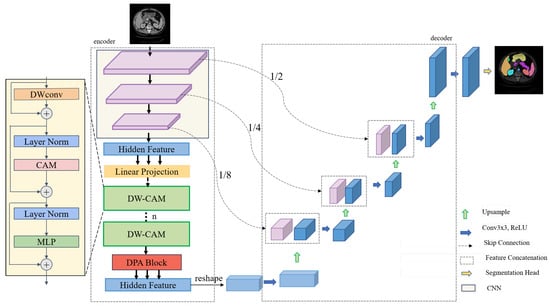
Figure 1.
The DC-TransDPANet framework utilizes an encoder–decoder architecture for medical image segmentation. The encoder extracts multi-scale features using convolutional neural networks, followed by transformer-based attention mechanisms, including the Depthwise Composite Attention Module (DW-CAM) and Depthwise Polarized Attention (DPA). The feature maps are downsampled to 1/2, 1/4, and 1/8 of the original resolution for multi-scale learning, integrating multi-scale information via the decoder. The final segmentation output is produced by the segmentation head.
3.1. Overview of the DC-TransDPANet Framework
DC-TransDPANet is a hybrid framework for medical image segmentation, designed to improve segmentation accuracy by combining convolutional and transformer-based mechanisms. The model takes an RGB image as input and produces a segmented output, where each pixel is classified into a specific category, thereby segmenting the image into meaningful regions (Figure 1).
The original transformer-based model relies heavily on the Multi-Scale Attention (MSA) layer to capture global dependencies. However, this approach cannot capture the depthwise spatial relationships needed for accurate medical image segmentation. This limitation may result in suboptimal feature representation, as the model struggles to balance preserving fine-grained spatial hierarchies with the need to leverage global context for accurate segmentation.
In DC-TransDPANet, the encoder first extracts local features using CNNs. These features are then processed by the Depthwise Composite Attention Module (DW-CAM), a hybrid block that combines depthwise convolutions with composite attention mechanisms. This combination significantly improves the model’s ability to capture both local and intermediate dependencies. The Depthwise Polarized Attention (DPA) block further refines feature fusion by addressing global dependencies in a more efficient manner, optimizing the model’s overall performance in segmentation tasks. This enhances the model’s robustness, particularly in segmenting complex structures within medical images. Finally, the decoder progressively restores the spatial resolution of the feature map through upsampling and skip connections, ensuring precise segmentation results.
Each of the core components—encoder, DW-CAM, DPA module, and decoder—plays a vital role in enhancing the segmentation accuracy, and they are described in greater detail in the following sections.
3.2. Encoder
As depicted in the left half of Figure 1, the encoder is a hybrid CNN-Transformer design. The CNN component extracts features via convolutional and downsampling layers, progressively reducing feature map size. After processing through the DW-CAM, the feature flows into the DPA: DW-CAM improves feature representation by integrating depthwise convolution with a Composite Attention Module (CAM), which combines Alterable Kernel Convolution (AKConv) [35], ParNet Attention (PA) [36], and Spatial and Channel Reconstruction Convolution (SCConv) [37] to capture multi-scale contextual information. The DPA improves global feature representation by combining spatial and channel attention. This hybrid approach balances local and global feature processing, optimizing encoder performance.
3.2.1. The Depthwise Composite Attention Module (DW-CAM)
In Figure 1, the DW-CAM is designed to address two primary limitations in hybrid models limited local feature extraction. The DW-CAM overcomes these challenges by incorporating the following:
- Depthwise convolution (DWConv): Enhances local feature extraction, capturing fine details more precisely.
- Composite Attention Module (CAM): Improves attention mechanisms and local feature extraction efficiency.
Depthwise convolution (DWConv): To enhance computational efficiency while preserving spatial information, we employ a depthwise convolution operation. Specifically, a depthwise convolution is utilized with a padding of 1 (stride = 1, padding = 1) to ensure that the spatial dimensions of the feature maps remain unchanged. The number of groups in the convolution is set equal to the number of input channels, allowing each channel to be processed independently. Compared to standard convolution, depthwise convolution significantly reduces the number of parameters and computational complexity while maintaining the integrity of spatial features, thus improving the overall computational efficiency of the model.
Composite Attention Module (CAM): As illustrated in Figure 2, the Composite Attention Module (CAM) represents an advanced attention mechanism designed to integrate spatial and channel information effectively. By incorporating geometric transformations, interactions between local and global features, and joint spatial–channel reconstruction, the CAM module dynamically enhances input features across multiple dimensions. Compared to conventional attention mechanisms, CAM demonstrates superior flexibility and adaptability and easily models features in complex scenarios while significantly reducing computational complexity, thus contributing to improved overall performance. The following is an introduction to AKConv, PA, and SCConv.
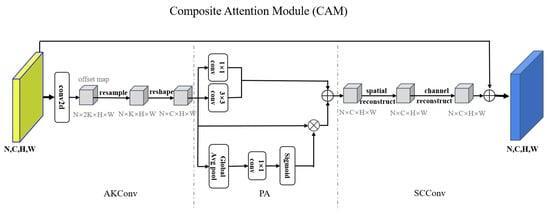
Figure 2.
Diagram of the proposed CAM architecture. It takes an input feature map and processes it by first using AKConv. Then, it applies attention to enhance feature focus. Next, it reconstructs spatial and channel features to reduce redundancy, producing an output feature map that is combined with the input via a residual connection.
Given an input feature map , where N is batch size, and H and W denote the height and width of the feature map, a 2D convolution is first applied to obtain an offset matrix. Before adjusting the sampling coordinates, an initial sampling coordinate set is generated, where K represents the number of sampling points (the kernel size), and each point is defined by two coordinate values (x,y). By adding the learned offsets to the initial sampling coordinates , the adjusted sampling coordinates are obtained as . Since these adjusted sampling coordinates may not align with integer grid positions, a resampling process is performed using bilinear interpolation to compute the feature values at the new locations. Finally, the resampled feature map is mapped to the standard channel dimension through a convolution, ensuring compatibility with subsequent network processing.
The output feature map is first processed using a convolution to extract channel-wise information and a convolution to capture spatial information; standard convolutions have stride = 1, padding = 0, and the convolution typically uses stride = 1 and padding = 1. Subsequently, global features are extracted by a global average pool (GAP), followed by a convolution for the reduction of dimensionality. A sigmoid activation function is then applied to generate attention weights for global attention computation. Next, attention-weighted refinement is performed by applying element-wise multiplication between the generated attention weights and the input feature map, thereby enhancing key regions. Finally, refined features are fused by aggregating the outputs of the convolution, convolution, and attention-weighted features through element-wise addition. This fusion process enhances target region information, improving the feature representation and its discriminative capability.
To optimize spatial and channel information, enhance feature representation capability, reduce redundant information, and improve computational efficiency, we introduce a Spatial Reconstruction Unit (SRU) and a Channel Reconstruction Unit (CRU). Given an input feature map , the SRU is first applied to separate primary information from redundant information and reconstruct the feature representation. Subsequently, the channel reconstruction unit is employed to further refine channel-wise information, thereby enhancing feature expressiveness. The final output is the optimized feature map .
The following describes the implementation steps of the SRU. To assess the significance of each channel, Group Normalization (GN) is employed to compute the weight of each channel:
In this process, represents the channel importance, which is utilized to distinguish between primary information and redundant information.
Based on the computed channel importance weights, the feature map is divided into two components:
where represents the primary information channels (high-weight features), while corresponds to the redundant information channels (low-weight features). The weight distribution satisfies to ensure the total information remains unchanged. and are weight masks derived from the channel importance vector (in Formula (1)). Element-wise multiplication (⊙) allows high-weight channels to retain more features while suppressing low-weight channels.
Finally, the spatial features are reconstructed by concatenating features from different sources to enhance information representation capability:
where ∪ represents channel concatenation, enabling the model to retain information from different perspectives simultaneously.
The spatially optimized feature map is fed into the CRU. Since the spatially optimized feature still contains redundant information, channel optimization is applied:
where represents the primary channel information (high-frequency features), and serves as the supplementary channel information (low-frequency features). The function divides the channels according to a split ratio.
Next, a transformation is performed using Group-Wise Convolution (GWC) and Point-Wise Convolution (PWC) to extract features:
where employs group-wise convolution to extract features and improve computational efficiency, while applies convolution to reduce channel dimensions. and represent the optimized channel features.
Finally, feature fusion is performed by computing Softmax weights to integrate the two types of channel information:
where and compute from global average pool, represented the importance scores of channel features, and compute from Softmax.
In summary, the CAM module effectively learns both local details and global contextual information from input features, thus enhancing the representational capacity of spatial and channel features. This multi-scale fusion approach significantly improves the model’s receptive field and the efficiency of information utilization. By adaptively generating weight distributions across spatial and channel dimensions based on the input content, the module dynamically models the relative importance of information. Through the integration of spatial reconstruction and channel reconstruction, the CAM module fully harnesses the potential of input features across multiple dimensions, ensuring comprehensive information fusion and substantially boosting the performance of downstream tasks.
3.2.2. The Depthwise Polarized Attention (DPA) Module
Figure 3 illustrates the Depthwise Polarized Attention (DPA) module, which is designed to effectively employ global context modeling to address the challenges of medical image segmentation. The module begins with a Depthwise Convolution (DWConv) layer, which allows the extraction of lightweight local features and preserves high-resolution details. Layer Normalization is applied in the channel attention branch () before the final sigmoid activation. Its purpose is to normalize the feature statistics across the channel dimension after the feature transformation ( conv) and potential reshaping/multiplication. This stabilizes training, prevents exploding/vanishing activations, and ensures the attention weights generated by the sigmoid function are in a well-behaved range, leading to more effective channel-wise feature refinement.
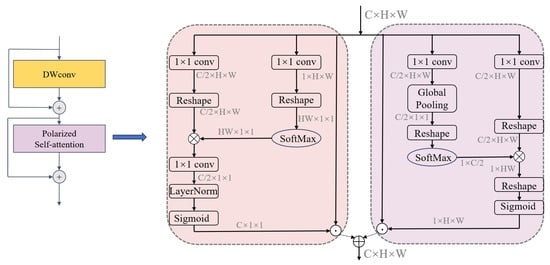
Figure 3.
Overall framework of the DPA module.
To further enhance feature representation, the DPA module employs a dual-branch mechanism that focuses on refining both spatial and channel information. This mechanism adaptively redistributes feature importance by dynamically learning the relevance of different spatial and channel dimensions. By combining these refined features, the DPA module achieves a comprehensive and robust representation of input features, balancing local detail preservation with global context integration.
In the channel dimension, the output features are represented as :
Here, , and are convolution layers, while and represent reshape operations. denotes the SoftMax function, the symbol “×” represents matrix multiplication, and represents the sigmoid function. The output channel dimension of , and is . Therefore, the output in the channel dimension is , and represents the multiplication operator in the channel dimension.
In the spatial dimension, the output features are represented as :
Here, and are both convolution layers, and and and are reshape operations. represents the Global Pooling operation. The symbol “×” indicates matrix multiplication. Thus, the output features in the spatial dimension are represented as , and denotes the multiplication operation in the spatial dimension. To reduce the risk of overfitting, we add Dropout layers after the channel partition and the spatial branch , respectively. The Dropout rate is set to 0.1, which means 10% of neurons are randomly discarded during training to enhance the adaptability of the model to complex medical images.
Fusion: To dynamically adjust the contribution of spatial and channel features, we introduce learnable weights and to perform weighted fusion. The output features from the two branches described above are fused in parallel:
Here, and are trainable parameters learned by the network with initial values set to 0.5 and optimized by backpropagation. The weights satisfy the constraint (implemented by softmax normalization) to ensure the stability of feature fusion.
Through its innovative design, the DPA module improves the segmentation accuracy by integrating multi-scale information and effectively enhancing critical features while suppressing redundant ones. This approach ensures precise and reliable delineation of anatomical structures in medical images, making it a powerful component in the proposed segmentation framework.
3.3. Decoder
Figure 1 illustrates the architecture of the decoder, where skip connections play a vital role in reconstructing image resolution from the feature maps generated by the encoder. The decoder employs a series of upsampling operations to progressively restore spatial resolution, doubling it at each stage while simultaneously decreasing the number of feature channels.
At each upsampling stage, skip connections facilitate the fusion of the upsampled feature maps with corresponding encoder feature maps. A subsequent convolutional layer refines these fused features, enhancing the extraction of segmentation-relevant information. The final refined feature map is processed by a segmentation head. This design effectively exploits high-dimensional encoder features, enabling the decoder to recover intricate image details and boundaries, thereby achieving superior segmentation performance.
4. Experiment
4.1. Dataset
Synapse multi-organ segmentation dataset (Synapse) [38]: This dataset contains 30 abdominal scans, totaling 3779 enhanced axial CT images, focusing on eight organs: liver, stomach, pancreas, left and right kidneys, aorta, spleen, and gallbladder.
CVC-ClinicDB dataset [39]: The CVC-ClinicDB dataset is widely used in medical image polyp segmentation tasks and serves as a benchmark for evaluating the performance of medical image segmentation tasks. This dataset consists of 612 static images extracted from colonoscopy videos, originating from 29 different sequences. Each image is accompanied by a groundtruth indicating the region covered by polyps within the image.
Kvasir-SEG dataset [40]: Kvasi-SEG is an open-access dataset and contains 1000 polyp images and their corresponding groudtruth, with a wide range of resolutions (from 332 × 487 to 1920 × 1072).
4.2. Implementation Settings
4.2.1. Experimental Details
Our model was developed using the PyTorch (version 1.13) framework and trained using an NVIDIA RTX 4090 GPU (NVIDIA Corporation, Santa Clara, CA, USA). To address overfitting, we employed data augmentation techniques, encompassing random image rotations within a range of −20 to +20 degrees and random horizontal flips applied with a probability of 0.5. The model was trained for 420 epochs on images with a resolution of , with a patch set to 16 (i.e., transformer) and a default batch size of 24.
Medical image segmentation datasets often exhibit class imbalance, where certain organs occupy significantly more pixels than others. To mitigate the potential bias towards dominant classes inherent in standard Cross-Entropy loss, we employed a compound loss function combining Dice loss and Cross-Entropy (CE) loss (Equation (13)). The Dice loss component is particularly effective in handling moderate class imbalance as it directly optimizes the overlap (Dice Similarity Coefficient—DSC) between the predicted segmentation and the groundtruth, treating foreground classes more equally regardless of their size. The CE loss component complements this by ensuring pixel-level classification accuracy. By equally weighting the Dice and CE components, we aimed to leverage the benefits of both: the robustness of Dice loss to class imbalance and the pixel-level precision encouraged by CE loss.
The optimization was performed using SGD with a momentum of 0.9 and a weight decay of . The training process was segmented into five cycles, employing multi-period cosine annealing with warm restarts, an initial learning rate of , and a minimum learning rate of . The loss function combined Dice loss and Cross-Entropy (CE loss) loss, defined as follows:
where T is the set of ground-truth pixels, and P is the set of predicted pixels.
where is the ground-truth label, and is the predicted probability for the class. Finally, the total loss is denoted as
4.2.2. Evaluation Metrics
We assessed the performance of DC-TransDPANet using the Dice Similarity Coefficient (also known as F1 score, DSC), Hausdorff Distance (HD), Intersection over Union (IoU), and Recall, which are standard metrics for medical image segmentation. DSC is utilized to quantify the overlap between the predicted and actual segmentation. The formula is presented as follows:
where T is the ground-truth, P is the predicted segmentation, and F gives the false positives. This coefficient measures how well the predicted segmentation matches the ground-truth by calculating the ratio of twice the intersection of the predicted and actual regions to the sum of their areas.
HD is used to measure the maximum distance between segmented and ground-truth boundaries. The formulas are shown below:
where T is the set of points in the ground-truth boundary, P is the set of points in the predicted boundary, t is a point in T, and p is a point in P.
IoU calculates the ratio of the intersection of the ground truth and predicted regions over their union.
Recall measures the proportion of the ground truth region correctly identified by the predicted region.
Performance metrics in Table 1, Table 2, Table 3, Table 4, Table 5 and Table 6 are reported as mean ± standard deviation.

Table 1.
Performance comparison of different models on the Synapse dataset.

Table 2.
Segmentation performance on CVC-ClinicDB dataset.

Table 3.
Segmentation performance on Kvasir-SEG dataset.

Table 4.
Performance comparison across multiple organs using different models.

Table 5.
Performance comparison of baseline and DW-CAM variants.

Table 6.
Performance comparison of different loss function combinations.
4.3. Comparison with State-of-the-Art (SOTA) Models
We evaluated DC-TransDPANet on the Synapse dataset against SOTA models, including U-Net [1], TransUNet [8], Swin-Unet [9], UNet++ [12], LeVit-Unet [42], Att-Unet [41], Residual U-Net [19], M2SNet [43], and DA-TransUNet [44]. As shown in Table 1, DC-TransDPANet achieves 81.15% and 19.63 mm in both DSC (%) and HD (mm), respectively. It improves upon TransUNet [8] by 3.67% in DSC and 12.06 mm in HD.
Our model shows a 2.02% increase in DSC compared to the pure transformer-based Swin-Unet [9]. While its HD metric is not the lowest, it demonstrates significant improvement over most models. Notably, our model achieves the highest results in Left Kidney (84.97%), Pancreas (62.46%), and so on, demonstrating exceptional performance in these categories. Compared to TransUNet [8], our model shows substantial improvements in several organs, particularly in Gallbladder (4.14%), Pancreas (6.6%), and Right Kidney (5.87%). These improvements highlight the superior performance of our model.
Figure 4 presented a comparison of our approach with various SOTA methods on the Synapse dataset. The results demonstrate while existing methods frequently suffer from overfitting or underfitting specific organs, our approach exhibits superior performance in visual segmentation. Notably, DC-TransDPANet accurately delineates organ boundaries, closely matching the ground-truth annotations.
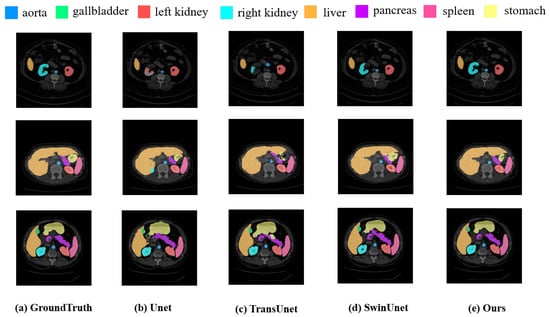
Figure 4.
Experimental segmentation results of methods on Synapse dataset.
Table 2 and Table 3 present the IoU, Dice scores (F1 scores), Recall, and HD for various segmentation models on the CVC-ClinicDB and Kasir-SEG datasets, with DC-TransDPANet achieving the highest performance in these metrics. In Figure 5 and Figure 6, we further evaluated our approach on the CVC-ClinicDB and Kasir-SEG datasets to compare DC-TransDPANet with several classical models. The comparative analysis reveals that DC-TransDPANet exhibits superior precision in handling edge details, with segmentation results closely aligning with the ground-truth masks, underscoring its robust segmentation capabilities. Compared to other models, DC-TransDPANet not only achieves high segmentation accuracy but also adapts effectively to varying polyp shapes, demonstrating a distinct advantage in polyp segmentation tasks.

Figure 5.
Comparison of polyp segmentation results on CVC-ClinicDB dataset using different methods.
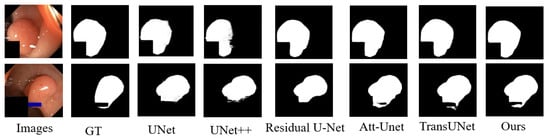
Figure 6.
Comparison of polyp segmentation results on Kvasir-SEG dataset using different methods.
4.4. Ablation Study
We performed ablation experiments to analyze the contribution of essential components within the DC-TransDPANet: DW-CAM, DPA, and their combination.
Effect of Different Modules: As shown in Table 4, both DW-CAM and DPA significantly improved performance over the baseline (77.48% DSC, 31.69 mm HD). DW-CAM increased DSC to 79.30% and reduced HD to 24.37 mm. This improvement stems from DW-CAM’s integration of Depthwise Convolution (DWConv), which enhances the extraction of fine-grained local details crucial for boundary definition, and the Composite Attention Module (CAM), which captures multi-scale contextual information and refines features through spatial and channel reconstruction, leading to better discrimination of complex structures. DPA raised DSC to 78.92% and lowered HD to 25.42 mm. This is attributed to the Depthwise Polarized Attention (DPA) mechanism, which employs Polarized Self-Attention to efficiently model long-range dependencies and global context across the feature map while preserving high-resolution spatial details. This addresses the limitation of CNNs in capturing global information effectively. Combining DW-CAM and DPA further boosted performance. This demonstrates the complementary strengths of the two modules: DW-CAM excels at refining local details and multi-scale features, while DPA efficiently integrates global context while preserving fine details. Their synergy allows the model to capture both intricate local structures and their broader spatial relationships, leading to more accurate and robust segmentation.
Effect of DW-CAM Layers: We evaluated the impact of varying the number of DW-CAM layers (see Table 5). Models with 12, 24, and 32 layers were compared against the baseline. The 12-layer model achieved the highest DSC (81.15%) and the lowest HD, indicating optimal boundary accuracy. The superior performance of the 12-layer model (Table 5) suggests it strikes the best trade-off between model capacity and complexity for the Synapse dataset. Fewer layers might underfit, while more layers (24, 32) likely lead to overfitting or diminishing returns, where the increased parameter count and depth do not translate to better generalization, potentially even hindering optimization or increasing sensitivity to noise. The 12-layer configuration appears sufficient to capture the necessary hierarchical features without becoming overly complex.
Effect of Loss Function Proportions: The loss function combined Cross-Entropy (CE loss) loss and Dice loss, defined as follows:
We investigated the effect of different (CE loss) and (Dice loss) combinations on performance (see Table 6). The baseline, with = 0.5 and = 0.5, achieved 81.94% DSC and 19.63 mm HD. Using only CE loss ( = 1.0, = 0) resulted in 77.45% DSC and 29.80 mm HD, while Dice loss alone ( = 0, = 1.0) improved DSC to 78.32% but still yielded 27.31mm HD. A higher weight on CE loss ( = 0.75, = 0.25) improved DSC to 79.19% and reduced HD to 24.82 mm, whereas a greater weight on Dice loss ( = 0.25, = 0.75) resulted in 79.89% DSC and 22.33 mm HD, closely matching the baseline. The combination with = 0.5 and = 0.5 was optimal for balancing accuracy and boundary precision in segmentation.
4.5. Statistical Significance Test
To verify whether the performance improvement of DC-TransDPANet over the baseline model is statistically significant, we used the Wilcoxon signed-rank test, which is a nonparametric statistical test method suitable for comparing paired non-normally distributed data. We extracted DSC and HD values for each of the 12 validation cases of the Synapse dataset, organ by organ—aorta, gallbladder, left kidney, right kidney, liver, pancreas, spleen, and stomach—to form paired data.
The procedure for the Wilcoxon signed-rank test is as follows: First, for each pair of matched data, compute the difference , where and represent the performance metrics (e.g., DSC or HD) of DC-TransDPANet and the baseline model TransUNet for the i-th validation case. Next, exclude pairs with a difference of zero (in this study, no zero differences were observed). Then, sort the absolute values of the differences in ascending order and assign ranks to each difference. Subsequently, calculate the positive rank sum (corresponding to positive differences) and the negative rank sum (corresponding to negative differences) based on the sign of each difference. Finally, take the smaller of the positive and negative rank sums as the test statistic W, and compute the two-tailed p-value using the exact distribution to assess the statistical significance of the performance difference between the two models.
Given that comparisons were conducted across eight organs, we applied the Bonferroni correction to control the overall Type I error rate. With a global significance level of , the corrected significance threshold is . A p-value below 0.00625 indicates a statistically significant difference in performance.
Table 7 and Table 8 presents the results of the Wilcoxon signed-rank test for per-organ DSC and HD values of DC-TransDPANet and the Baseline on the Synapse dataset. For DSC, the Wilcoxon signed-rank test results further verify the superior performance of DC-TransDPANet on the Synapse dataset, especially showing significant improvement on complex organs such as the gallbladder, right kidney, and pancreas (p < 0.00625). These significant results indicate that the combination of DW-CAM and DPA modules effectively improves the model’s ability to segment small and complex structures, especially on organs with fuzzy anatomical boundaries, such as the pancreas. However, no significant differences were observed on the aorta and liver (p > 0.00625), possibly because the segmentation task of these organs is relatively simple, TransUNet is close to the upper performance limit, or the statistical test is not powerful enough to detect small performance differences due to the limited number of validation cases (only 12 cases).

Table 7.
Comparison of Baseline DSC and Ours DSC across different organs, with improvement, p-value, and significance.

Table 8.
Comparison of Baseline HD and Ours HD across different organs, with differences, p-values, and significance.
For HD, DC-TransDPANet showed statistically significant differences (p = 0.0020) on all organs except the aorta (p = 0.0732). However, Ours has higher HD values than Baseline on most organs (e.g., gallbladder: 16.8874 vs. 26.8150, difference −9.9276), indicating that the boundary accuracy of Ours improved, but HD values increased on the right kidney (+9.1762) and aorta (+0.9447), possibly due to large boundary errors in some cases.
In both DSC and HD tests, p-values for multiple organs are identical (e.g., 0.0020), a phenomenon attributed to the small sample size (12 cases) and highly similar rank distributions of the paired differences (e.g., all positive or all negative differences). Future studies with a larger validation set could provide more robust statistical power to detect differences.
4.6. Uncertainty Quantification
To assess the prediction stability of DC-TransDPANet on the Synapse dataset, we employed the Monte Carlo Dropout (MC Dropout) method for uncertainty quantification. MC Dropout is a Bayesian approximation technique that estimates prediction uncertainty by simulating the posterior distribution of model parameters through multiple forward passes with Dropout enabled during inference.
The uncertainty analysis procedure is as follows: For each validation case, we enable Dropout during inference (with a Dropout probability p = 0.1 and T = 50) and perform 50 forward passes. For each organ, we compute the DSC value for each forward pass, forming a sequence of DSC values, and calculate its standard deviation as the uncertainty metric. A smaller standard deviation indicates greater prediction stability, while a larger standard deviation suggests higher prediction uncertainty. Additionally, we record the HD95 values for each forward pass and compute their mean to ensure consistency with the per-case HD95 results. The MC Dropout implementation was performed using the PyTorch framework, with Dropout layers activated during inference.
In Table 9, the results indicate that DC-TransDPANet exhibits the lowest prediction uncertainty on the aorta (DSC standard deviation = 0.0080) and liver (DSC standard deviation = 0.0085), suggesting stable segmentation performance, which aligns with their high DSC values (aorta: 0.8854, liver: 0.9591). In contrast, the gallbladder (DSC standard deviation = 0.0260) and pancreas (DSC standard deviation = 0.0250) show higher uncertainty, reflecting greater variability in predictions, consistent with their relatively lower DSC values (gallbladder: 0.6727, pancreas: 0.6246). The right kidney has a moderate uncertainty (DSC standard deviation = 0.0180) and a high DSC (0.8289), but its elevated HD (40.6209) suggests that boundary prediction errors may contribute to the observed uncertainty. Overall, organs with lower uncertainty typically correspond to higher DSC values, while those with higher uncertainty, such as the gallbladder and pancreas, face greater segmentation challenges due to their anatomical complexity.

Table 9.
Per-organ uncertainty table: Mean DSC, DSC Std Dev (Uncertainty), and Mean HD.
4.7. Computational Cost Analysis
Table 10 presents the computational cost comparison between DC-TransDPANet (Ours) and TransUNet (baseline), based on 224 × 224 resolution, batch_size = 24, and setting T = 50 forward propagation consistent with the uncertainty quantization setting. The results show that the FLOPs of DC-TransDPANet increase by 13.5%, reflecting the additional computational complexity introduced by DW-CAM and DPA, which together increase the computational overhead. A significant 45.5% reduction in memory usage indicates a substantial improvement in memory efficiency during inference. Inference time increased by 14.3%, consistent with the increase in FLOPs. These results show that DW-CAM and DPA introduce less computational overhead while significantly improving memory efficiency. Inference time (0.064 s/sample) is well below subsecond requirements and memory requirements (682.4 MB) are also modest.

Table 10.
Computational cost comparison of DC-TransDPANet (Ours) and TransUNet (Baseline) (224 × 224, T = 50).
5. Discussion
In this study, we introduced DC-TransDPANet, a novel hybrid CNN-Transformer framework designed to overcome limitations in existing medical image segmentation models, particularly in effectively balancing local feature extraction and global context modeling. Our extensive experiments on diverse benchmark datasets (Synapse multi-organ CT, CVC-ClinicDB and Kvasir-SEG polyp endoscopy) demonstrate that DC-TransDPANet achieves highly competitive performance, and showing statistically significant improvements in key metrics compared to baseline on the Synapse dataset.
Despite the promising results and comprehensive analyses, this study has limitations. While we included statistical tests against the baseline, direct statistical comparisons against all SOTA methods were not performed due to the unavailability of per-case results for all cited works. Performance on certain organs (e.g., Stomach) still indicates room for improvement, suggesting potential benefits from task-specific optimizations or more advanced data augmentation. Future work will focus on extending the framework to 3D volumetric data (requiring significant architectural adaptation as discussed in our previous response), exploring further optimizations for specific challenging organs, developing lightweight variants for resource-constrained environments, and conducting validation on larger, more diverse clinical datasets.
6. Conclusions
In this paper, we introduced DC-TransDPANet, a novel transformer-based framework designed to enhance medical image segmentation accuracy by effectively integrating local and global features. By incorporating the proposed Depthwise Composite Attention Module (DW-CAM) for improved local and multi-scale feature extraction, and the Depthwise Polarized Attention (DPA) module for efficient global context modeling while preserving details, our framework addresses key limitations of existing hybrid models. Extensive experiments on diverse datasets demonstrated statistically significant improvements over the baseline and state-of-the-art or highly competitive performance overall, showcasing robustness in segmenting complex structures. Analyses of uncertainty and computational cost provide further insights into the model’s reliability and efficiency trade-offs. DC-TransDPANet represents a significant step towards more accurate and reliable automated medical image analysis, with future work planned to address statistical validation against more SOTA methods, computational efficiency, and broader applicability including 3D data.
Author Contributions
All authors contributed to the conception and design of the study. Conceptualization, J.X.; methodology, M.Z.; supervision, W.L.; writing—original draft, M.Z.; writing—review and editing, W.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The data supporting the findings of this study are available from the corresponding author(s) upon reasonable request.
Acknowledgments
The authors would like to thank all contributors and reviewers for their insightful comments and suggestions that helped to improve this manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the 18th International Conference on Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015, Munich, Germany, 5–9 October 2015. [Google Scholar]
- Lakhani, P.; Sundaram, B. Deep learning at chest radiography: Automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology 2017, 284, 574–582. [Google Scholar] [CrossRef]
- Min, J.K.; Kwak, M.S.; Cha, J.M. Overview of deep learning in gastrointestinal endoscopy. Gut Liver 2019, 13, 388. [Google Scholar] [CrossRef] [PubMed]
- Würfl, T.; Hoffmann, M.; Christlein, V.; Breininger, K.; Huang, Y.; Unberath, M.; Maier, A.K. Deep learning computed tomography: Learning projection-domain weights from image domain in limited angle problems. IEEE Trans. Med. Imaging 2018, 37, 1454–1463. [Google Scholar] [CrossRef] [PubMed]
- Lell, M.M.; Kachelrieß, M. Recent and upcoming technological developments in computed tomography: High speed, low dose, deep learning, multienergy. Investig. Radiol. 2020, 55, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention is all you need. In Proceedings of the 31st Conference on Neural Information Processing Systems (NIPS 2017), Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. Transunet: Transformers make strong encoders for medical image segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-unet: Unet-like pure transformer for medical image segmentation. In Proceedings of the European Conference on Computer Vision, Tel Aviv, Israel, 23–27 October 2022. [Google Scholar]
- Guo, Y.; Li, Y.; Wang, L.; Rosing, T. Depthwise convolution is all you need for learning multiple visual domains. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019. [Google Scholar]
- Shelhamer, E.; Long, J.; Darrell, T. Fully convolutional networks for semantic segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2016, 39, 640–651. [Google Scholar] [CrossRef]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Proceedings of the DLMIA and ML-CDS Workshops at MICCAI 2018, Granada, Spain, 20 September 2018. [Google Scholar]
- Xiao, X.; Lian, S.; Luo, Z.; Li, S. Weighted res-unet for high-quality retina vessel segmentation. In Proceedings of the International Conference on Information Technology in Medicine and Education, Hangzhou, China, 19–21 October 2018. [Google Scholar]
- Cai, S.; Tian, Y.; Lui, H.; Zeng, H.; Wu, Y.; Chen, G. Dense-UNet: A novel multiphoton in vivo cellular image segmentation model based on a convolutional neural network. Quant. Imaging Med. Surg. 2020, 10, 1275. [Google Scholar] [CrossRef]
- Ma, S.; Tang, J.; Guo, F. Multi-task deep supervision on attention R2U-net for brain tumor segmentation. Front. Oncol. 2021, 11, 704850. [Google Scholar] [CrossRef]
- Ibtehaz, N.; Kihara, D. Acc-unet: A completely convolutional unet model for the 2020s. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Vancouver, BC, Canada, 8–12 October 2023. [Google Scholar]
- Chen, J.; Chen, R.; Wang, W.; Cheng, J.; Zhang, L.; Chen, L. TinyU-Net: Lighter Yet Better U-Net with Cascaded Multi-receptive Fields. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Marrakesh, Morocco, 6–10 October 2024. [Google Scholar]
- Hussain, T.; Shouno, H. MAGRes-UNet: Improved medical image segmentation through a deep learning paradigm of multi-attention gated residual U-Net. IEEE Access 2024, 12, 40290–40310. [Google Scholar] [CrossRef]
- Diakogiannis, F.I.; Waldner, F.; Caccetta, P.; Wu, C. ResUNet-a: A deep learning framework for semantic segmentation of remotely sensed data. ISPRS J. Photogramm. Remote. Sens. 2020, 162, 94–114. [Google Scholar] [CrossRef]
- Datta, S.K.; Shaikh, M.A.; Srihari, S.N.; Gao, M. Soft attention improves skin cancer classification performance. In Proceedings of the IMIMIC and TDA4MedicalData Workshops at MICCAI 2021, Strasbourg, France, 27 September 2021. [Google Scholar]
- Guo, C.; Szemenyei, M.; Yi, Y.; Wang, W.; Chen, B.; Fan, C. Sa-unet: Spatial attention u-net for retinal vessel segmentation. In Proceedings of the International Conference on Pattern Recognition, Virtual, 10–15 January 2021. [Google Scholar]
- Zhu, W.; Chen, X.; Qiu, P.; Farazi, M.; Sotiras, A.; Razi, A.; Wang, Y. Selfreg-unet: Self-regularized unet for medical image segmentation. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2024, Proceedings of the 27th International Conference, Marrakesh, Morocco, 6–10 October 2024; Springer Nature: Cham, Switzerland, 2024; pp. 601–611. [Google Scholar]
- Rahman, M.M.; Munir, M.; Marculescu, R. Emcad: Efficient multi-scale convolutional attention decoding for medical image segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 17–21 June 2024. [Google Scholar]
- Hussain, I.; Nataliani, Y.; Ali, M.; Hussain, A.; Mujlid, H.M.; Almaliki, F.A.; Rahimi, N.M. Weighted Multiview K-Means Clustering with L2 Regularization. Symmetry 2024, 16, 1646. [Google Scholar] [CrossRef]
- Hussain, T.; Shouno, H.; Mohammed, M.A.; Marhoon, H.A.; Alam, T. DCSSGA-UNet: Biomedical image segmentation with DenseNet channel spatial and semantic guidance attention. Knowl.-Based Syst. 2025, 314, 113233. [Google Scholar] [CrossRef]
- Touvron, H.; Cord, M.; Douze, M.; Massa, F.; Sablayrolles, A.; Jégou, H. Training data-efficient image transformers & distillation through attention. In Proceedings of the International Conference on Machine Learning, Virtual, 18–24 July 2021. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Virtual, 11–17 October 2021. [Google Scholar]
- Graham, B.; El-Nouby, A.; Touvron, H.; Stock, P.; Joulin, A.; Jégou, H.; Douze, M. Levit: A vision transformer in convnet’s clothing for faster inference. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Virtual, 11–17 October 2021. [Google Scholar]
- Jamali, A.; Roy, S.K.; Li, J.; Ghamisi, P. TransU-Net++: Rethinking attention gated TransU-Net for deforestation mapping. Int. J. Appl. Earth Obs. Geoinf. 2023, 120, 103332. [Google Scholar] [CrossRef]
- Lin, A.; Chen, B.; Xu, J.; Zhang, Z.; Lu, G.; Zhang, D. Ds-transunet: Dual swin transformer u-net for medical image segmentation. IEEE Trans. Instrum. Meas. 2022, 71, 4005615. [Google Scholar] [CrossRef]
- Heidari, M.; Kazerouni, A.; Soltany, M.; Azad, R.; Aghdam, E.K.; Cohen-Adad, J.; Merhof, D. Hiformer: Hierarchical multi-scale representations using transformers for medical image segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 3–7 January 2023. [Google Scholar]
- Li, Z.; Li, Y.; Li, Q.; Wang, P.; Guo, D.; Lu, L.; Jin, D.; Zhang, Y.; Hong, Q. Lvit: Language meets vision transformer in medical image segmentation. IEEE Trans. Med. Imaging 2023, 43, 96–107. [Google Scholar] [CrossRef]
- Hao, Z.; Quan, H.; Lu, Y. EMF-Former: An Efficient and Memory-Friendly Transformer for Medical Image Segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Marrakesh, Morocco, 6–10 October 2024. [Google Scholar]
- Hussain, T.; Shouno, H.; Hussain, A.; Hussain, D.; Ismail, M.; Mir, T.H.; Hsu, F.R.; Alam, T.; Akhy, S.A. EFFResNet-ViT: A fusion-based convolutional and vision transformer model for explainable medical image classification. IEEE Access 2025, 13, 54040–54068. [Google Scholar] [CrossRef]
- Zhang, X.; Song, Y.; Song, T.; Yang, D.; Ye, Y.; Zhou, J. AKConv: Convolutional kernel with arbitrary sampled shapes and arbitrary number of parameters. arXiv 2023, arXiv:2311.11587. [Google Scholar]
- Goyal, A.; Bochkovskiy, A.; Deng, J.; Koltun, V. Non-deep networks. In Proceedings of the Advances in Neural Information Processing Systems, New Orleans, NY, USA, 27 November–9 December 2022. [Google Scholar]
- Li, J.; Wen, Y.; He, L. Scconv: Spatial and channel reconstruction convolution for feature redundancy. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 18–22 June 2023. [Google Scholar]
- Becker, C.; Ali, K.; Knott, G.; Fua, P. Learning context cues for synapse segmentation. IEEE Trans. Med. Imaging 2013, 32, 1864–1877. [Google Scholar] [CrossRef]
- Jha, D.; Riegler, M.A.; Johansen, D.; Halvorsen, P.; Johansen, H.D. Doubleu-net: A deep convolutional neural network for medical image segmentation. In Proceedings of the International Symposium on Computer-Based Medical Systems, Virtual, 28–30 June 2020. [Google Scholar]
- Jha, D.; Smedsrud, P.H.; Riegler, M.A.; Halvorsen, P.; De Lange, T.; Johansen, D.; Johansen, H.D. Kvasir-seg: A segmented polyp dataset. In Proceedings of the International Conference on Multimedia Modeling, Daejeon, South Korea, 5–8 January 2020. [Google Scholar]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention U-Net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Xu, G.; Zhang, X.; He, X.; Wu, X. Levit-unet: Make faster encoders with transformer for medical image segmentation. In Proceedings of the Chinese Conference on Pattern Recognition and Computer Vision, Xiamen, China, 13–15 October 2023. [Google Scholar]
- Zhao, X.; Jia, H.; Pang, Y.; Lv, L.; Tian, F.; Zhang, L.; Sun, W.; Lu, H. M2SNet: Multi-scale in multi-scale subtraction network for medical image segmentation. arXiv 2023, arXiv:2303.10894. [Google Scholar]
- Sun, G.; Pan, Y.; Kong, W.; Xu, Z.; Ma, J.; Racharak, T.; Nguyen, L.M.; Xin, J. DA-TransUNet: Integrating spatial and channel dual attention with transformer U-net for medical image segmentation. Front. Bioeng. Biotechnol. 2024, 12, 1398237. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).