Abstract
This work develops a new method for vector data augmentation. The proposed method applies principal component analysis (PCA), determines the eigenvectors of a set of training vectors for a machine learning (ML) method and uses them to generate the distilled vectors. The training and PCA-distilled vectors have the same dimension. The user chooses the number of vectors to be distilled and augmented to the set of training vectors. A statistical approach determines the lowest number of vectors to be distilled such that when augmented to the original vectors, the extended set trains an ML classifier to achieve a required accuracy. Hence, the novelty of this study is the distillation of vectors with the PCA method and their use to augment the original set of vectors. The advantage that comes from the novelty is that it increases the statistics of ML classifiers. To validate the advantage, we conducted experiments with four public databases and applied four classifiers: a neural network, logistic regression and support vector machine with linear and polynomial kernels. For the purpose of augmentation, we conducted several distillations, including nested distillation (double distillation). The latter notion means that new vectors were distilled from already distilled vectors. We trained the classifiers with three sets of vectors: the original vectors, original vectors augmented with vectors distilled by PCA and original vectors augmented with distilled PCA vectors and double distilled by PCA vectors. The experimental results are presented in the paper, and they confirm the advantage of the PCA-distilled vectors increasing the classification statistics of ML methods if the distilled vectors augment the original training vectors.
1. Introduction
One fundamental problem in the field of data classification with supervised machine learning (ML) is a shortage of or imbalanced training data [1,2,3]. For example, the training set of the public image database ISIC2020 [4] contains only 584 malignant images and approximately 33,126 benign images. Such a large class imbalance leads to overfitting [5], which impacts the prediction accuracy of a classifier. This holds because the model is biased toward the class with the larger set of samples. In the example given above, the ML classifier may produce a large number of false negatives (type I error), which will indicate poor performance of the model.
A natural way to tackle the problem of imbalanced classes and data shortage is to fill up the gap with additional data, but in many practical cases, the process of obtaining additional real data is time-consuming and expensive. Hence, other sources of and approaches to data generation should be explored and used. One approach is to artificially produce data which will have the same characteristics as the original data, and after augmenting the training data with the artificial data, the entire set will increase the statistical outcomes of the ML method. An approach that produces artificial data is to apply geometric transformations on a certain part of the training images and augment the original training set with the transformed set of images. Typical geometric image transformations consist of rotation, translation, flipping [6] or cropping [7]. Another set of techniques that generate images for the purpose of training data augmentation includes Gaussian blur, image sharpening, smoothing [8], and noise injection [6].
In recent years, a method was developed to embed a vector field (VF) into an image [9]. This method augments the set of image features with VF features like singular points and trajectories. Augmenting the set of features in every image of an image database increases the learning capabilities of the ML methods and leads to an increase in its classification abilities. One may observe that the method listed above produce new data on the basis of existing data. Also, the VF embedding methods [9] augment the set of image features with VF features, but none of them change the main theme of (keeping the subjects that belong to) the images in the case of image data.
On the other hand, data distilation methods based on NNs [10] and infinite NNs [11] generate new data or images which do not resemble the original sets at all but are still useful for training data augmentation. Another useful way to generate new images on the basis of existence, for the purpose of training data augmentation, is to use a generative adversarial network (GAN). A review of this type of NN is given in [12]. In [13], the authors used the GAN architecture to generate synthetic images and resolve the problem of limited data availability for three different classes. For the purpose of augmentation, the authors of [14] developed and implemented a CNN to determine the likelihood of an object category being present inside a box that encompasses a given neighborhood. Further, the method found suitable locations in images to place new objects.
A comprehensive survey on the recent methods of data distillation, including the application of NNs, is given in [15]. The authors provide detailed descriptions of multiple data distillation techniques based on the concepts of “meta-model”, “gradient”, “distribution”, “trajectory” and “factorization” matching. Additional papers reviewing state-of-the-art augmentation techniques are available [16,17,18].
In [19], the principal component analysis (PCA) [20] method was used to distill the photo response non-uniformity reference by removing the interference noise. In the present paper, we propose a novel idea to extend the well-known PCA [21] method for vector data distillation from existing training data. Our scientific conjecture, regarding an extension of PCA that may lead to enhancement of the classification statistics of the ML methods, is that the PCA method lies in the eigenvectors of the data feature correlation matrix. Also, this matrix maps every eigenvector to a vector on the same straight line. This implies that we may receive (distill) different vectors which possess feature correlations similar to the original vectors. To validate that extended PCA distills vectors whose augmentation, relative to the original training vectors, increases the ML classification statistics, we conducted a set of experiments with four publicly available databases. We applied on them three classifiers: an NN, a support vector machine (SVM) and logistic regression. The experimental results are shown in this paper, and they confirm the advantage that augmenting the training set with PCA-distilled vectors leads to an increase in the classification statistics.
The rest of this paper is organized as follows. In its first subsection, Section 2 describes the new method for data distillation with PCA, while the second subsection introduces a statistical method [22] that determines the lower bound of the number of distilled vectors necessary to augment the set of training vectors in order to provide certain accuracy of classification. Section 3 presents the four classifiers implemented to validate the new distillation method, and the next section describes the original datasets along with the augmented vector datasets. Finally, Section 5 and Section 6 describe our results and discuss the novelties and advantages of the new vector distillation method.
2. Vector Distilation with Principal Component Analysis (PCA)
PCA [20,21] is a commonly employed technique for decreasing the dimensionality of given data. This is achieved through mapping a high-dimensional dataset into a lower dimensional space while preserving the important features of the dataset. PCA has numerous applications to data visualization, feature extraction and data compression. In the present paper, we propose a new application of PCA and employ the method to generate new data ordered from top to bottom according to their importance. To augment the original data, we select the most representative vectors from the top. We apply a statistical method to estimate the minimum number of vectors to be selected.
2.1. Principal Components and the Projection Matrix
Assume we are given a set of training vectors , where . Hence, every training vector consists of k variables, and we will also call features for and . Therefore, the matrix of the database . Then, we denote the vector of the jth feature with and introduce it as . Furthermore, we denote the mean of the jth feature (variable) vector , which is a column of , with and the rank of with :
- 1.
- Data normalization is the first step of the calculation, and we subtract the mean of each feature vector from the corresponding vector entries. This is performed to ensure that the data are centered around zero. This means that the mean of every feature vector is zero. The equation for data normalization, with the help of the feature vector mean, iswhere is the i-th feature value, in which for the j-th feature (variable) vector and , while is the mean of this feature vector.
- 2.
- The covariance matrixmeasures the statistical relationship between the features in the entire dataset [21]. For this purpose, we developed the equation below and calculated the entry of the features’ covariance matrix of the training data matrix :It follows from Equation (2) that the covariance matrix of the features is of the formNow, we calculate the eigenvectors and the eigenvalues of the covariance matrix of the database features as follows:where we denote the identity with .
- 3.
- For projection, in the present step, we construct the matrix , whose columns are the normalized eigenvectors of the covariance matrix of the dataset features:where every eigenvector and r is the rank of . Also, let us arrange the vector columns from left to right according to the decreasing eigenvalues of the corresponding eigenvectors. We consider the largest one of them at the leftmost position if we think of the eigenvalues as a decreasing sequence of consecutive numbers. The matrix is applied to project the training dataset onto the new subspace spanned by the eigenvectors . The equation that conducts the projection is [21]where is the new low-dimensional representation of the ith training sample , in which is a vector row in whose rank .We consider that an eigenvector is stronger than another one if the former has a greater eigenvalue than the latter. Hence, we selected from the strongest components (eigenvectors) , which appeared as the top m rows of the matrix. Using the strongest components as vector columns, we constructed the matrix which transformed (mapped) the original dataset into , which we call the transformed data:
2.2. Finding the Distilled Vectors
We conducted the inverse projection with the matrix , which mapped the transformed data , where
into the modified original dataset such that . Note that the modified original data belong to the original feature space and have the same cardinality n as the initial original data. Now, among the modified training data set , we determine the images of the m strongest components (eigenvectors) we selected from . In order to accomplish this, we used the consecutive numbers of their positions in Equation (4). We call the set of m modified vectors in the set of distillated vectors from the original training vectors . We augmented the set of initial original training vectors with the set of distilled vectors and used the union as a training set of an ML classifier. One immediate advantage of this kind of augmentation is enlarging a class in case of a massive disproportion between two classes.
2.3. Minimum Number of Training Samples (MNTS)
The present subsection provides an algorithm to determine the minimum number of training samples [22] m that need to be distilled out of the total number of training samples n such that the original samples augmented with the distilled samples train a classifier to provide an accuracy a:
- 1.
- Assume we are looking for an accuracy . It follows that and the confidence level ;
- 2.
- Using the confidence level C, we determine from the standard normal distribution table [23];
- 3.
- We select the success and failure rates, denote them as p and q, respectively, and calculate m from
- 4.
- We consider a training set with two classes (the number of benign samples and the number of malignant samples ), where , which is the total number of samples;
- 5.
- We calculate , ;
- 6.
- Here, and are the number of malignant and number of benign distilled vectors, respectively, while , such that .
3. Classifiers Applied for Experimental Validation
In the present section, we describe three classifiers we use to validate our method for distillation and augmentation and the advantage that comes from them. The novelty is that by using the training vectors of an ML algorithm, the PCA method generates (distills) vectors which increase the classification statistics of the ML method if the original training set is augmented with the distilled set.
3.1. Logistic Regression (LR)
Logistic regression is a statistical technique employed to model the relation between dependent variables. In the case of binary classification, it models the probability of an observation belonging to a particular class. Given a dataset with n observations and k independent variables where , the logistic regression model can be represented as follows:
where Y is the binary dependent variable with outcomes “0” or “1”, is the training data matrix with n rows (samples of training vectors) and k columns (variables), are the coefficients or parameters estimated by the model and is the natural logarithm of the odds ratio of the probability of to the probability of . The logistic function or sigmoid function is used to convert the linear combination of independent variables and coefficients into a probability between 0 and 1:
To evaluate the coefficients of the model, the maximum likelihood estimation algorithm [24] was used. The goal was to maximize the likelihood of observing the dataset if the model was given. This could be accomplished by minimizing the negative log-likelihood:
where is the ground truth label for the training vector , while is the predicted probability of for this vector. The parameters for can be estimated using optimization algorithms such as gradient descent [25] or Newton’s method.
3.2. Support Vector Machine (SVM)
The SVM is a useful ML tool for vector classification [26,27]. The method was developed and gained popularity during the 1990s in the 20th century [26] and is applied mainly for binary classification, but multi-class SVMs could be built as well. Unlike NNs, which provide accurate classification if thousands or hundreds of thousands of training samples are used, SVMs are effective classifiers if hundreds of training samples are available. In the present study, we employ a binary SVM to classify medical vector data. For the purpose of SVM training, we use two kinds of data: original data and original data augmented with PCA-distilled data.
Following the concepts from [26,27], we define the binary SVM as the function given below:
In Equation (13), the symbol denotes the label of the ith training sample , is the unknown sample subject to classification and is the Lagrange multiplier associated with the ith support vector such that , while is a bias. Furthermore, with , we denote the kernel of the SVM. Examples of the most used kernels are the linear, polynomial, radial basis function (RBF), Gaussian and sigmoid kernels. For the purpose of our experimental validation of the advantage of this study, we will use the linear and the polynomial kernels. The latter is defined below:
where . Hence, we difine an SVM with a linear (SVML) kernel for and an SVM with a polynomial (SVMP) kernel for .
3.3. Neural Network (NN)
In recent years, the NN became the most used classifier because of its reliability and repeatability as well as the opportunity to greatly increase performance statistics. Note that the image is a 2D signal and can be presented in the form of a 1D signal or as a sequence. Despite this, we could roughly divide NNs into those which are designed to work with images [6,8] and those designed to work with signals and sequences of numbers [28,29]. A well-established representative of the first kind is the convolutional NN (CNN) [6,8], while examples of the second kind include long short-term memory (LSTM) NNs [28,29]. The main hyperparameters of an NN consist of the architecture, input layers, hidden layers, output layers, activation functions, loss function, optimization method and training process. The architecture of an NN typically consists of multiple interconnected layers, each containing a set of neurons or nodes. The arrangement and number of layers, as well as the number of neurons in each layer, vary depending on the specific problem and desired model complexity [8,12,28,29]. The NN model we use consists of two hidden layers and an output layer. Each hidden layer has 512 and 256 neurons and uses the ReLU activation function. The output layer has one neuron with the sigmoid activation function. The equations used in the two hidden layers are described as follows:
Here, and represent the matrices of weights for the first and second hidden layer, respectively, and denotes the matrix of the input data. The terms and represent matrices of the bias terms for the first and second hidden layers, respectively, while and denote the pre-activation calculations of the two hidden layers. Furthermore, and represent the ReLU-calculated outputs of the first and the second layers, respectively.
Concerning the output layer that calculates the vector of predicted values, we implement the following equations:
In Equation (17), represents the vector of weights for the output layer, denotes the vector of biases, represents the pre-activation output of the output layer and represents the vector of the predicted output calculated by the sigmoid activation function .
During the training process, we apply the Adam optimizer to update the matrix and vector parameters , , , , and . The loss function used is binary cross-entropy, and the model’s accuracy is measured using the accuracy metric.
4. Validation Datasets and Training of the Models
4.1. Original Datasets
In the present section, we describe the four datasets of feature vectors we used to validate that augmenting the training dataset with PCA-distilled vectors improves the statistical outcomes of an ML classifier:
- The first one we call the skin lesion (SL) dataset, which comprises 162 observations (samples S) of 5D feature vectors () extracted by an active contour in [30] from skin lesion images with a ground truth [31]. The SL dataset contains 100 benign samples (skin lesion feature vectors labeled B) and 62 malignant observations (labeled M).
- The diabetes (D) dataset [32] consists of 768 feature vectors with a dimension of 8 (). The vectors are distributed in two sets labeled P if the corresponding vector indicates diabetes. Otherwise, the vector is labeled N, which means that the vector indicates no diabetes. Class P, which suggests diabetes, consists of 268 vectors, while class N contains 500 vectors with a dimension of 8.
- The heart disease (HD) dataset [33] consists of 1025 feature vectors with a dimension of 13. The data samples are distributed in two classes: (N) and (P). The N samples in the HD datasets total 499, while the P samples total 526.
- The breast cancer (BC) dataset [34] contains total of 569 feature vectors (observations) with a dimension of 30. Out of them, 347 are labeled as benign B, while the remaining 212 were determined by medical experts to be malignant M.
A read of the above databases’ information shows that all databases are binary. In three of the cases—, D and —the two classes are unbalanced. The benign (negative class) contains almost twice as many feature vectors as the malignant (positive) class.
4.2. Augmentation of the Original Training Data
For the validating experiments, we used the following split of the original data: 90% for training and 10% for testing. Then, we distilled the vectors from the selected training set and augmented this set with the distilled vectors. Hence, we constructed three kinds of augmentations. For the purpose of distillation, we applied the method from Section 2.3 to determine the minimum number of training samples (MNTS) to be distilled and used this to augment the original training samples such that they trained a classifier to provide an accuracy of . Table 1 presents the experimental results of classifying the 10% testing data with the three models introduced above and trained by the augmented data, as well as the same models trained by the original data.

Table 1.
Comparison of accuracy in classification of the SL data. Each column shows the average of 10-fold experiments. In bold are the highest values.
Augmented Dataset 1 was created from the SL dataset using a testing/training split of 10%/90%, where we kept the proportion the same as it was in the original SL dataset. Therefore, we had 90 B and 56 M vectors for training as well as 10 B and 6 M for testing. Next, using PCA, we distilled the 26 vectors from the selected 90% (90 B and 56 M) training original vectors such that the proportion was preserved. The cardinality of the distilled vectors was 26 and was determined by the MNST approach, consisting of 16 B and 10 M. Next, we added the distilled vectors to the original 90 B and 56 M training samples. Then, we trained our classifiers to learn from the augmented training set and tested the models on the remaining 10% B and 10% M original vectors.
Augmented Dataset 2 was for the SL dataset as well. In this augmentation, we randomly selected 90% of the entire original data for training and used the remaining 10% of original data for testing. This means that in the selected training vectors, the proportion did not need to be as in the entire original SL dataset. Furthermore, in the distilled set of 26 vectors, the ratio equals the ratio of the randomly selected training vectors.
Augmented Dataset 3 analogous to the case of the SL dataset, used 90% randomly selected samples for training from each class, and we distilled from the training samples, 101, 151 and 201 new vectors for each of the datasets D, HD and BC, respectively. Note that in the distilled sets, the proportion M/B or P/N was the same as in the original dataset. Then, for each of these datasets, we created augmented training sets of the form Original + 101-PCA-d, Original + 151-PCA-d and Original + 201-PCA-d:. With Original, we denoted the randomly selected 90% original vector samples, which consisted of 90% of the M (P) and 90% of the B (N) samples. The sets of 101, 151 and 201 were distilled from the 90% original vectors with the help of the PCA method described in Section 2. Therefore, for each of the original datasets D, HD and BC, we developed three augmented datasets for training. Then, we trained the models and tested them on the remaining 10% of the original data.
Augmented Dataset 4 involved distilling 51, 76 and 101 samples from the 101, 151 and 201 samples, respectively, which were distilled earlier with the help of the same PCA method described in Section 2. In the double distilled three sets of vectors, the proportion M/B or P/N was kept the same as in the initially distilled dataset. With the help of these new and nested distillations, we created three augmented training sets of the form Original + (101 + 51)-PCA-d, Original + (151 + 76)-PCA-d and Original + (201 + 101)-PCA-d for each of the original datasets D, HD, and BC. Therefore, we developed nine training augmented datasets and tested them on the remaining 10% of the original data.
4.3. Model Training
Since the SL data were quite small, we designed a slightly different NN than the one described in Section 3.3. The modified NN was designed with the Keras Sequential API and contained two fully connected layers. The first layer comprised 10 neurons and implemented the ReLU activation function, while the second layer consisted of 1 neuron with the sigmoid activation function. The model used the Adam optimizer for 100 epochs with a batch size of 32 in order to be trained with the rather small sample sets of 100 and 106 samples.
As for the classification of the other three datasets, we applied the neural network presented in Section 3.3. The model also included two dropout layers with a rate of 0.5 to reduce overfitting. The input dimension was set to eight. The model was compiled with the Adam optimizer and used the binary cross-entropy loss function.
Recall that the advantage of this study is that augmenting the training data with PCA-distilled vectors increases the classification statistics. The next model we used to validate this advantage was the logistic regression model described in Section 3.1. We defined this classifier again with the scikit-learn library as a logistic regression object. The model used a maximum of 1000 iterations to ensure convergence.
The last model we implemented to validate our approach was the support vector machine (SVM). We designed this classifier with the help of the scikit-learn library and made the SVM works with two kernels: linear and polynomial degree 2. Furthermore, we set the machine’s regularization strength C = 1.
For the NN model, the accuracy score was calculated using the evaluate() method with the testing data. For the logistic regression and SVM models, the accuracy score was calculated using the score() method with the testing data. The confusion matrix was also calculated for each model using the confusion_matrix() function from the scikit-learn library.
4.4. Cross-Validation
To fairly validate the classification statistics of the three classifiers, we implemented the Monte Carlo cross-validation approach. It randomly split the entire set of samples into training and test samples and repeated the splitting m times. For each split, a sample appeared in exactly one of the sets of training or testing. The mean of the m experiments was calculated for each evaluation statistic. In our experiments, we set m = 10 and show the results in Table 1, Table 2, Table 3, Table 4, Table 5, Table 6, Table 7, Table 8, Table 9, Table 10, Table 11, Table 12 and Table 13 for the SL and HD datasets. The advantages of applying Monte Carlo cross-validation come from the fact that the approach decreases the variance of the split sample error estimate, and the proportion of the training-test random splits do not depend on the selected number m.
4.5. Classification Metrics
Accuracy, sensitivity and specificity are commonly used metrics for evaluating the effectiveness of a classification model. Below is a description of each metric along with their equations. The notations used are TP = true positive, TN = true negative, FP = false positive and FN = false negative:
- Accuracy measures the overall correctness of the model’s predictions by calculating the ratio of correctly predicted instances to the total number of instances:
- Sensitivity, also known as recall or the true positive rate, measures the model’s ability to correctly identify positive instances out of all the actual positive instances:
- Specificity (true negative rate) calculates the model’s ability to correctly identify negative instances out of all the actual negative instances:
5. Experimental Results
To validate that augmenting a training set of vectors with vectors distilled from them by the PCA method increases the classification statistics, we conducted experiments applying NN, SVM and LR classifiers on the four databases of feature vectors: skin lesion SL [30,31], diabetes D [32], heart disease HD [33] and breast cancer BC [34]. For this purpose, we designed different set-ups for the training datasets as described in Section 4.2. The classification results obtained are presented in various tables throughout the present section. In the reporting tables, we show two types of accuracy for each experiment. The first percentage represents the model’s accuracy for one iteration, while the second percentage indicates the mean accuracy of the Monte Carlo cross-validation approach for 10 experiments. The bold percentages in all the tables indicate the best results. We present our results in three separate tables for the classification metrics mentioned in Section 4.5 for each dataset.
In Figure 1 are shown the curves of the loss function for the NN model on the SL data. One may observe that the curves of the loss functions for the training processes with the original dataset and Augmented Dataset 1 resemble each other. On the other hand, the curves of the loss functions for the two validation processes are quite different. The curve for the validation process when the NN was trained with the original vectors is convex from below. This indicates possible divergence after the 100 epochs, while the curve for validation when the NN was trained with Augmented Dataset 1 steadily decreases, which suggests a classification improvement if more than 100 epochs are conducted.
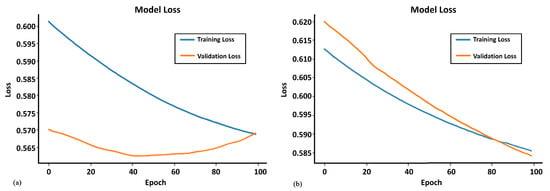
Figure 1.
(a) Training and validation loss curves of the original skin lesion data. (b) Training and validation loss curves of Augmented Dataset 1.
In Table 1, we show a comparison of the classification accuracies of different classifiers for the SL dataset and its augmentations mentioned in Section 4.2. The total number of samples in the original dataset was 162, and we split it into 90% vs. 10% sets for training and testing, respectively. By studying Table 1, one can tell that the mean accuracy of the LR significantly increased from 71% to 82% for Augmented Dataset 2.Failure in the remaining classifiers may have occurred due to the fact that the total sample size was too small. Hence, we continued the experimental validation with relatively larger feature vector databases: D, which contained 768 feature vectors of 8D with a P/N ratio of 268/500 [32]; heart disease HD, which contained 1025 feature vectors of 13D and a P/N ratio of 499/526 [33], and breast cancer BC, which contained 569 feature vectors of 30D with a P/N ratio of 212/347 [34].
The results from D data classification with the four classifiers are presented in Table 2, Table 3 and Table 4. We applied the NN, LR and SVM classifiers with linear and polynomial degree 2 kernels, denoted as SVML and SVMP, respectively. The classifiers were trained with the Original, Original + -PCA-d and Original + -PCA-d datasets, which are described in Section 4.2. One may notice that the training with the augmented data improved the classification accuracy if compared with the training by the original data. For the NN and SVMP classifiers, the highest increase came for the 10-fold experiments with the single augmentation by 101 distilled vectors. For the LR classifier, the highest results came from the double augmentation of and distilled vectors, while for the SVML classifier, the highest outcome was obtained with distilled vectors. On the other hand, the outcomes for sensitivity and specificity showed high results for the former statistic and results twice as small for the latter. This tells us that the augmentations did not balance the classification models.




Table 2.
Comparison of classification accuracies of four classifiers trained on 90% of the original D data and various augmentations, tested on 10% of the original D data. Each column shows the average of 10-fold experiments. Highest values are shown in bold.
Table 2.
Comparison of classification accuracies of four classifiers trained on 90% of the original D data and various augmentations, tested on 10% of the original D data. Each column shows the average of 10-fold experiments. Highest values are shown in bold.
| Training Data | NN | LR | SVML | SVMP |
|---|---|---|---|---|
| Original | 75% | 70% | 70% | 70% |
| Original + 101-PCA-d | 82% | 77% | 74% | 82% |
| Original + (101 + 51)-PCA-d | 79% | 78% | 78% | 78% |
| Original + 151-PCA-d | 77% | 73% | 74% | 80% |
| Original + (151 + 76)-PCA-d | 79% | 78% | 64% | 67% |
| Original + 201-PCA-d | 75% | 67% | 73% | 73% |
| Original + (201 + 101)-PCA-d | 75% | 70% | 70% | 73% |

Table 3.
Comparison of classification sensitivities of different classifiers trained on 90% of the original D data and various augmentations, tested on 10% of the original D data. Each column shows the average of the 10-fold experiments. In bold is given the highest value of a column.
Table 3.
Comparison of classification sensitivities of different classifiers trained on 90% of the original D data and various augmentations, tested on 10% of the original D data. Each column shows the average of the 10-fold experiments. In bold is given the highest value of a column.
| Training Data | NN | LR | SVML | SVMP |
|---|---|---|---|---|
| Original | 52% | 56% | 68% | 67% |
| Original + 101-PCA-d | 77% | 48% | 30% | 37% |
| Original + (101 + 51)-PCA-d | 81% | 56% | 26% | 56% |
| Original + 151-PCA-d | 52% | 41% | 30% | 56% |
| Original + (151 + 76)-PCA-d | 74% | 52% | 19% | 22% |
| Original + 201-PCA-d | 44% | 22% | 33% | 33% |
| Original + (201 + 101)-PCA-d | 52% | 41% | 41% | 22% |

Table 4.
Comparison of classification specificities of different classifiers trained on 90% of the original D data and various augmentations, tested on 10% of the original D data. Each column shows the average of 10-fold experiments. In bold are the highest values.
Table 4.
Comparison of classification specificities of different classifiers trained on 90% of the original D data and various augmentations, tested on 10% of the original D data. Each column shows the average of 10-fold experiments. In bold are the highest values.
| Training Data | NN | LR | SVML | SVMP |
|---|---|---|---|---|
| Original data | 93% | 83% | 72% | 72% |
| Original + 101-PCA-d | 88% | 92% | 94% | 94% |
| Original + (101 + 51)-PCA-d | 86% | 90% | 88% | 90% |
| Original + 151-PCA-d | 94% | 90% | 98% | 94% |
| Original + (151 + 76)-PCA-d | 86% | 92% | 88% | 92% |
| Original + 201-PCA-d | 96% | 94% | 94% | 94% |
| Original + (201 + 101)-PCA-d | 90% | 86% | 98% | 90% |
As mentioned above, we conducted distillation-augmentation experiments using the heart disease HD [33] dataset as well. Recall that it contained 1025 feature vectors with a dimension of 13. Out of them, there were 499 positive (P) vectors and 526 negative (N) vectors. The former vectors indicate heart disease, while the latter indicate heathy samples. The four classifiers were trained again with the following types of sets: Original, Original + -PCA-d and Original + -PCA-d. The number denotes the number of vectors distilled from the original training vectors, while denotes the number of vectors distilled from the already distilled vectors. The experimental results, with 90% P and 90% N randomly selected samples for training, are shown in Table 5, Table 6 and Table 7. One may tell from there that the average accuracy increased for all classifiers when they were trained with the augmented set Original + -PCA-d, which contained double distillation. The sensitivity increased only for the SVMP classifier because it was already at the maximum for the original training data for the other classifiers. The specificity increased (significantly for the LR, SVML and SVMP classifiers) for all classifiers if trained with augmented sets that contained single and double distillated data. Another important achievement obtained with the HD dataset and the used distillation augmentations is that all classifiers were balanced according to the sensitivity/specificity ratio.




Table 5.
Comparison of classification accuracies of different classifiers trained on 90% of the original HD data and various augmentations, tested on 10% of the original HD data. Each column shows the average of 10-fold experiments. In bold are the highest values.
Table 5.
Comparison of classification accuracies of different classifiers trained on 90% of the original HD data and various augmentations, tested on 10% of the original HD data. Each column shows the average of 10-fold experiments. In bold are the highest values.
| Training Data | NN | LR | SVML | SVMP |
|---|---|---|---|---|
| Original | 95% | 86% | 83% | 83% |
| Original + 101-PCA-d | 96% | 87% | 88% | 86% |
| Original + (101 + 51)-PCA-d | 96% | 90% | 91% | 91% |
| Original + 151-PCA-d | 95% | 86% | 86% | 84% |
| Original + (151 + 76)-PCA-d | 95% | 83% | 83% | 83% |
| Original + 201-PCA-d | 89% | 86% | 85% | 83% |
| Original + (201 + 101)-PCA-d | 90% | 83% | 83% | 83% |

Table 6.
Comparison of classification sensitivities of different classifiers trained on 90% of the original HD data and various augmentations, tested on 10% of the original HD data. Each column shows the average of the 10-fold experiments. In bold is given the highest value in a column.
Table 6.
Comparison of classification sensitivities of different classifiers trained on 90% of the original HD data and various augmentations, tested on 10% of the original HD data. Each column shows the average of the 10-fold experiments. In bold is given the highest value in a column.
| Training Data | NN | LR | SVML | SVMP |
|---|---|---|---|---|
| Original | 100% | 96% | 98% | 69% |
| Original + 101-PCA-d | 100% | 94% | 94% | 94% |
| Original + (101 + 51)-PCA-d | 96% | 94% | 96% | 85% |
| Original + 151-PCA-d | 100% | 85% | 89% | 87% |
| Original + (151 + 76)-PCA-d | 98% | 85% | 83% | 87% |
| Original + 201-PCA-d | 100% | 96% | 96% | 98% |
| Original + (201 + 101)-PCA-d | 91% | 87% | 92% | 89% |

Table 7.
Comparison of classification specificities of different classifiers trained on 90% of the original HD data and various augmentations, tested on 10% of the original HD data. Each column shows the average of the 10-fold experiments. In bold is given the highest value in a column.
Table 7.
Comparison of classification specificities of different classifiers trained on 90% of the original HD data and various augmentations, tested on 10% of the original HD data. Each column shows the average of the 10-fold experiments. In bold is given the highest value in a column.
| Training Data | NN | LR | SVML | SVMP |
|---|---|---|---|---|
| Original | 96% | 79% | 71% | 64% |
| Original + 101-PCA-d | 94% | 80% | 82% | 78% |
| Original + (101 + 51)-PCA-d | 98% | 86% | 86% | 88% |
| Original + 151-PCA-d | 94% | 88% | 84% | 78% |
| Original + (151 + 76)-PCA-d | 98% | 80% | 84% | 90% |
| Original + 201-PCA-d | 86% | 76% | 74% | 68% |
| Original + (201 + 101)-PCA-d | 92% | 76% | 74% | 78% |
With the help of the heart disease (HD) [33] dataset, we conducted a second set of experiments, decreasing the number of randomly selected original vectors for training to 70%. Then, the testing vectors were the remaining 30% of the feature vectors. All other activities such as distilations and augmentations were the same as in the case of the 90% selected original feature vectors for training. The obtained results for classification with the four chosen classifiers are shown in Table 8, Table 9 and Table 10.




Table 8.
Comparison of classification accuracies of different classifiers trained on 70% of the original HD data and various augmentations, tested on 30% of the original HD data. Each column shows the average of 10-fold experiments. In bold are the highest values.
Table 8.
Comparison of classification accuracies of different classifiers trained on 70% of the original HD data and various augmentations, tested on 30% of the original HD data. Each column shows the average of 10-fold experiments. In bold are the highest values.
| Training Data | NN | LR | SVML | SVMP |
|---|---|---|---|---|
| Original | 92% | 84% | 81% | 81% |
| Original + 101-PCA-d | 86% | 82% | 82% | 82% |
| Original + (101 + 51)-PCA-d | 91% | 80% | 80% | 86% |
| Original + 151-PCA-d | 93% | 85% | 85% | 85% |
| Original + (151 + 76)-PCA-d | 88% | 84% | 83% | 82% |
| Original + 201-PCA-d | 91% | 83% | 83% | 87% |
| Original + (201 + 101)-PCA-d | 90% | 83% | 84% | 82% |

Table 9.
Comparison of classification sensitivities of different classifiers trained on 70% of the original HD data and various augmentations, tested on 30% of the original HD data. Each column shows the average of the 10-fold experiments. In bold is given the highest value in a column.
Table 9.
Comparison of classification sensitivities of different classifiers trained on 70% of the original HD data and various augmentations, tested on 30% of the original HD data. Each column shows the average of the 10-fold experiments. In bold is given the highest value in a column.
| Training Data | NN | LR | SVML | SVMP |
|---|---|---|---|---|
| Original | 92% | 93% | 56% | 76% |
| Original + 101-PCA-d | 93% | 89% | 91% | 89% |
| Original + (101 + 51)-PCA-d | 96% | 88% | 90% | 90% |
| Original + 151-PCA-d | 97% | 94% | 95% | 96% |
| Original + (151 + 76)-PCA-d | 91% | 92% | 93% | 93% |
| Original + 201-PCA-d | 93% | 87% | 86% | 91% |
| Original + (201 + 101)-PCA-d | 94% | 92% | 94% | 89% |

Table 10.
Comparison of classification specificities of different classifiers trained on 70% of the original HD data and various augmentations, tested on 30% of the original HD data. Each column shows the average of the 10-fold experiments. In bold is given the highest value in a column.
Table 10.
Comparison of classification specificities of different classifiers trained on 70% of the original HD data and various augmentations, tested on 30% of the original HD data. Each column shows the average of the 10-fold experiments. In bold is given the highest value in a column.
| Training Data | NN | LR | SVML | SVMP |
|---|---|---|---|---|
| Original | 96% | 82% | 36% | 63% |
| Original + 101-PCA-d | 85% | 74% | 73% | 75% |
| Original + (101 + 51)-PCA-d | 88% | 71% | 81% | 81% |
| Original + 151-PCA-d | 91% | 77% | 73% | 74% |
| Original + (151 + 76)-PCA-d | 88% | 76% | 72% | 71% |
| Original + 201-PCA-d | 90% | 79% | 81% | 83% |
| Original + (201 + 101)-PCA-d | 91% | 73% | 74% | 75% |
A study of the results in the above tables shows that selecting 70% of the original HD feature vectors for training, distillation and augmentation kept the same trends of increasing the classification statistics as in the case of 90% selection. Moreover, one may observe that with the 70/30% split, the specificities for all classifiers increased when trained with the Original+151-PCA-d dataset. In summary, the augmentation of the original training set with distilled vectors from this set increased the classification statistics and made the classifiers balanced. Only the classification statistics in the 70% split were smaller than the classification statistics in the 90% case.
We conducted the final set of experiments with the BC dataset, which contained 569 feature vectors of 30D with a P/N ratio of 212/347 [34]. For training, we randomly selected 90% of the P and 90% of the negative samples 10 times. Every time, from every selection of training samples, we distilled 101, 151 and 201 vectors with same dimensions as the original training vectors. From every distilled set, we distilled 51, 76, and 101 vectors, respectively, as a second distillation. The experimental results with the original set and the augmented Original + -PCA-d and Original + -PCA-d training sets are reported in Table 11, Table 12 and Table 13. Recall that with , we denote a set of vectors distilled during the first distillation, while denotes a set of vectors from the second distillation.
A study of the results shows a significant increase in the classification accuracy of the NN classifier for the augmented set Original+101-PCA-d and for the SVML classifier with the augmented set Original + 151-PCA-d. Concerning the LR and SVMP classifiers, their highest results with the latter augmented set were the same as the accuracy of classification with the original training data set. The reason for this is that the accuracies of the latter data set were high enough, and there was no room for further increases. The same observation and conclusion hold for the specificity of the NN and the sensitivities of the LR and SVMP classifiers. The last two statistics exhibited a significant increase for the remaining classifiers when trained with augmented datasets. Moreover, the sensitivity/specificity ratio was balanced well, leading to the highest balanced accuracy = (sensitivity + specificity)/2.




Table 11.
Comparison of classification accuracies of different classifiers trained on 90% of the original BC data and various augmentations, tested on 10% of the original BC data. Each column shows the average of 10-fold experiments. In bold are the highest values.
Table 11.
Comparison of classification accuracies of different classifiers trained on 90% of the original BC data and various augmentations, tested on 10% of the original BC data. Each column shows the average of 10-fold experiments. In bold are the highest values.
| Training Data | NN | LR | SVML | SVMP |
|---|---|---|---|---|
| Original | 87% | 95% | 65% | 98% |
| Original + 101-PCA-d | 97% | 84% | 88% | 86% |
| Original + (101 + 51)-PCA-d | 90% | 84% | 86% | 88% |
| Original + 151-PCA-d | 81% | 95% | 98% | 98% |
| Original + (151 + 76)-PCA-d | 95% | 81% | 86% | 90% |
| Original + 201-PCA-d | 94% | 95% | 97% | 97% |
| Original + (201 + 101)-PCA-d | 84% | 93% | 97% | 93% |

Table 12.
Comparison of classification sensitivities of different classifiers trained on 90% of the original BC data and various augmentations, tested on 10% of the original BC data. Each column shows the average of the 10-fold experiments. In bold is given the highest value in a column.
Table 12.
Comparison of classification sensitivities of different classifiers trained on 90% of the original BC data and various augmentations, tested on 10% of the original BC data. Each column shows the average of the 10-fold experiments. In bold is given the highest value in a column.
| Training Data | NN | LR | SVML | SVMP |
|---|---|---|---|---|
| Original | 100% | 79% | 47% | 94% |
| Original + 101-PCA-d | 100% | 100% | 100% | 100% |
| Original + (101 + 51)-PCA-d | 100% | 95% | 95% | 95% |
| Original + 151-PCA-d | 100% | 100% | 100% | 100% |
| Original + (151 + 76)-PCA-d | 100% | 100% | 100% | 100% |
| Original + 201-PCA-d | 100% | 95% | 95% | 95% |
| Original + (201 + 101)-PCA-d | 100% | 100% | 100% | 100% |

Table 13.
Comparison of classification specificities of different classifiers trained on 90% of the original BC data and various augmentations, tested on 10% of the original BC data. Each column shows the average of the 10-fold experiments. In bold is given the highest value in a column.
Table 13.
Comparison of classification specificities of different classifiers trained on 90% of the original BC data and various augmentations, tested on 10% of the original BC data. Each column shows the average of the 10-fold experiments. In bold is given the highest value in a column.
| Training Data | NN | LR | SVML | SVMP |
|---|---|---|---|---|
| Original | 83% | 100% | 73% | 100% |
| Original + 101-PCA-d | 100% | 75% | 81% | 78% |
| Original + (101 + 51)-PCA-d | 89% | 77% | 80% | 83% |
| Original + 151-PCA-d | 83% | 92% | 97% | 97% |
| Original + (151 + 76)-PCA-d | 94% | 69% | 78% | 83% |
| Original + 201-PCA-d | 94% | 94% | 97% | 97% |
| Original + (201 + 101)-PCA-d | 81% | 89% | 94% | 89% |
6. Discussion
The present paper developed a method for vector data augmentation through distillation. The method is based on principal component analysis (PCA) [20,21]. Its difference with the extension we developed for vector data distillation is that the former method maps the set of original vectors to a set of vectors with a smaller dimension and the same cardinality. On the other hand, the extended PCA method maps the original set of vectors to a set of vectors with the same dimension as the original set but with a smaller cardinality.
The extended PCA method for distillation is unproductive if the matrix in Equation (7) is 1-orthogonal. Note that by definition, the matrix is 1-orthogonal if and only if , where is the identity matrix. Therefore, if is 1-orthogonal, then Equations (7) and (8) will map the original training dataset onto itself, and distilled vectors will not be generated. To remedy the problem, we selected strongest vectors instead of m, as is shown above in Equation (7).
The novelty of the present study is the development and use of the extended PCA method for distillation of vectors from given set of vectors such that the distilled vectors have the same dimension as the original vectors.
The advantage that comes from the proposed novelty is that by adding the PCA-distilled vectors to the original vectors, from which the former ones were distilled, we received a new training set. This new set trains a classifier such that its model has statistics higher than the statistics of the model trained only with the original dataset.
We validated the advantage by applying four classifiers—NN, LR, SVML, SVMP—on four different datasets of vectors: SL, D, HD, and BC. Every experimental result was produced by Monte-Carlo 10-fold cross-validation, where we randomly selected 10% of the samples for testing 10 times while the remaining 90% were used for training. Then, the average was taken. For every selection, the augmented vectors were distilled from the set of selected training vectors. We conducted a second (nested) distillation from every set of vectors generated during the first distillation. The experimental results are shown in Table 1, Table 2, Table 3, Table 4, Table 5, Table 6, Table 7, Table 8, Table 9, Table 10, Table 11, Table 12 and Table 13, where we compared the outcomes after training with the original vectors only and the outcomes after training with augmented datasets. It is evident that the latter outcomes were higher and better balanced if compared with the former.
In Table 1, one may observe that the NN accuracy was lower than the LR accuracy. It is known that NNs are intrinsically more prone to overfitting than LR. While being a universal approximator, it is expected that the NNs will outperform LRs in some instances. However, these models will not always be superior, especially when a rather small amount of training data is available. Recall that we used 90% of the SL data for training. This gave 145 training vectors, which is a quite insufficient amount for the efficient training of an NN and is the main reason for the lower NN results if compared with LR.
In [19], the PCA method was used to distill a photo response non-uniformity reference by removing the interference noise, which is another example of applying this method to the field of data enhancement. However, a direct comparison is not possible because our method distills vectors, while the method in [19] distills images. Our tests on a laptop with an Intel (13th Gen) i5-1335U processor at 1.30 GHz and 16.0 GB of RAM show that on average (averaged over 10 runs), the time required for distillation with PCA and augmentation of the dataset was approximately 0.1 milliseconds per vector.
GANs [12] and VAEs are typically used to generate or augment image data, and they achieve extraordinary results in this domain. On the contrary, the extended PCA method distills vector data. However, 1D data generation and augmentation are more difficult to achieve, and the results of generative or variational networks are much more modest. Aside from that, these networks require a significant amount of training data in the first place to produce good results. We are tackling the problem that no more data are available for training.
Our future work continues with enlarging classes which have small cardinalities. In certain datasets, a big difference exists between the number of samples in the different classes. Hence, we will conduct distillations from the samples of the class with the smallest cardinality and will augment this class to increase its cardinality. For this purpose, we will investigate the number of consecutive distillations which provide meaningful and useful sample vectors.
Author Contributions
Conceptualization, N.M.S., T.S. and A.N.; methodology, N.M.S. and A.N.; software, T.S. and A.N.; validation, T.S.; formal analysis, N.M.S., T.S. and A.N.; data curation, T.S.; writing, N.M.S.; writing—review and editing, T.S. and A.N.; supervision, N.M.S. and A.N. All authors have read and agreed to the published version of the manuscript.
Funding
Research reported in this publication was supported in part by the National Institutes of Health under award numbers R01HL151421, R01HD105205 and UG3NS130202. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Data Availability Statement
The link below provides access to a web page, where the reader may find the following: original datasets of diabetes, heart disease and breast cancer; the code for the NN, LR, SVM-linear and SVM-polynomial 2 classifiers to classify the heart disease data augmented with 151 PCA-distilled vectors; the Excel file that contains the 151 vectors distilled from a selection of 90% original vectors for training; the Matlab code that generated the 151 distilled vectors. https://github.com/nakhmani/PCA-augmentation/wiki (accesed on 9 December 2023).
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| ML | machine learning |
| VF | vector field |
| NN | neural network |
| CNN | convolutional neural network |
| LSTM | long short-term memory |
| MNTS | minimum number of training samples |
| SVM | support vector machine |
| SVML | support vector machine with a linear kernel |
| SVMP | support vector machine with a second-degree polynomial kernel |
| LR | logistic regression |
| SL | skin lesion |
| D | diabetes |
| HD | heart disease |
| BC | breast cancer |
| B | benign |
| M | malignant |
| P | positive |
| N | negative |
| TP | true positive |
| TN | true negative |
| FP | false positive |
| FN | false negative |
References
- Leevy, J.L.; Khoshgoftaar, T.M.; Bauder, R.A.; Seliya, N. A survey on addressing high-class imbalance in big data. J. Big Data 2018, 1, 1–30. [Google Scholar] [CrossRef]
- Victoria, L.; Fernández, A.; García, S.; Palade, V.; Herrera, F. An insight into classification with imbalanced data: Empirical results and current trends on using data intrinsic characteristics. Inf. Sci. 2013, 250, 113–141. [Google Scholar]
- Qiong, G.; Cai, Z.; Zhu, L.; Huang, B. Data mining on imbalanced data sets. In Proceedings of the 2008 International Conference on Advanced Computer Theory and Engineering, Washington, DC, USA, 20–22 December 2008; pp. 1020–1024. [Google Scholar]
- International Skin Imaging Collaboration. SIIM-ISIC 2020 Challenge Dataset. Internat. Skin Imaging Collaboration. Available online: https://challenge2020.isic-archive.com/ (accessed on 1 May 2023).
- Wang, B.; Klabjan, D. Regularization for Unsupervised Deep Neural Nets. arXiv 2016, arXiv:1608.04426. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Zhong, Z.; Zheng, L.; Kang, G.; Li, S.; Yang, Y. Random erasing data augmentation. In Proceedings of the AAAI Conference on Artificial Intelligence (AAAI), New York, NY, USA, 7–12 February 2020. [Google Scholar]
- Sajjad, M.; Khan, S.; Muhammad, K.; Wu, W.; Ullah, M.; Baik, S.W. Multi-grade brain tumor classification using deep CNN with extensive data augmentation. J. Comput. Sci. 2019, 30, 174–182. [Google Scholar] [CrossRef]
- Chen, M.; Sirakov, N.M. Poisson Equation Solution and its Gradient Vector Field to Geometric Features Detection. In Proceedings of the International Conference on Theory and Practice of Natural Computing, Dublin, Ireland, 12–14 December 2018; pp. 36–48. [Google Scholar]
- Radosavovic, I.; Dollár, P.; Girshick, R.; Gkioxari, G.; He, K. Data Distillation: Towards Omni-Supervised Learning. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake, UT, USA, 18–22 June 2018; pp. 4119–4128. [Google Scholar]
- Nguyen, T.; Novak, R.; Xiao, L.; Lee, J. Dataset Distillation with InfinitelyWide Convolutional Networks. arXiv 2022, arXiv:2107.13034v3. [Google Scholar]
- Durgadevi, K.S.; Generative Adversarial Network (GAN). A general review on different variants of GAN and applications. In Proceedings of the 2021 6th International Conference on Communication and Electronics Systems (ICCES), Coimbatre, India, 8–10 July 2021; pp. 1–8. [Google Scholar] [CrossRef]
- Karakanis, S.; Leontidis, G. Lightweight deep learning models for detecting COVID-19 from chest X-ray images. Comput. Biol. Med. 2021, 130, 104181. [Google Scholar] [CrossRef] [PubMed]
- Dvornik, N.; Mairal, J.; Schmid, C. Modeling Visual Context is Key to Augmenting Object Detection Datasets. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 364–380. [Google Scholar]
- Sachdeva, N.; McAuley, J. Data Distillation: A Survey. arXiv 2021, arXiv:2301.04272. [Google Scholar]
- Khosla, C.; Saini, B.S. Enhancing performance of deep learning models with different data augmentation techniques: A survey. In Proceedings of the 2020 International Conference on Intelligent Engineering and Management (ICIEM), London, UK, 17–19 June 2020; pp. 79–85. [Google Scholar]
- Mumuni, A.; Fuseini, M. Data augmentation: A comprehensive survey of modern approaches. Array 2022, 16, 100258. [Google Scholar] [CrossRef]
- Kiran, M.; Mondal, S.; Nemade, B. A review: Data pre-processing and data augmentation techniques. Glob. Transitions Proc. 2022, 3, 91–99. [Google Scholar]
- Li, J.; Liu, Y.; Ma, B.; Wang, C.; Qin, C.; Wu, X.; Li, S. A Novel PCA-Based Method for PRNU Distillation to the Benefit of Source Camera Identification. Appl. Sci. 2023, 13, 6583. [Google Scholar] [CrossRef]
- Hotelling, H. Analysis of a complex of statistical variables into principa. J. Educ. Psychol. 1933, 24, 417. [Google Scholar] [CrossRef]
- Abdi, H.; Williams, L.J. Principal component analysis. Wires Comput. Stat. 2010, 2, 433–459. [Google Scholar] [CrossRef]
- Manal, A. Estimating Sample Size and Confidence Interval. Master’s Thesis, Texas A & M University-Commerce, Commerce TX, USA, March 2013. [Google Scholar]
- Standard Normal Distribution Table. Available online: https://www.simplypsychology.org/z-table.html (accessed on 17 October 2023).
- Meng, X.L.; Rubin, D.B. Maximum likelihood estimation via the ECM algorithm: A general framework. Biometrika 1993, 80, 267–278. [Google Scholar] [CrossRef]
- Gradient Descend Method. Available online: https://www.geeksforgeeks.org/gradient-descent-algorithm-and-its-variants/ (accessed on 20 October 2023).
- Vapnik, V. The Nature of Statistical Learning Theory; Springer: New York, NY, USA, 1995. [Google Scholar]
- Mete, M.; Sirakov, N.M. Dermoscopic Diagnosis of Melanoma in a 4D Feature Space Constructed by Active Contour Extracted Features. Comput. Med. Imaging Graph. 2012, 36, 572–579. [Google Scholar] [CrossRef] [PubMed]
- Kandhare, P.G.; Ambalavanan, N.; Travers, C.P.; Carlo, W.A.; Sirakov, N.M.; Nakhmani, A. Comparison metrics for multi-step prediction of rare events in vital sign signals. Biomed. Signal Process. Control. 2023, 80, 2. [Google Scholar] [CrossRef]
- Kandhare, P.G.; Nakhmani, A.; Sirakov, N.M. Deep learning for location prediction on noisy trajectories. Pattern Anal. Appl. 2023, 26, 107–122. [Google Scholar] [CrossRef]
- Sirakov, N.M.; Mete, M.; Selvaggi, R.; Luong, M. New accurate automated melanoma diagnosing systems. In Proceedings of the 2015 International Conference on Healthcare Informatics (ICHI), Dallas, TX, USA, 21–23 October 2015; pp. 374–379. [Google Scholar]
- Argenziano, G.; Soyer, H.P.; De Giorgi, V. Dermoscopy: A Tutorial; Edra Medical Publishing, New Media: Milan, Italy, 2000. [Google Scholar]
- Kahn, M. Diabetes. UCI Machine Learning Repository. Available online: https://archive.ics.uci.edu/dataset/34/diabetes (accessed on 27 October 2023).
- Detrano, R.; Jánosi, A.; Steinbrunn, W.; Pfisterer, M.; Schmid, J.; Sandhu, S.; Guppy, K.; Lee, S.; Froelicher, R. Heart Disease. UCI Machine Learning Repository. 1998. Available online: https://archive.ics.uci.edu/dataset/45/heart+disease (accessed on 27 October 2023).
- Street, W.; Wolberg, W.; MangasarianBreast, O. Cancer Wisconsin (Diagnostic). UCI Machine Learning Repository. 1995. Available online: https://archive.ics.uci.edu/dataset/17/breast+cancer+wisconsin+diagnostic (accessed on 27 October 2023).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).