Skin Lesion Segmentation through Generative Adversarial Networks with Global and Local Semantic Feature Awareness
Abstract
1. Introduction
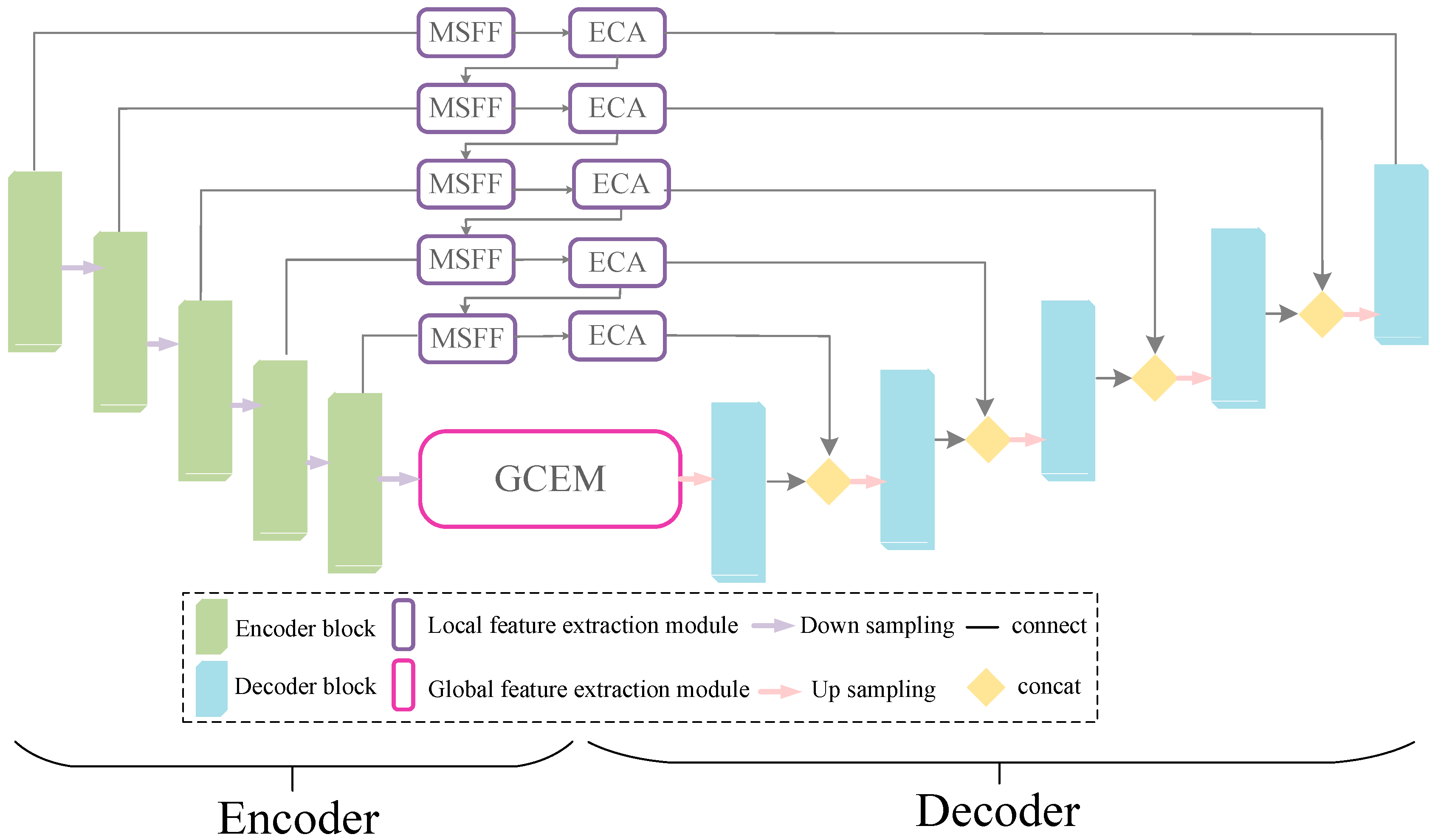
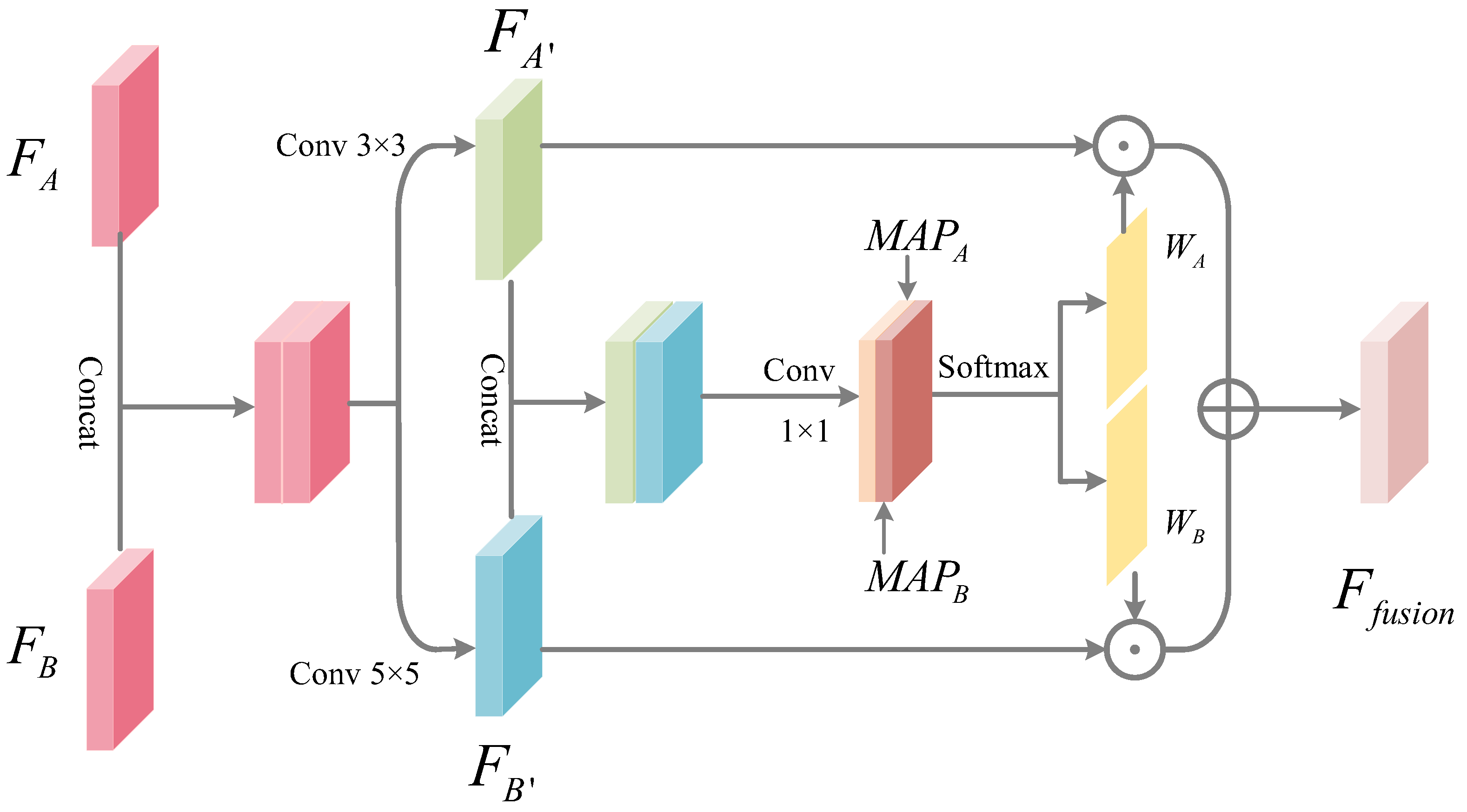
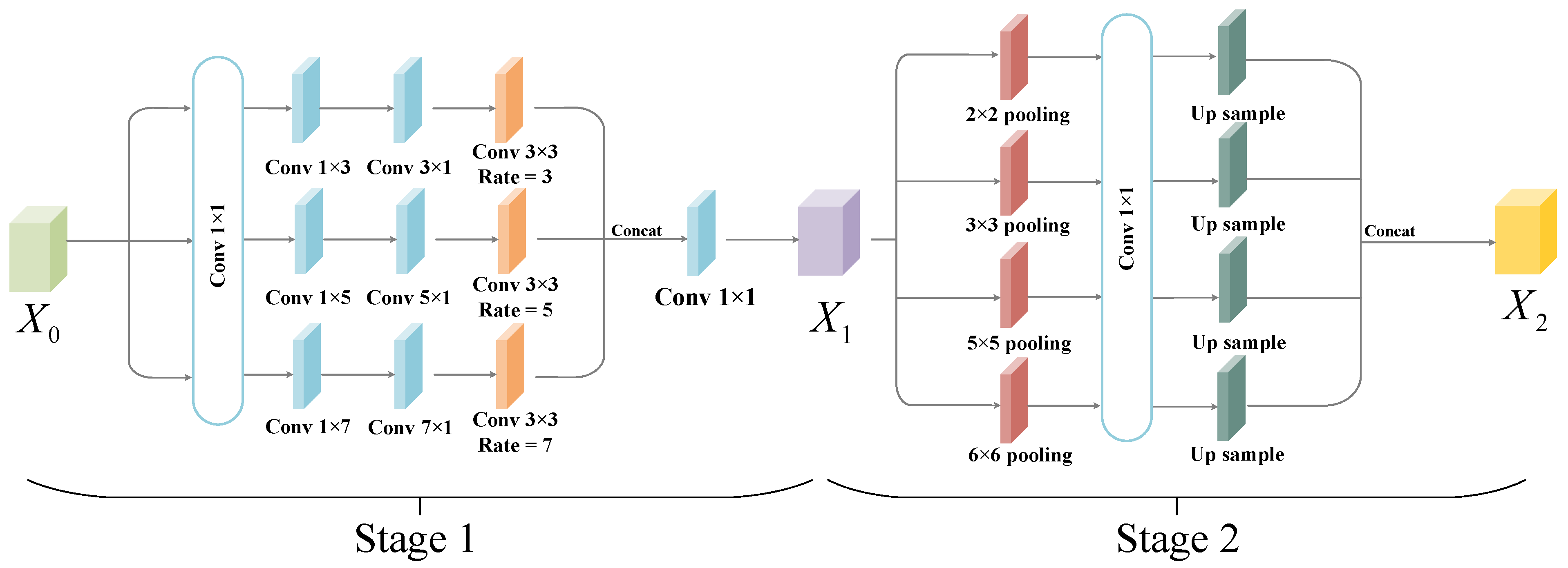
- The multi-scale localized feature fusion module is designed in this work to fuse different scales of features. The generator acquires the multi-scale local detailed information of the skin lesion area through skip connections with local feature information modules, thus preserving the rich details and local features of the target area.
- The encoder loses some of the information during the downsampling process due to the constant downsampling as well as the convolution operation. For this reason, this study proposes the global context extraction module to capture more global semantic features as well as spatial information, thereby enabling the segmentation network to achieve the accurate localization of the target region.
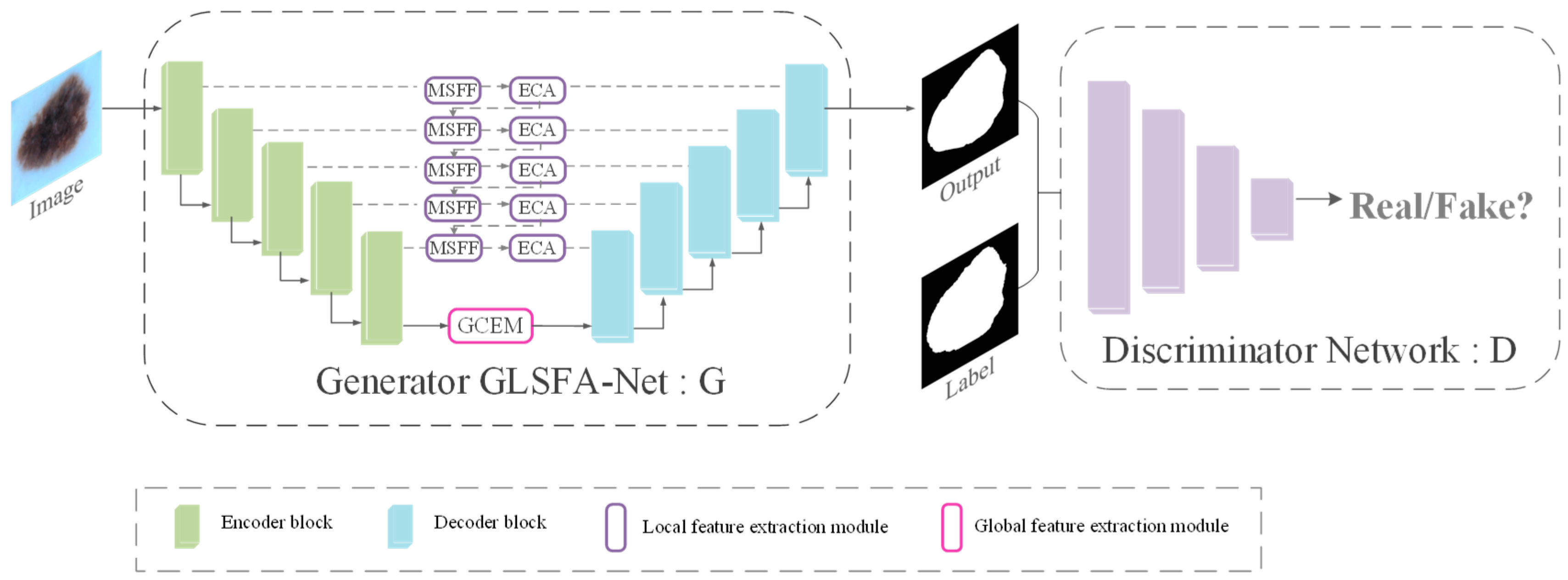
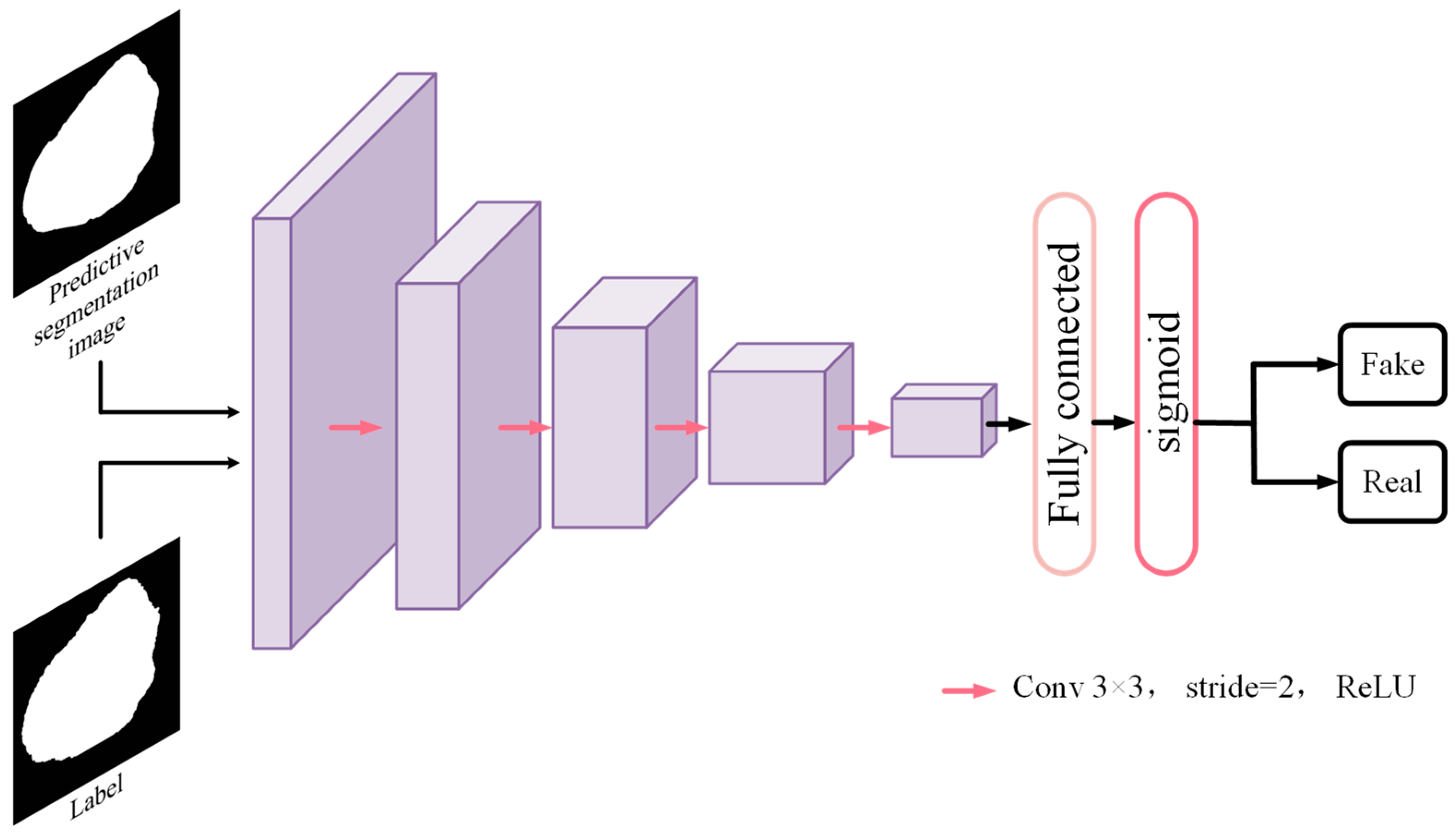
- GLSFA-GAN involves a generator GLSFA-Net (the segmentation network) and a discriminator. An adversarial training strategy is used to make the discriminator discriminate between the generated labels as well as the segmentation prediction maps, prompting the generator to yield more accurate segmentation results.
2. Related Work
2.1. Segmentation of Skin Lesion
2.2. Generative Adversarial Networks
3. Methods
3.1. Overall Architecture of the Proposed Model
3.2. Generator GLSFA-Net
3.2.1. Multi-Scale Local Feature Fusion Module
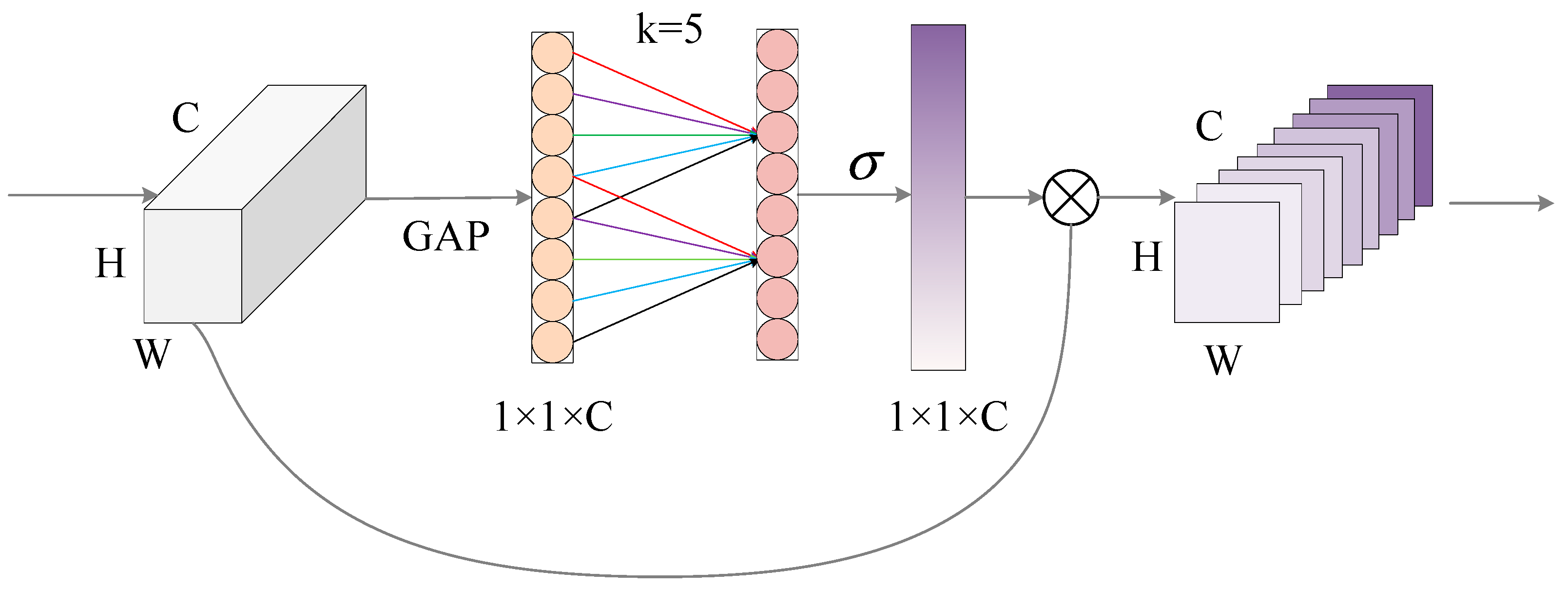
3.2.2. Efficient Channel Attention Module
3.2.3. Global Context Information Extraction Module
3.3. Discriminator Module
3.4. The Loss Function
4. Datasets and Implemented Setting
4.1. Dataset Descriptions
4.2. Implemented Settings
4.3. Evaluation Criterion
5. Experimental Results
5.1. Comparison to the State-of-the-Art Models
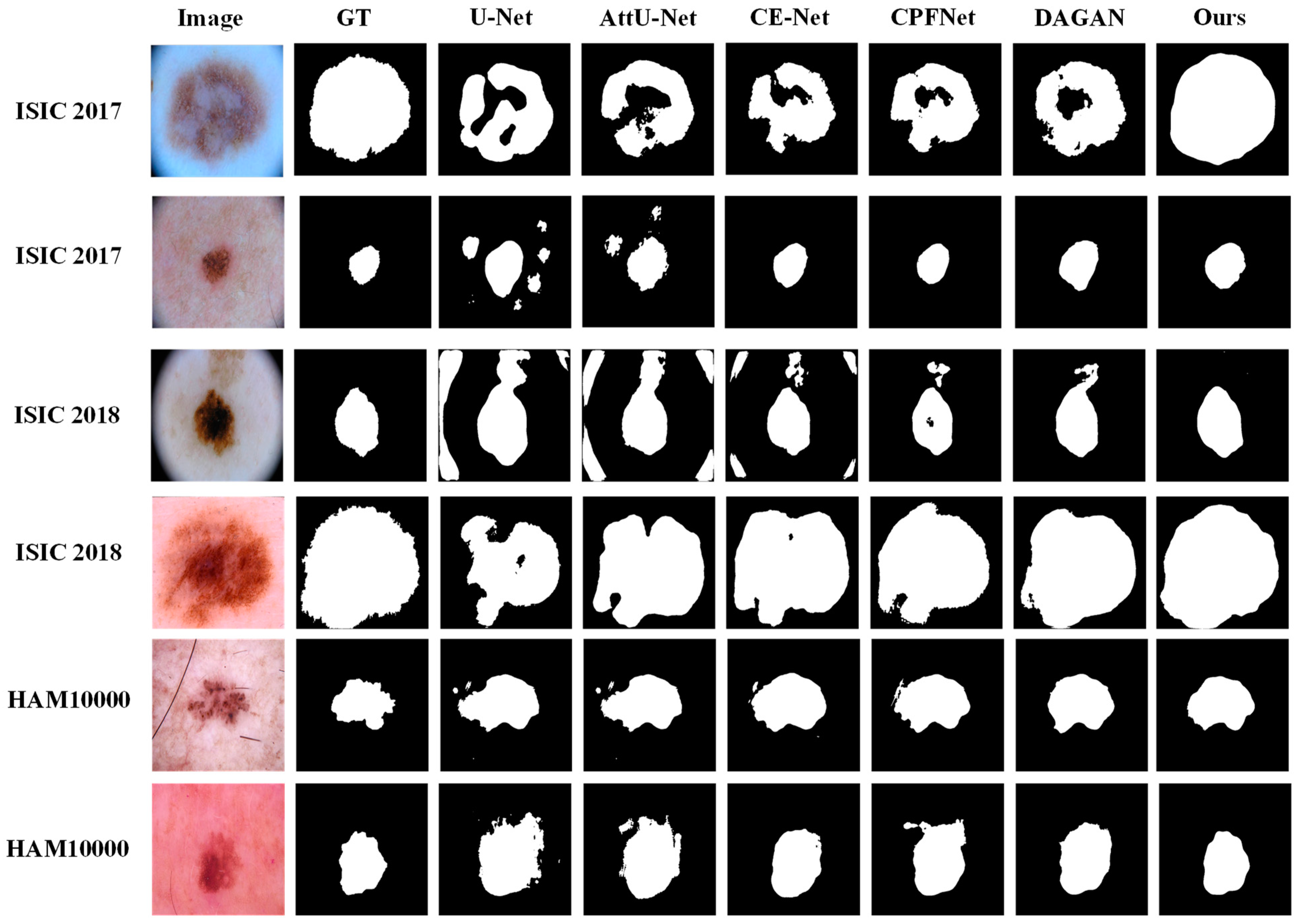
5.1.1. Qualitative Visual Comparison
5.1.2. Performance Comparison of the ISIC 2017 Dataset
5.1.3. Performance Comparison on the ISIC 2018 Dataset
5.1.4. Performance Comparison on HAM10000 Dataset
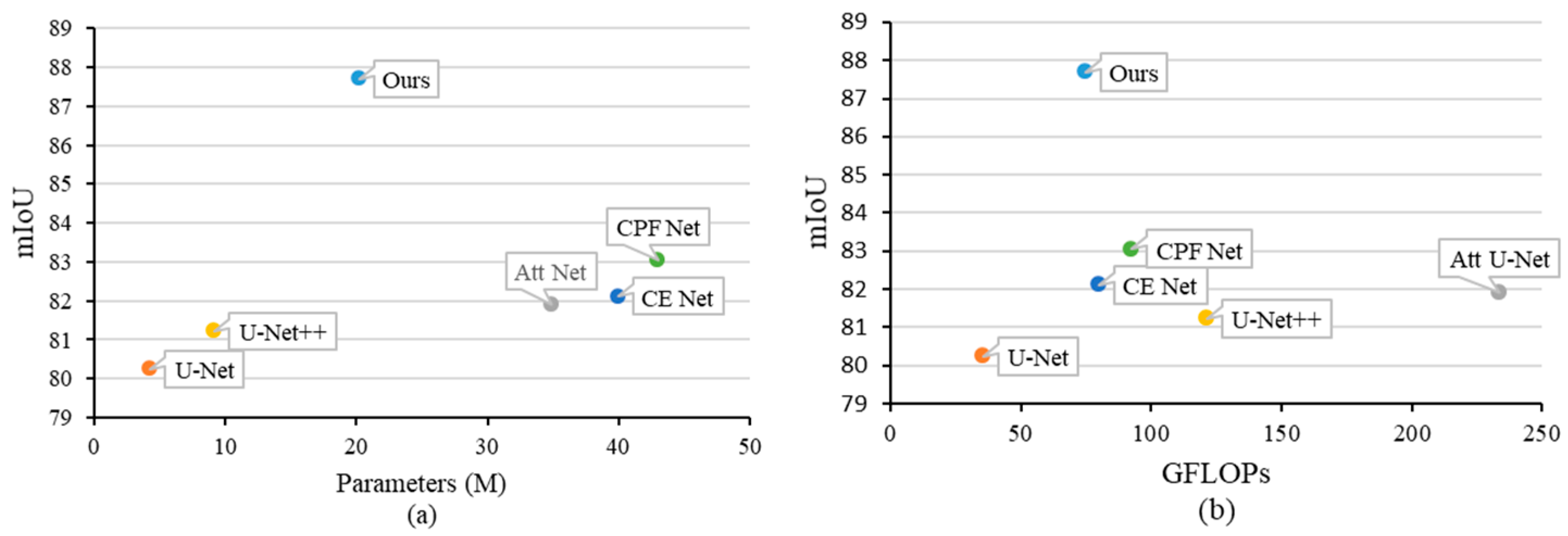
5.2. Ablation Research
6. Discussion
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Cullen, J.K.; Simmons, J.L.; Parsons, P.G.; Boyle, G.M. Topical treatments for skin cancer. Adv. Drug Deliv. Rev. 2020, 153, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Huang, Q.; Alkhayyat, A. A computer aided system for skin cancer detection based on Developed version of the Archimedes Optimization algorithm. Biomed. Signal Process. Control 2024, 90, 105870. [Google Scholar] [CrossRef]
- Leiter, U.; Keim, U.; Garbe, C. Epidemiology of skin cancer: Update 2019. Adv. Exp. Med. Biol. 2020, 1268, 123–139. [Google Scholar] [PubMed]
- Blazek, K.; Furestad, E.; Ryan, D.; Damian, D.; Fernandez-Penas, P.; Tong, S. The impact of skin cancer prevention efforts in New South Wales, Australia: Generational trends in melanoma incidence and mortality. Cancer Epidemiol. 2022, 81, 102263. [Google Scholar] [CrossRef]
- Gershenwald, J.E.; Scolyer, R.A.; Hess, K.R.; Sondak, V.K.; Long, G.V.; Ross, M.I.; Lazar, A.J.; Faries, M.B.; Kirkwood, J.M.; McArthur, G.A. Melanoma staging: Evidence-based changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J. Clin. 2017, 67, 472–492. [Google Scholar] [CrossRef]
- Lallas, A.; Apalla, Z.; Lazaridou, E.; Ioannides, D. Dermoscopy. In Imaging in Dermatology; Elsevier: Amsterdam, The Netherlands, 2016; pp. 13–28. [Google Scholar]
- Apalla, Z.; Lallas, A.; Argenziano, G.; Ricci, C.; Piana, S.; Moscarella, E.; Longo, C.; Zalaudek, I. The light and the dark of dermatoscopy in the early diagnosis of melanoma: Facts and controversies. Clin. Dermatol. 2013, 31, 671–676. [Google Scholar] [CrossRef]
- Warsi, M.F.; Chauhan, U.; Gupta, S.N.; Tiwari, P. A comparative analysis of melanoma detection methods based on computer aided diagnose system. Mater. Today Proc. 2022, 57, 1962–1968. [Google Scholar] [CrossRef]
- Dayananda, C.; Yamanakkanavar, N.; Nguyen, T.; Lee, B. AMCC-Net: An asymmetric multi-cross convolution for skin lesion segmentation on dermoscopic images. Eng. Appl. Artif. Intell. 2023, 122, 106154. [Google Scholar] [CrossRef]
- Qin, H.; Deng, Z.; Shu, L.; Yin, Y.; Li, J.; Zhou, L.; Zeng, H.; Liang, Q. Portable Skin Lesion Segmentation System with Accurate Lesion Localization Based on Weakly Supervised Learning. Electronics 2023, 12, 3732. [Google Scholar] [CrossRef]
- Garnavi, R.; Aldeen, M.; Celebi, M.E.; Varigos, G.; Finch, S. Border detection in dermoscopy images using hybrid thresholding on optimized color channels. Comput. Med. Imaging Graph. 2011, 35, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wang, X.; Wang, W.; Huang, W. PHCU-Net: A parallel hierarchical cascade U-Net for skin lesion segmentation. Biomed. Signal Process. Control 2023, 86, 105262. [Google Scholar] [CrossRef]
- Feng, K.; Ren, L.; Wang, G.; Wang, H.; Li, Y. SLT-Net: A codec network for skin lesion segmentation. Comput. Biol. Med. 2022, 148, 105942. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zhang, R.; Zhang, Y.; Bao, F.; Lv, H.; Li, L.; Zhang, C. Pact-Net: Parallel CNNs and Transformers for medical image segmentation. Comput. Methods Programs Biomed. 2023, 242, 107782. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Luo, W.; Shi, Q. Res-CDD-net: A network with multi-scale attention and optimized decoding path for skin lesion segmentation. Electronics 2022, 11, 2672. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Zhang, Z.; Liu, Q.; Wang, Y. Road extraction by deep residual u-net. IEEE Geosci. Remote Sens. Lett. 2018, 15, 749–753. [Google Scholar] [CrossRef]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. Unet++: Redesigning skip connections to exploit multiscale features in image segmentation. IEEE Trans. Med. Imaging 2019, 39, 1856–1867. [Google Scholar] [CrossRef]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention u-net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Milletari, F.; Navab, N.; Ahmadi, S.-A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Huang, H.; Lin, L.; Tong, R.; Hu, H.; Zhang, Q.; Iwamoto, Y.; Han, X.; Chen, Y.-W.; Wu, J. Unet 3+: A full-scale connected unet for medical image segmentation. In Proceedings of the ICASSP 2020—2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4–8 May 2020; pp. 1055–1059. [Google Scholar]
- Gu, Z.; Cheng, J.; Fu, H.; Zhou, K.; Hao, H.; Zhao, Y.; Zhang, T.; Gao, S.; Liu, J. Ce-net: Context encoder network for 2d medical image segmentation. IEEE Trans. Med. Imaging 2019, 38, 2281–2292. [Google Scholar] [CrossRef]
- Lei, B.; Xia, Z.; Jiang, F.; Jiang, X.; Ge, Z.; Xu, Y.; Qin, J.; Chen, S.; Wang, T.; Wang, S. Skin lesion segmentation via generative adversarial networks with dual discriminators. Med. Image Anal. 2020, 64, 101716. [Google Scholar] [CrossRef]
- Zhang, J.; Yu, L.; Chen, D.; Pan, W.; Shi, C.; Niu, Y.; Yao, X.; Xu, X.; Cheng, Y. Dense GAN and multi-layer attention based lesion segmentation method for COVID-19 CT images. Biomed. Signal Process. Control 2021, 69, 102901. [Google Scholar] [CrossRef]
- Kazeminia, S.; Baur, C.; Kuijper, A.; van Ginneken, B.; Navab, N.; Albarqouni, S.; Mukhopadhyay, A. GANs for medical image analysis. Artif. Intell. Med. 2020, 109, 101938. [Google Scholar] [CrossRef] [PubMed]
- Baur, C.; Albarqouni, S.; Navab, N. Generating highly realistic images of skin lesions with GANs. In Proceedings of the OR 2.0 Context-Aware Operating Theaters, Computer Assisted Robotic Endoscopy, Clinical Image-Based Procedures, and Skin Image Analysis: First International Workshop, OR 2.0 2018, 5th International Workshop, CARE 2018, 7th International Workshop, CLIP 2018, Third International Workshop, ISIC 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 16–20 September 2018; Proceedings 5. pp. 260–267. [Google Scholar]
- Mirikharaji, Z.; Abhishek, K.; Bissoto, A.; Barata, C.; Avila, S.; Valle, E.; Celebi, M.E.; Hamarneh, G. A survey on deep learning for skin lesion segmentation. Med. Image Anal. 2023, 88, 102863. [Google Scholar] [CrossRef] [PubMed]
- Green, A.; Martin, N.; Pfitzner, J.; O’Rourke, M.; Knight, N. Computer image analysis in the diagnosis of melanoma. Melanoma Res. 1994, 31, 958–964. [Google Scholar] [CrossRef] [PubMed]
- Erkol, B.; Moss, R.H.; Joe Stanley, R.; Stoecker, W.V.; Hvatum, E. Automatic lesion boundary detection in dermoscopy images using gradient vector flow snakes. Skin Res. Technol. 2005, 11, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Emre Celebi, M.; Alp Aslandogan, Y.; Stoecker, W.V.; Iyatomi, H.; Oka, H.; Chen, X. Unsupervised border detection in dermoscopy images. Skin Res. Technol. 2007, 13, 454–462. [Google Scholar] [CrossRef]
- Gomez, D.D.; Butakoff, C.; Ersboll, B.K.; Stoecker, W. Independent histogram pursuit for segmentation of skin lesions. IEEE Trans. Biomed. Eng. 2007, 55, 157–161. [Google Scholar] [CrossRef]
- Zortea, M.; Skrøvseth, S.O.; Schopf, T.R.; Kirchesch, H.M.; Godtliebsen, F. Automatic segmentation of dermoscopic images by iterative classification. Int. J. Biomed. Imaging 2011, 2011, 972648. [Google Scholar] [CrossRef]
- Han, Q.; Qian, X.; Xu, H.; Wu, K.; Meng, L.; Qiu, Z.; Weng, T.; Zhou, B.; Gao, X. DM-CNN: Dynamic Multi-scale Convolutional Neural Network with uncertainty quantification for medical image classification. Comput. Biol. Med. 2024, 168, 107758. [Google Scholar] [CrossRef]
- Feng, S.; Zhao, H.; Shi, F.; Cheng, X.; Wang, M.; Ma, Y.; Xiang, D.; Zhu, W.; Chen, X. CPFNet: Context pyramid fusion network for medical image segmentation. IEEE Trans. Med. Imaging 2020, 39, 3008–3018. [Google Scholar] [CrossRef]
- Zhang, C.; Pan, X.; Li, H.; Gardiner, A.; Sargent, I.; Hare, J.; Atkinson, P.M. A hybrid MLP-CNN classifier for very fine resolution remotely sensed image classification. ISPRS J. Photogramm. Remote Sens. 2018, 140, 133–144. [Google Scholar] [CrossRef]
- Sushma, B.; Pulikala, A. AAPFC-BUSnet: Hierarchical encoder–decoder based CNN with attention aggregation pyramid feature clustering for breast ultrasound image lesion segmentation. Biomed. Signal Process. Control 2024, 91, 105969. [Google Scholar]
- Shu, X.; Wang, J.; Zhang, A.; Shi, J.; Wu, X.-J. CSCA U-Net: A channel and space compound attention CNN for medical image segmentation. Artif. Intell. Med. 2024, 150, 102800. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial nets. arXiv 2018, arXiv:1406.2661. [Google Scholar]
- Zhu, J.-Y.; Park, T.; Isola, P.; Efros, A.A. Unpaired image-to-image translation using cycle-consistent adversarial networks. In Proceedings of the IEEE international Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2223–2232. [Google Scholar]
- Bi, L.; Feng, D.; Fulham, M.; Kim, J. Improving skin lesion segmentation via stacked adversarial learning. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 1100–1103. [Google Scholar]
- Han, C.; Kitamura, Y.; Kudo, A.; Ichinose, A.; Rundo, L.; Furukawa, Y.; Umemoto, K.; Li, Y.; Nakayama, H. Synthesizing diverse lung nodules wherever massively: 3D multi-conditional GAN-based CT image augmentation for object detection. In Proceedings of the 2019 International Conference on 3D Vision (3DV), Quebec City, QC, Canada, 16–19 September 2019; pp. 729–737. [Google Scholar]
- Bansal, N.; Sridhar, S. Hexa-gan: Skin lesion image inpainting via hexagonal sampling based generative adversarial network. Biomed. Signal Process. Control 2024, 89, 105603. [Google Scholar] [CrossRef]
- Salvi, M.; Branciforti, F.; Veronese, F.; Zavattaro, E.; Tarantino, V.; Savoia, P.; Meiburger, K.M. DermoCC-GAN: A new approach for standardizing dermatological images using generative adversarial networks. Comput. Methods Programs Biomed. 2022, 225, 107040. [Google Scholar] [CrossRef]
- Wang, Q.; Wu, B.; Zhu, P.; Li, P.; Zuo, W.; Hu, Q. ECA-Net: Efficient channel attention for deep convolutional neural networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 11534–11542. [Google Scholar]
- Yu, F.; Koltun, V. Multi-scale context aggregation by dilated convolutions. arXiv 2015, arXiv:1511.07122. [Google Scholar]
- Jha, D.; Riegler, M.A.; Johansen, D.; Halvorsen, P.; Johansen, H.D. Doubleu-net: A deep convolutional neural network for medical image segmentation. In Proceedings of the 2020 IEEE 33rd International Symposium on Computer-Based Medical Systems (CBMS), Rochester, MN, USA, 28–30 July 2020; pp. 558–564. [Google Scholar]
- Radford, A.; Metz, L.; Chintala, S. Unsupervised representation learning with deep convolutional generative adversarial networks. arXiv 2015, arXiv:1511.06434. [Google Scholar]

| Method | mIoU | Accuracy | Precision | Recall | Specificity |
|---|---|---|---|---|---|
| DCGAN [49] | 63.41 | 88.39 | 84.21 | 76.14 | 93.58 |
| U-Net [18] | 73.91 | 92.07 | 88.49 | 83.83 | 95.36 |
| U-Net++ [20] | 68.12 | 90.12 | 86.46 | 76.29 | 95.17 |
| Att U-Net [21] | 74.28 | 93.04 | 91.52 | 83.97 | 95.69 |
| CE Net [24] | 75.55 | 93.18 | 89.04 | 84.18 | 97.04 |
| CPF Net [36] | 77.22 | 93.78 | 89.61 | 84.86 | 96.22 |
| DAGAN [25] | 77.13 | 93.51 | 89.73 | 84.54 | 96.35 |
| Ours | 79.87 | 95.14 | 90.37 | 86.21 | 97.06 |
| Method | mIoU | Accuracy | Precision | Recall | Specificity |
|---|---|---|---|---|---|
| DCGAN [49] | 71.71 | 89.53 | 81.89 | 86.32 | 90.37 |
| U-Net [18] | 80.25 | 95.21 | 88.33 | 90.62 | 96.35 |
| U-Net++ [20] | 81.23 | 95.47 | 89.93 | 90.66 | 97.22 |
| Att U-Net [21] | 81.92 | 95.44 | 91.52 | 89.07 | 97.64 |
| CE Net [24] | 82.11 | 95.79 | 86.43 | 89.28 | 97.52 |
| CPF Net [36] | 83.05 | 96.13 | 86.32 | 90.05 | 97.77 |
| DAGAN [25] | 83.97 | 96.73 | 91.72 | 92.17 | 97.74 |
| Ours | 86.79 | 96.84 | 91.97 | 91.56 | 97.63 |
| Method | mIoU | Accuracy | Precision | Recall | Specificity |
|---|---|---|---|---|---|
| DCGAN [49] | 78.89 | 91.95 | 83.49 | 83.29 | 92.36 |
| U-Net [18] | 83.34 | 94.78 | 86.25 | 90.62 | 95.43 |
| U-Net++ [20] | 85.77 | 94.35 | 89.93 | 88.47 | 95.87 |
| Att U-Net [21] | 86.46 | 94.93 | 90.67 | 89.35 | 96.35 |
| Double U-Net [48] | 86.64 | 94.47 | 91.26 | 88.32 | 96.79 |
| CPF Net [36] | 87.39 | 95.67 | 89.99 | 92.44 | 96.82 |
| DAGAN [25] | 87.74 | 95.33 | 91.14 | 89.73 | 97.06 |
| Ours | 88.63 | 95.79 | 91.33 | 90.25 | 97.11 |
| Model | Generator | Discriminator | Metrics | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Baseline | MSFF | ECA | GCEM | mIoU | Acc | Pre | Rec | Spe | ||
| I | ✓ | 80.25 | 95.21 | 88.33 | 90.62 | 96.35 | ||||
| II | ✓ | ✓ | 82.43 | 95.57 | 89.14 | 90.71 | 96.72 | |||
| III | ✓ | ✓ | ✓ | 83.58 | 95.84 | 90.41 | 90.92 | 97.11 | ||
| IV | ✓ | ✓ | ✓ | 85.27 | 96.39 | 90.47 | 91.21 | 97.46 | ||
| V | ✓ | ✓ | ✓ | ✓ | 86.11 | 96.47 | 91.44 | 91.44 | 97.57 | |
| VI | ✓ | ✓ | ✓ | ✓ | ✓ | 86.79 | 96.84 | 91.56 | 91.56 | 97.63 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, R.; Zhang, J.; Wu, Y. Skin Lesion Segmentation through Generative Adversarial Networks with Global and Local Semantic Feature Awareness. Electronics 2024, 13, 3853. https://doi.org/10.3390/electronics13193853
Zou R, Zhang J, Wu Y. Skin Lesion Segmentation through Generative Adversarial Networks with Global and Local Semantic Feature Awareness. Electronics. 2024; 13(19):3853. https://doi.org/10.3390/electronics13193853
Chicago/Turabian StyleZou, Ruyao, Jiahao Zhang, and Yongfei Wu. 2024. "Skin Lesion Segmentation through Generative Adversarial Networks with Global and Local Semantic Feature Awareness" Electronics 13, no. 19: 3853. https://doi.org/10.3390/electronics13193853
APA StyleZou, R., Zhang, J., & Wu, Y. (2024). Skin Lesion Segmentation through Generative Adversarial Networks with Global and Local Semantic Feature Awareness. Electronics, 13(19), 3853. https://doi.org/10.3390/electronics13193853






