1. Introduction
Cervical cancer is the fourth most prevalent cancer globally, as well as one of the leading causes of female mortality attributed to cancer. In the year 2020 alone, approximately 604,127 new cases of cervical cancer were diagnosed, accompanied by about 341,831 lives lost [
1]. There are many factors that contribute to cervical cancer. The most significant factor is the Human Papillomavirus (HPV) [
2]. But, this does not mean that individuals with HPV will definitely develop cervical cancer, as the development of cervical cancer due to HPV’s influence typically takes a long time [
3]. Other risk factors such as smoking, HIV infection, organ transplantation, and so on, may increase the probability of cervical cancer to varying degrees [
4]. Although cervical cancer can lead to a high mortality rate if not treated, it is worth emphasizing that it is also one of the most treatable forms of cancer when detected early [
5]. As a matter of fact, nearly
of cervical cancer-related fatalities occur in low-income countries with insufficient medical resources [
6]. Currently, cervical cancer diagnosis predominantly hinges on biopsy results obtained through colposcopy. This approach encounters resistance among women due to the discomfort involved, and it imposes a significant burden on medical resources. Consequently, it becomes less accessible to individuals with lower incomes. Therefore, there is an urgent need for a cost-effective early cervical cancer diagnostic method.
In recent years, with the rapid advancement of machine learning (ML) technologies, automated cancer diagnosis by ML methods has become a prominent research direction. For example, in the field of lung cancer, Fermino et al. [
7] utilized a cascade support vector machine (CSVM) for the prediction of cancer, while Setio et al. [
8] introduced a novel computer-aided diagnostic system based on a multi-view convolutional network. In the domain of breast cancer, Abdel-Zaher et al. [
9] developed a CAD system based on Deep Belief Networks (DBN), while Mughal B et al. [
10] proposed a classification model based on Backpropagation Neural Networks (BPNN) for automated breast cancer diagnosis. Additionally, Ben-Cohen A et al. [
11] presented a liver segmentation and liver metastases detection model using a fully convolutional network (FCN), and Rau et al. [
12] developed a predictive model for liver cancer in individuals with Type II diabetes by employing both an Artificial Neural Network (ANN) and Logistic Regression (LR). In addition, there are numerous other cases [
13,
14,
15,
16,
17,
18].
The above examples demonstrate that ML can indeed provide significant assistance in automated cancer diagnosis. However, there are still some issues that need to be addressed. ML methods typically require a sufficiently large number of samples to avoid overfitting and ensure generalization. Moreover, they do not necessitate an excess of features, to avoid the curse of dimensionality [
19]. However, tasks related to disease diagnosis often involve a large number of features. Among these features, redundant or irrelevant ones will increase computational costs and even lead to decreased prediction accuracy [
20]. Furthermore, since some features are derived from medical examinations at hospitals, retaining useless features can lead to an unnecessary increase in financial burden on patients. The process of reducing features in machine learning (ML) methods is known as feature selection (FS). In this study, we extend research in this direction by proposing a new FS method designed to eliminate useless features in the context of ML-based automatic diagnosis of cervical cancer.
The remaining structure of this paper is as follows:
Section 2 introduces related works and the contributions of this paper.
Section 3 provides a detailed presentation of the proposed method. The experiments and discussions are shown in
Section 4. Finally,
Section 5 concludes this paper.
2. Related Work
Currently, the most commonly used ML-based approaches for cervical cancer diagnosis is utilizing pap-smear images [
21]. For example, Liu et al. [
22] proposed a novel CVM-Cervix model for cervical cancer diagnosis by combining the vision transformer (VIT) [
23] and DeiT Model [
24]. Pramanik et al. [
25] in their work employ an ensemble approach for cervical cancer detection, which achieving higher accuracy compared to using InceptionV2 [
26], MobileNetV2 [
27], and ResNetV2 [
28] individually. In addition, an exemplar pyramid deep feature extraction-based method was utilized by Yaman et al. [
29] for predicting cervical cancer. Furthermore, Shi et al. [
30] employed graph neural networks to predict cervical cancer based on pap-smear images, and Tripathi et al. [
31] utilized ResNet-152 for cervical cancer classification.
On the other hand, aided by the advancement of computer-assisted technologies, alternative approaches such as molecular dynamics simulation techniques have provided new insights into the research on cervical cancer identification. Through comprehensive molecular and integrative analysis, researchers have been able to uncover numerous novel genomic and proteomic characteristics specific to different subtypes of cervical cancer [
32]. Previous researchers have discovered that a significant quantity of microRNAs (miRNAs) exhibit abnormal expression in cervical cancer tissues, contributing to tumorigenesis, progression, and metastasis. This has subsequently led to the identification of multiple miRNA sequences as potential diagnostic biomarkers for cervical cancer [
33]. Additionally, long non-coding RNAs (lncRNAs) have emerged as biomarkers for cervical cancer [
34]. DNA methylation, a pivotal epigenetic mechanism, holds significant sway in biological processes [
35]. It was verified that methylation markers exhibit higher sensitivity than protein markers in cancer diagnosis [
36]. Subsequently, numerous studies have unveiled methylation biomarkers specific to cervical cancer [
37]. All the methods mentioned above have shown considerable advancements in the field of computer-aided diagnosis for cervical cancer. However, these approaches still face certain challenges, including limited flexibility and substantial associated costs, complicating their deployment in low-income regions. A viable solution is to utilize the risk factors identified in early screenings to predict the presence of cervical cancer. In 2017, the University of California released a relevant dataset on the UCI database, which greatly propelled research in this regard [
38]. Subsequently, researchers have conducted a series of studies on the dataset. For one, the dataset exhibits significant class imbalance, which can severely impact the accuracy of model predictions. To deal with the problem, Newaz et al. [
39] proposed a novel data balancing method by combining techniques such as SMOTE [
40], the Condensed Nearest Neighbor Rule (CNN) [
41], and the Edited Nearest Neighbor Rule (ENN) [
42]. Experimental results demonstrated that the proposed approach outperformed using SMOTE, CNN, or ENN individually, yielding a higher accuracy. Additionally, as different classifiers have varying preferences in extracting information from the data, Lu et al. attempted to use an ensemble method that incorporates five different classifiers in order to surpass the performance of a single classifier [
43]. The above two works attempted to enhance the predictive accuracy of this dataset from different perspectives.
As mentioned before, FS is also a crucial step when using ML-based methods. Significant improvements in both predictive accuracy and speed of the model can be achieved by removing irrelevant or redundant features. The FS methods can be categorized into three main types: filter-based, wrapper-based, and embedded-based methods. Firstly, filter-based methods employ statistical measures to assess the relevance of features with respect to a class label. These methods fall into two primary categories: ranking-based (univariate) and search-space-based (multivariate). In the ranking-based category, features with higher ranks are selected using a predefined threshold value. These ranks are determined based on the associations between each feature and the specified class label, aiming to eliminate the least pertinent features. Conversely, the search-space-based approaches consider inter-feature relationships; thus, they are capable of eliminating both irrelevant and redundant features [
44]. Secondly, wrapper-based methods rely on the evaluation of classifiers to selected features. Consequently, they are capable of selecting a feature subset that can yield optimal results. Wrapper-based methods comprise three main components: a search algorithm, a classifier, and a fitness function [
45]. Finally, embedded methods automatically enhance classification performance by selecting features as an integral part of the learning process [
20]. Each method comes with its own set of pros and cons. Filter-based methods are known for their lower computational complexity and reduced risk of overfitting compared to other two methods, while wrapper-based methods enhance accuracy beyond what filter methods offer; they do so at the cost of increased computational time [
46]. In addition, embedded methods are notably affected by the choice of classifiers and hyper-parameters, which can make them difficult to apply effectively. Nithya et al. in their work explored the utilization of these three FS techniques to determine the significance of various risk factors in cervical cancer diagnosis [
47]. It was demonstrated that wrapper-based methods outperformed other approaches in terms of performance, albeit requiring more time. Moreover, it is worth mentioning that the previously introduced two works by Newaz et al. and Lu et al. also employed wrapper-based methods by default for FS. In fact, wrapper-based methods are the most commonly used FS methods for such datasets. And variants of heuristic algorithms like the Genetic Algorithm (GA) [
48], the Differential Evolution Algorithm (DE) [
49], and the Particle Swarm Optimization Algorithm (PSO) [
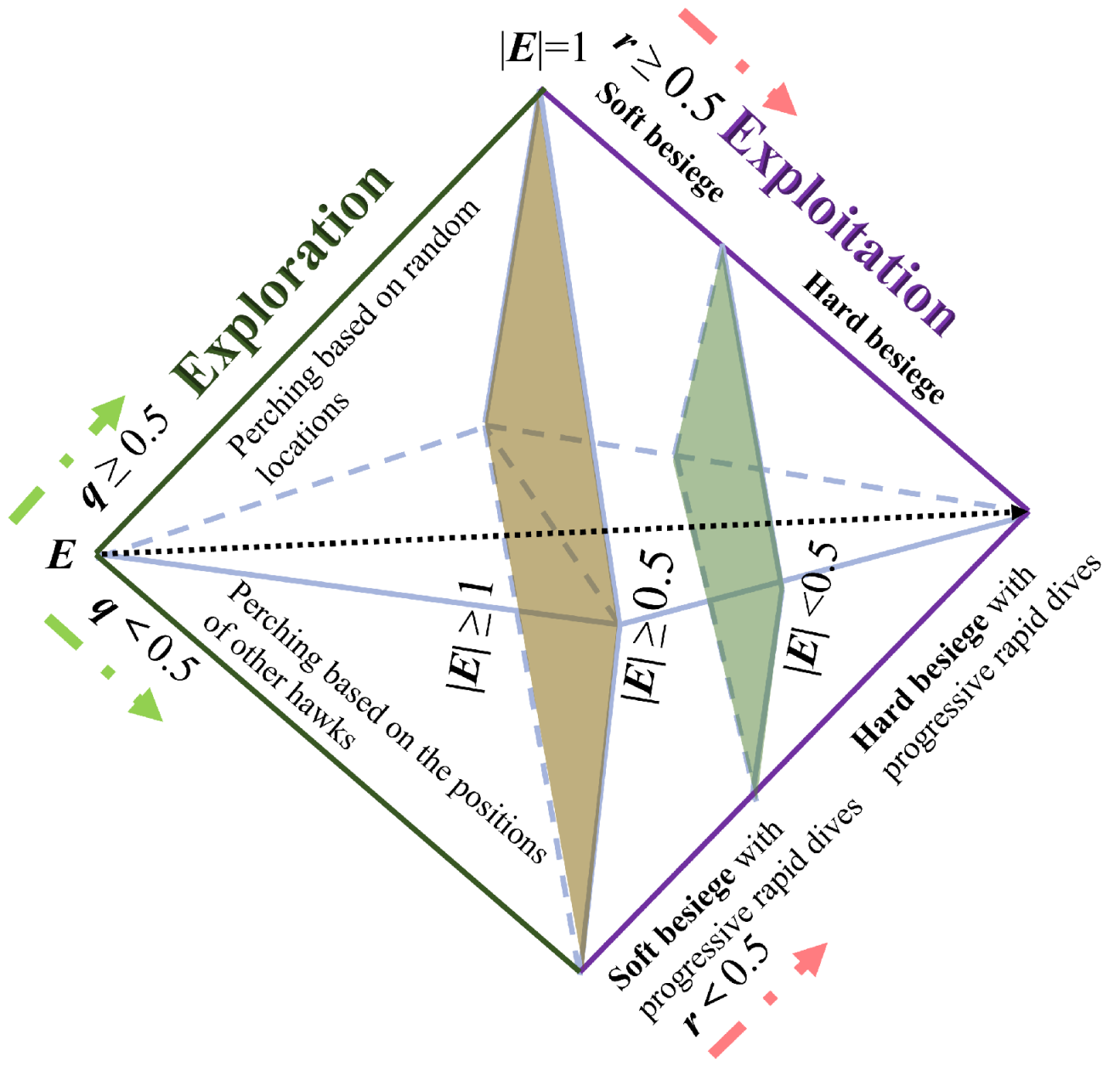
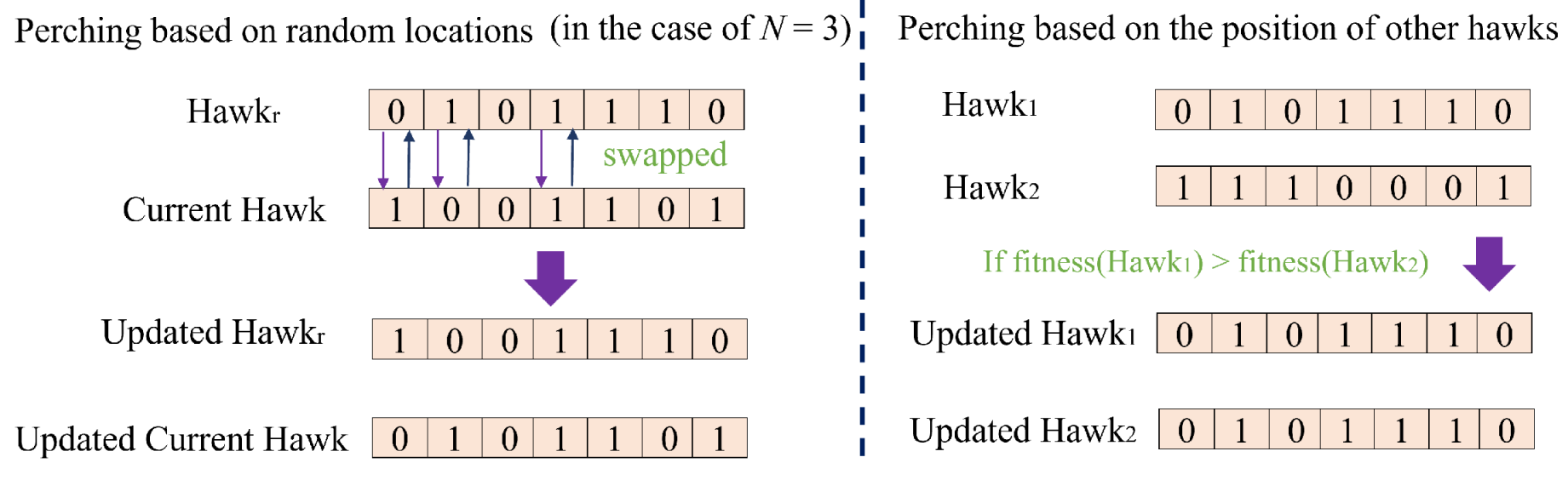
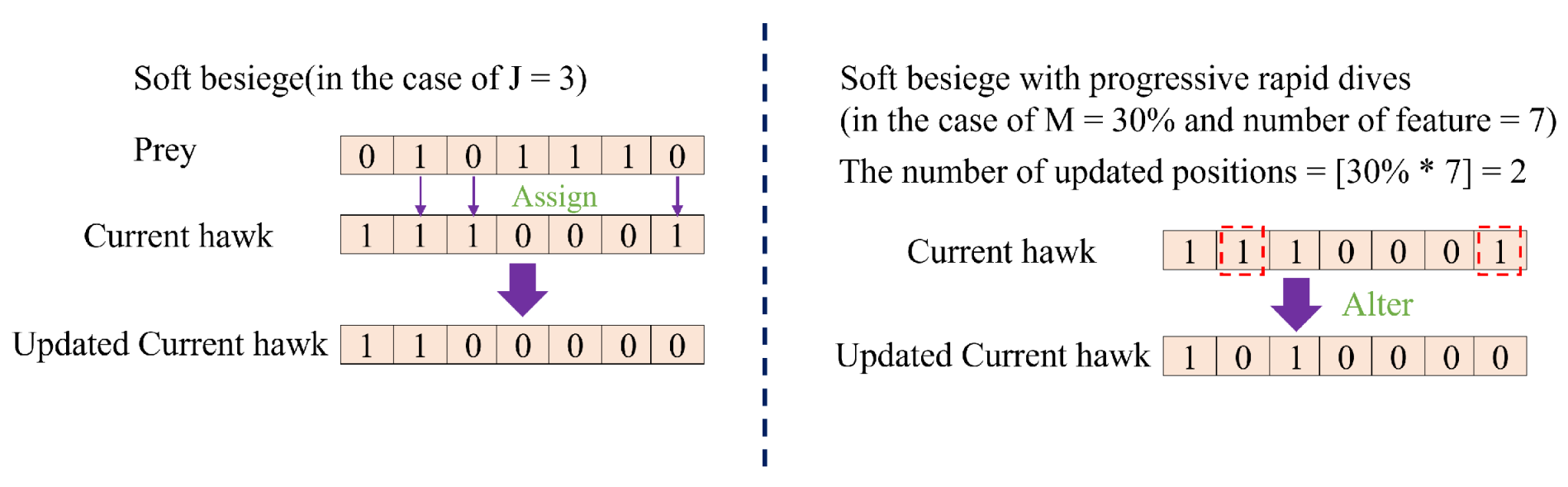
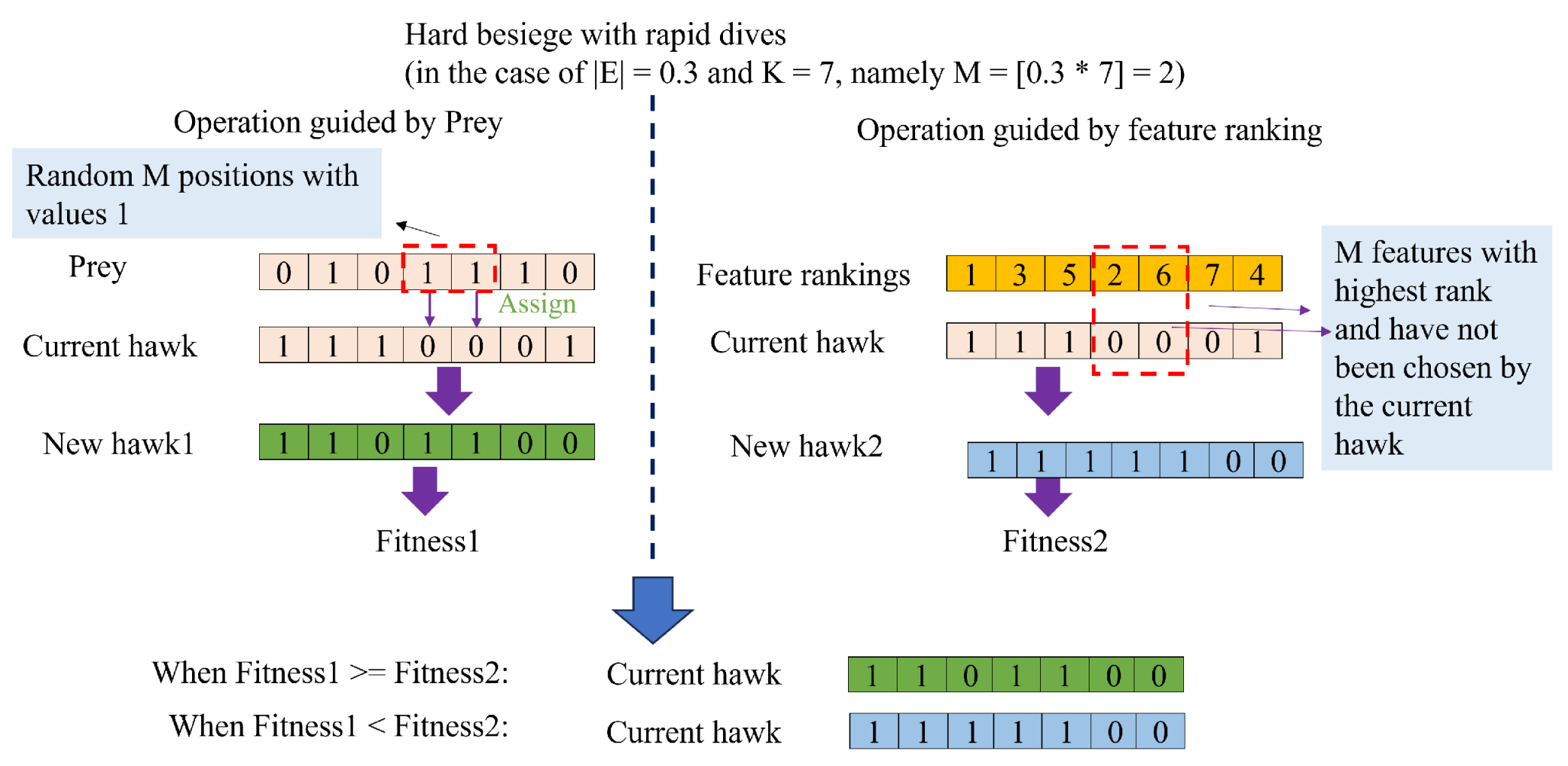
50] are frequently employed as the primary tools of wrapper-based methods. However, these methods all suffer from performance instability. This is attributed to the fact that most current heuristic algorithms possess hyper-parameters that requiring manual adjustment. Furthermore, the performance of these algorithms is easily influenced by these hyper-parameters when applied to a specific tasks. To address this issue, this paper proposes a novel wrapper-based FS method named the Binary Harris Hawk Optimization (BHHO) algorithm. Compared to other algorithms, BHHO has the smallest number of hyper-parameters, which significantly reduces the extent to which it is affected by hyper-parameters, thereby providing more stable performance. BHHO is developed from the Harris Hawk Optimization algorithm (HHO) [
51]. The HHO algorithm is a heuristic algorithm designed for solving continuous numerical problems and is not inherently suitable for addressing binary numerical problems such as FS. In this study, we have extended it into BHHO that is capable of handling binary numerical problems. Additionally, we have introduced a novel rank-based selection mechanism in order to make BHHO more suitable for specific tasks like cervical cancer prediction by considering various ranking approaches. The main contributions of this paper are as follows:
(1) Based on the HHO algorithm, a novel BHHO algorithm is proposed for FS of cervical cancer data. The BHHO algorithm has fewer hyper-parameters and better stability compared to other wrapper-based FS algorithms.
(2) In the BHHO algorithm, we have introduced an rank-based selection mechanism. This mechanism directs the generation of new solutions based on the feature ranking, thereby further enhancing the algorithm’s performance.
(3) We compared the proposed BHHO algorithm with commonly used wrapper-based and filter-based methods on the cervical cancer dataset, verifying the superiority of the proposed BHHO algorithm.
(4) To assess the generality of the proposed BHHO algorithm, we conducted experiments on three additional disease datasets apart from the cervical cancer dataset. The results indicate that BHHO performs remarkably well even on other datasets.
(5) On the cervical cancer dataset, the proposed BHHO algorithm was further integrated with filter-based feature selection methods to reduce computational costs while maintaining or enhancing performance.
In the next section, we will provide a detailed explanation of the proposed BHHO algorithm.
4. Results and Discussion
In this study, seven classifiers include the Support Vector Machine (SVM) [
57], the Random Forest (RF) [
58], the Naive Bayes (NB) [
59], the Adaptive Boosting (Adaboost) [
60], the Discriminant Analysis (DA) [
61], the Logistic Regression (LR) [
62], and the k-Nearest Neighbors (KNN) [
63] are used in this paper. In addition, the commonly used wrapper-based methods including GA algorithm, PSO algorithm, and DE algorithm were used for comparison. The flowchart of experiments is illustrated in
Figure 6. We commence by preprocessing datasets, which eliminating missing values within datasets. Subsequently, we employ a 5-fold cross-validation scheme to partition the data. In each fold, we conduct an FS operation on the training set. Since the dataset is severely imbalanced, the SMOTE [
40] algorithm is used to balancing the training set for a better performance. Then, both the training set and testing set undergo normalization. Finally, we utilize the training set for model training and employ the testing set for performance evaluation. All experiments were conducted on a personal PC with an Intel(R) Core
,
GHz, and 16 GB memory using Python.
4.1. Dataset
4.1.1. Cervical Cancer Dataset
The cervical cancer dataset is available at the UCI repository and was originally collected from the Hospital Universitario de Caracas in Venezuela. It was utilized in this study. There are 858 records, each characterized by 32 features that encompass habits, sexual history, demographic information, and more. The original dataset includes 55 positive cases and 803 negative cases. Due to privacy concerns, some patients choose not to answer some questions. Hence, the data contains some missing values. Two features, “STDs: Time since first diagnosis” and “STDs: Time since the last diagnosis”, have a total number of 787 missing values. As the number of missing values in the two features exceeds
, we have chosen to remove them. Additionally, there are 105 patients with missing values across 18 features. These patients were excluded from the dataset since more than half of their feature information was incomplete. The rest of the missing values were filled by the Multivariate Imputation by Chained Equation (MICE) method [
64]. MICE is a highly flexible imputation method that more accurately measures the uncertainty of missing values compared to other imputation techniques. It replaces missing data by running multiple regression models, where each missing value is imputed based on the other variables in the dataset. To prevent data leakage, we train the MICE algorithm model solely on the training set, and then use the trained model to impute the missing values in the entire dataset. As a result, we have obtained a dataset consisting of information from 753 patients with 30 features. The dataset includes 53 positive cases and 700 negative cases.
4.1.2. Other Datasets
To better illustrate the performance of the proposed BHHO algorithm, we conducted additional experiments on three other disease datasets. These three datasets include the Cleveland dataset [
65], the Z-Alizadeh Sani dataset [
66], and the Parkinson dataset [
67]. The Cleveland dataset contains 303 cases, with 139 positive cases and 164 negative cases. Although this dataset originally comprises 75 features, it is common for most researchers to utilize only 13 of these features. In this study, we also only selected these 13 features as the input to our model. The Z-Alizadeh Sani dataset encompasses 303 cases, consisting of 216 positive cases and 87 negative cases, with a total of 55 features. The Parkinson dataset encompasses 240 cases, consisting of 120 positive cases and 120 negative cases, with a total of 46 features. The objective of both the Cleveland dataset and the Z-Alizadeh Sani dataset is to ascertain whether individuals have heart disease, while the Parkinson dataset aims to identify the presence of Parkinson’s disease in individuals. All three datasets are two classification problems.
4.2. Performance Metrics
In this paper, six metrics including
,
,
,
,
-
, and
-
(
G-
) are employed to evaluate the performance of the proposed method. The equations of these six metrics are defined as follows:
where
represents the correct identification of a diseased person as sick;
signifies the correct classification of a healthy person as being healthy;
denotes the incorrect classification of a healthy person as diseased; and
refers to the incorrect classification of a diseased person as healthy. It is noted that the
-
is used as the objective function when employing wrapper-based methods, with the macro average applied due to the imbalanced nature of the data.
4.3. Validation on the Cervical Cancer Dataset
In this part, we validate the performance of proposed BHHO algorithm on the cervical cancer dataset.
Table 1 presents the experimental results without an FS method. The results of proposed BHHO methods are demonstrated in
Table 2. In addition, the results of GA method, PSO method, and DE method are shown in
Table 3. Since we employed a 5-fold strategy to partition the dataset, all the table presents the experimental results in the form of “mean ± variance”.
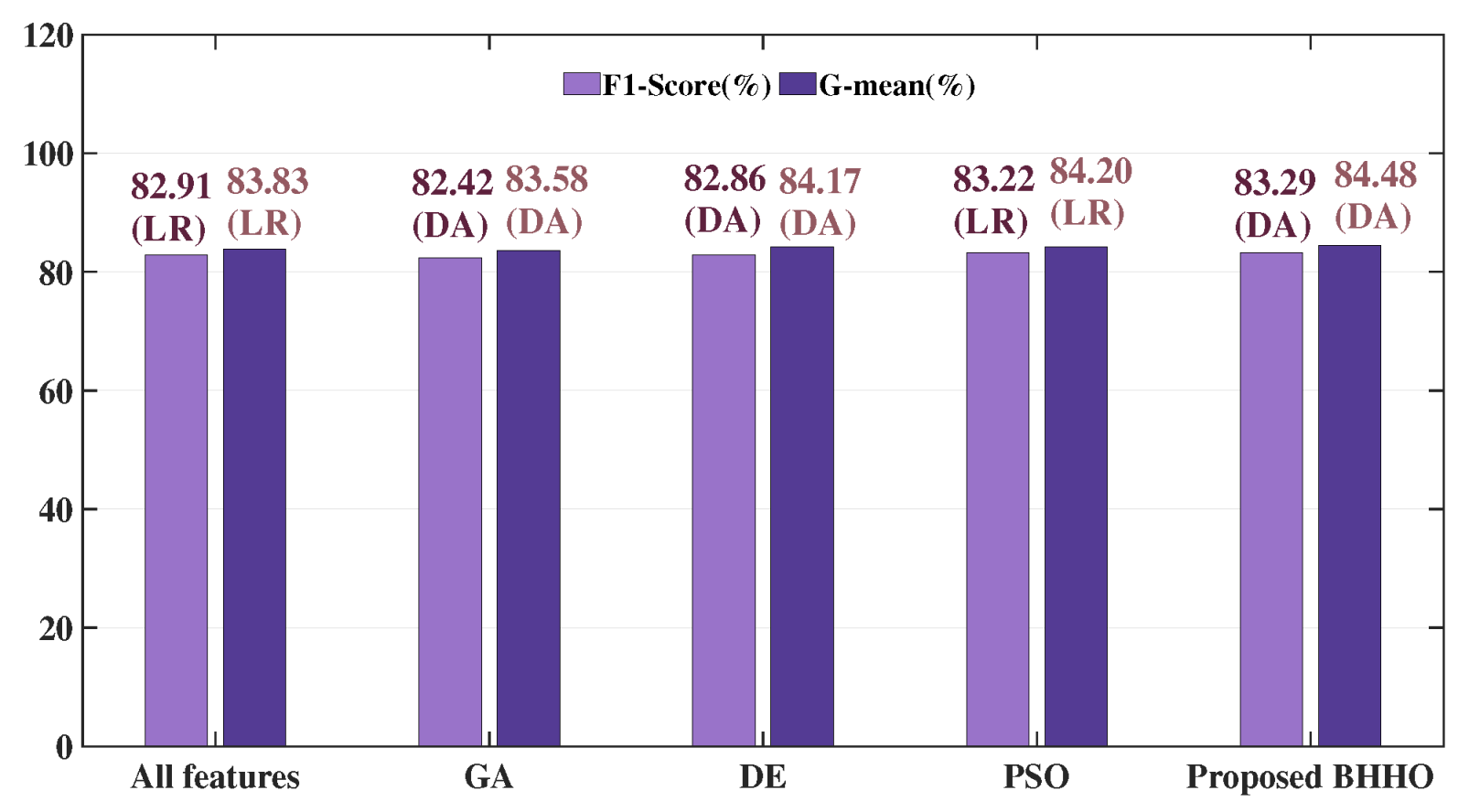
Clearly, all the experimental results exhibit high and but low and . This is because the dataset belongs to the category of highly imbalanced datasets, namely, the majority class vastly outnumbers the minority class. In the real world, the diagnosis of cervical cancer is precisely such a situation where negative results overwhelmingly outnumber positive results. In such a situation, using simple metrics like or may not accurately assess the performance since these two metrics can easily yield high value when the majority class greatly outnumbers the minority class. In the case of cervical cancer, the consequences of incorrectly predicting the minority class are much more severe than incorrectly predicting the majority class. Therefore, we pay more attention to the results of and . However, this does not mean that and should be ignored. To comprehensively evaluate the performance, F1-score and G-mean are used as the main reference indicators in our experiments. So, we have highlighted the best F1-score and G-mean in bold on each table.
From the three tables, it can be observed that when using the wrapper-based methods, most results tend to be better than those obtained using all features, which represents that using wrapper-based FS algorithms can capturing the implicit relationships between features, effectively reducing the number of features while improving performance. From
Table 2, it can be observed that among three BHHO variants, VTBHHO achieved the highest
F1-
score of
when using Adaboost as the classifier and the highest
G-mean of
when using LR as the classifier. Furthermore, it can be found that the highest result obtained by proposed BHHO algorithm surpasses all outcomes using the GA method, PSO method, and DE method.
Figure 7 visually demonstrated such a situation. In
Figure 7, the best
F1-
score and
G-mean of each wrapper-based method are selected as representatives. The specific values and the classifiers that achieved these results are annotated above each bar. Clearly, the best results from all the wrapper-based methods surpassed the best results obtained using all features, and the best results from proposed BHHO methods surpassed other commonly used wrapper-based methods.
Table 4,
Table 5,
Table 6 show the experimental results after employing Rf method, VT method, and MI method. The top
,
,
, and
of features are selected as the features subset. It can be observed that when using Rf method, most classifiers did not surpass the performance of the original feature set, while when using VT method or MI method, a majority of the classifiers managed to exceed the performance of the original feature set, while there were still some classifiers that experienced a decline in performance. It is worth emphasizing that only in MI method (using top
of features), the performance of DA completely surpassed the results of all classifiers using the all feature set. But this result is still less than majority of wrapper-based method. It is evident that not all FS methods yield positive results. While filter-based FS methods can reduce redundant features and enhance accuracy to some extent, their effectiveness is quite limited. In some cases, this method may even degrade the performance. This is because the importance of features in a dataset cannot be judged solely from a statistical perspective. The relationships between features must also be considered. Some features, which might seem statistically insignificant on their own, can significantly improve performance when used in conjunction with other features. Filter-based methods often overlook this aspect. Therefore, in practical applications, most researchers do not rely solely on filter-based methods.
Based on the above validation, we further attempted a hybrid method that first uses filter-based methods to eliminate a small portion of redundant features and then applies proposed BHHO methods to process the remaining features. Specifically, we used the top
ranked features from the Rf, VT, and MI methods separately as the input for the VTBHHO method. The results of the experiment are shown in
Table 7. We bold the best
F1-
score and
G-mean among all results. In the combination of Rf and VTBHHO, the highest
F1-
score (
) was obtained when using SVM as the classifier. In the combination of MI and VTBHHO, the highest
G-mean (
) was achieved when using Adaboost as the classifier. It can be observed that, compared to using VTBHHO alone, the hybrid approaches achieved a higher
F1-
score but obtained a lower
G-mean. This is because we used the
F1-
score as the objective function for the wrapped-based methods, so the methods are more inclined to favor feature subsets that can improve the
F1-
score. When using wrapper-based methods alone, some features with negative effects impacted the quality of the final solution. However, by first using filter-based methods to eliminate those features that were most likely completely useless, the quality of all feature subsets was improved, thereby increasing the likelihood of obtaining better solutions.
Finally, we analyze the selected feature sets obtained when combining Rf and VTBHHO (using SVM as the classifier), that is, the situation that achieves the highest
F1-
score. The selected feature results are shown in
Table 8. Obviously, when the distribution of the dataset varies, even using the same FS method and classifier, the selected features can vary significantly. In the five different subsets of the dataset, the number of features used ranged from a maximum of 18 features to a minimum of 8 features among the 30 features. The table summarizes the usage of all features, and it can be observed that the last feature “DX” has been used in all subsets. This suggests that past diagnostic results play a crucial role in the current decision-making process. Additionally, “Hormonal Contraceptives (year)”, “STDs”, “STDs:syphilis”, and “STDs:molluscum contagiosum” have been used four times, indicating that these risk factors significantly increase the likelihood of cervical cancer detection. On the other hand, “STDs:condylomatosis” and “STDs:cervical condylomatosis” have been shown to have no association with the discovery of cervical cancer. These statistical findings can provide valuable recommendations to the general population. When highly relevant risk factors appear in daily life, we should be vigilant and seek further medical diagnosis.
4.4. Validation on Other Datasets
In this part, we validate the proposed BHHO algorithm on the other three disease datasets. The experimental results are shown in
Table 9,
Table 10,
Table 11,
Table 12,
Table 13 and
Table 14 that are presented in the form of
, and the best
F1-
score and
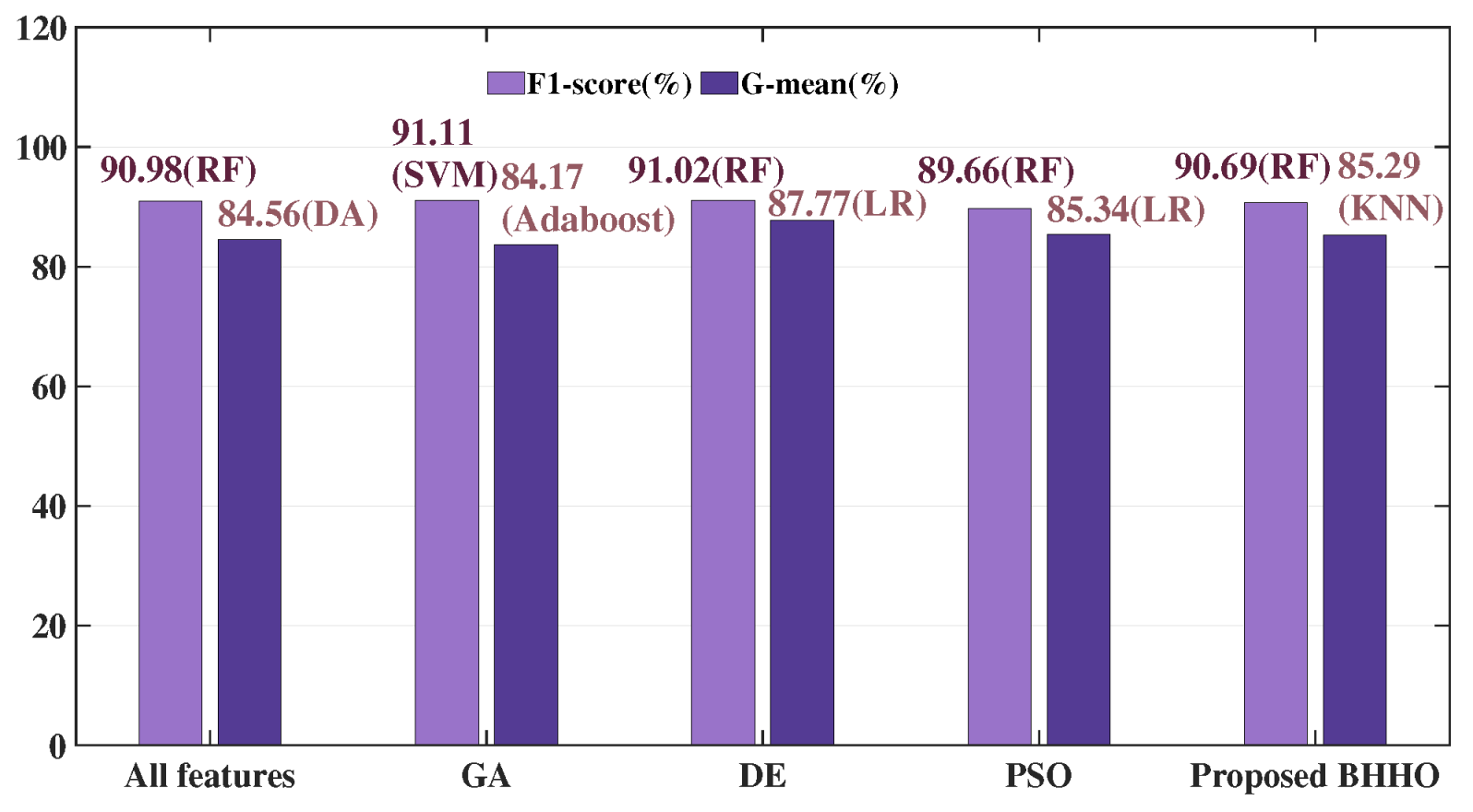
G-mean of each method are highlighted in bold. On the Cleveland dataset, VTBHHO’s DA achieved the best
F1-
score (
) and
G-mean (
). On the Z-Alizadeh Sani dataset, GA’s SVM achieved the highest
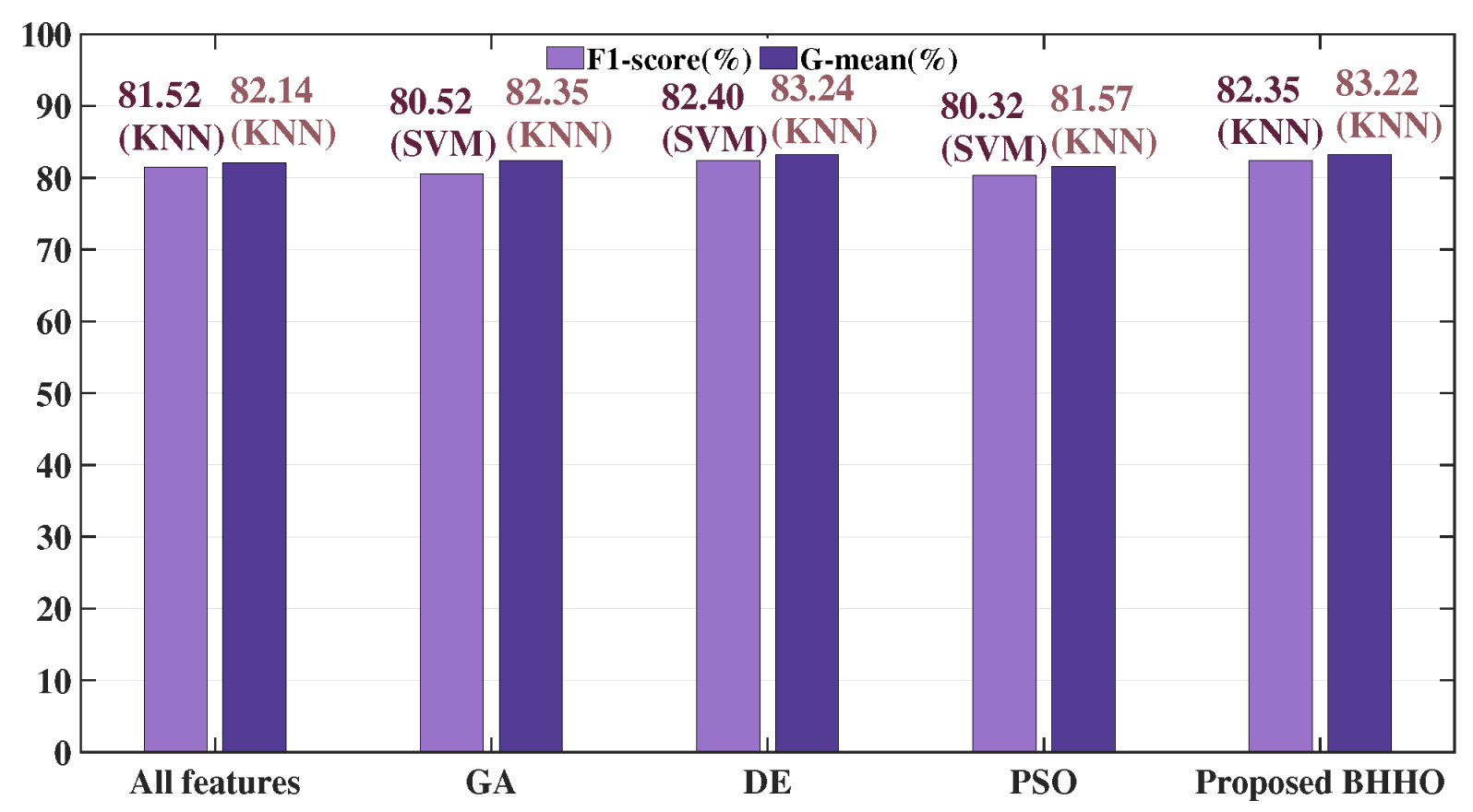
F1-
score (
), while DE’s LR obtained the highest
G-mean (
). On the Parkinson’s dataset, MIBHHO’s KNN achieved the best
F1-
score (
) and the highest
G-mean (
).
Figure 8,
Figure 9 and
Figure 10 illustrate the experimental conditions for these three datasets using barcharts. The proposed BHHO achieved the best results in two out of the three datasets. Although it did not achieve the best result on the Z-Alizadeh Sani dataset, it still performed close to the optimal outcome. It demonstrates that the proposed BHHO method is not only effective for cervical cancer dataset but also has a certain degree of generalizability to other disease datasets. Furthermore, observing the results of the three BHHO variants on different datasets reveals that different ranking strategies play varying roles depending on the dataset. The VTBHHHO variant, which performed best on the cervical cancer dataset, may not necessarily be the optimal choice for other datasets. Therefore, we can adjust the ranking strategy according to the actual tasks to achieve higher performance. In other words, the ranking strategy incorporated into BHHO allows it to flexibly adapt to different environments. Although the combination of the proposed BHHO with RF, VT, and MI did not achieve the best results on the Z-Alizadeh Sani dataset, it can be expected that combining it with other ranking strategies could yield higher accuracy. In summary, the experimental results prove that proposed BHHO method can offer a competitive performance than other commonly used methods.