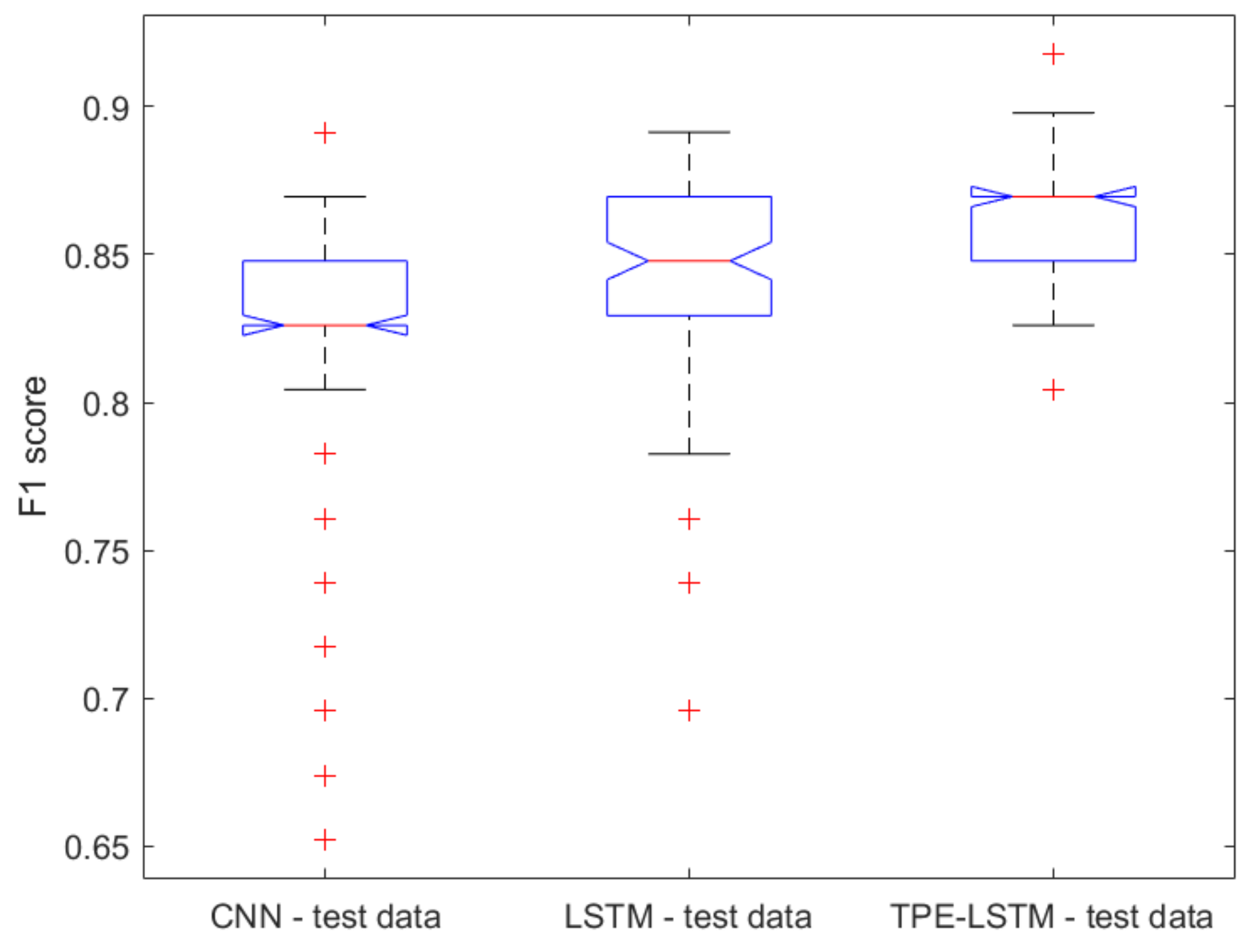
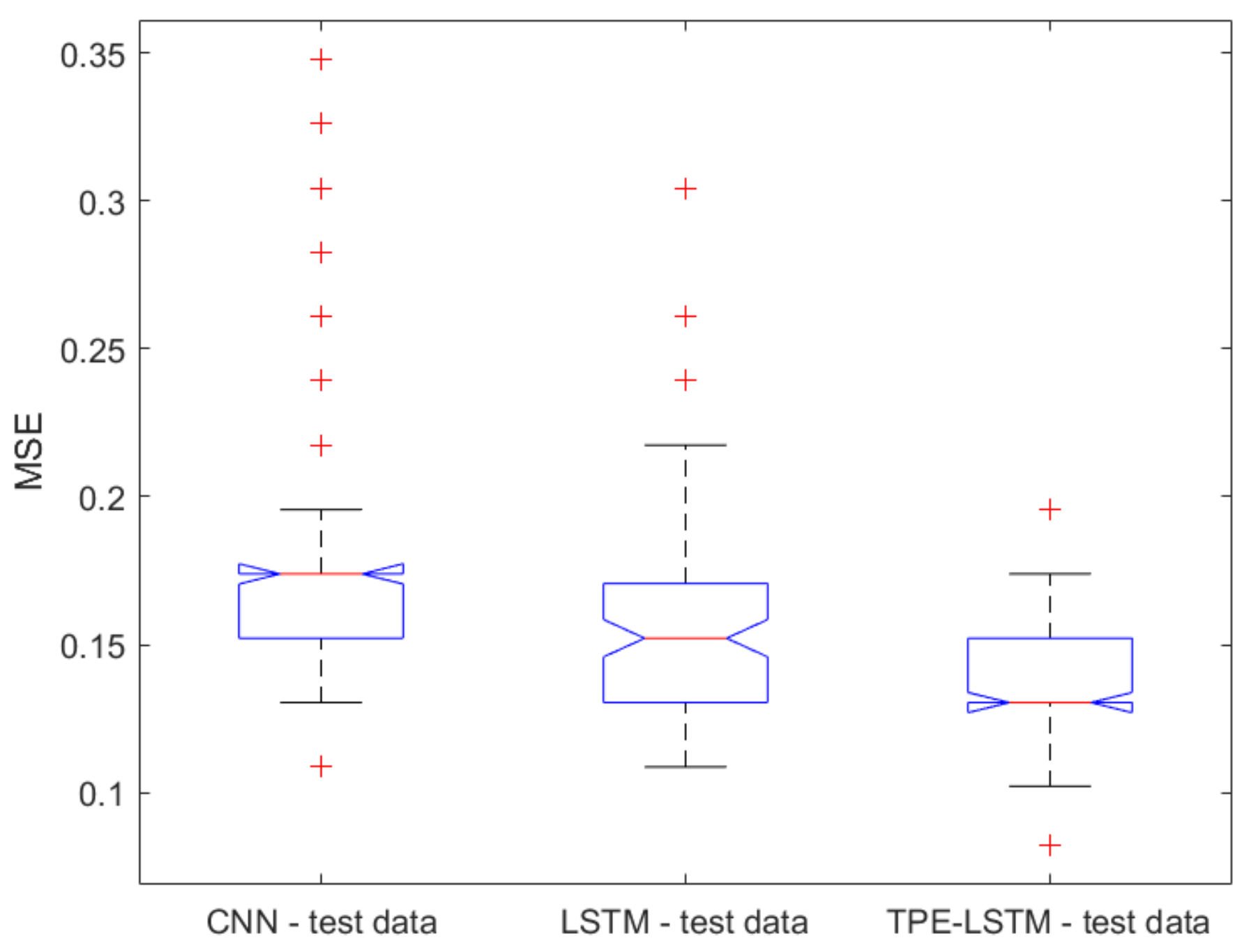
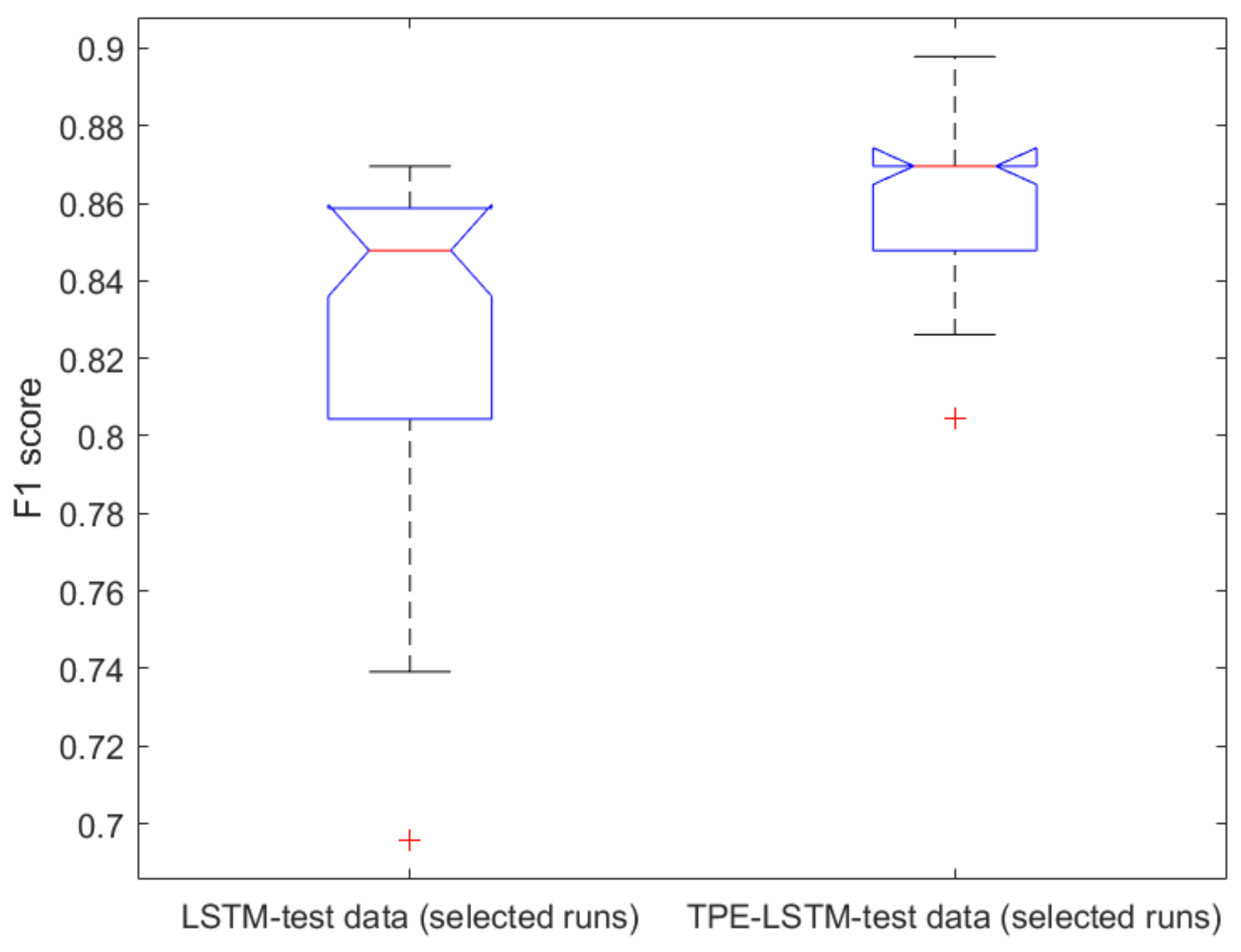
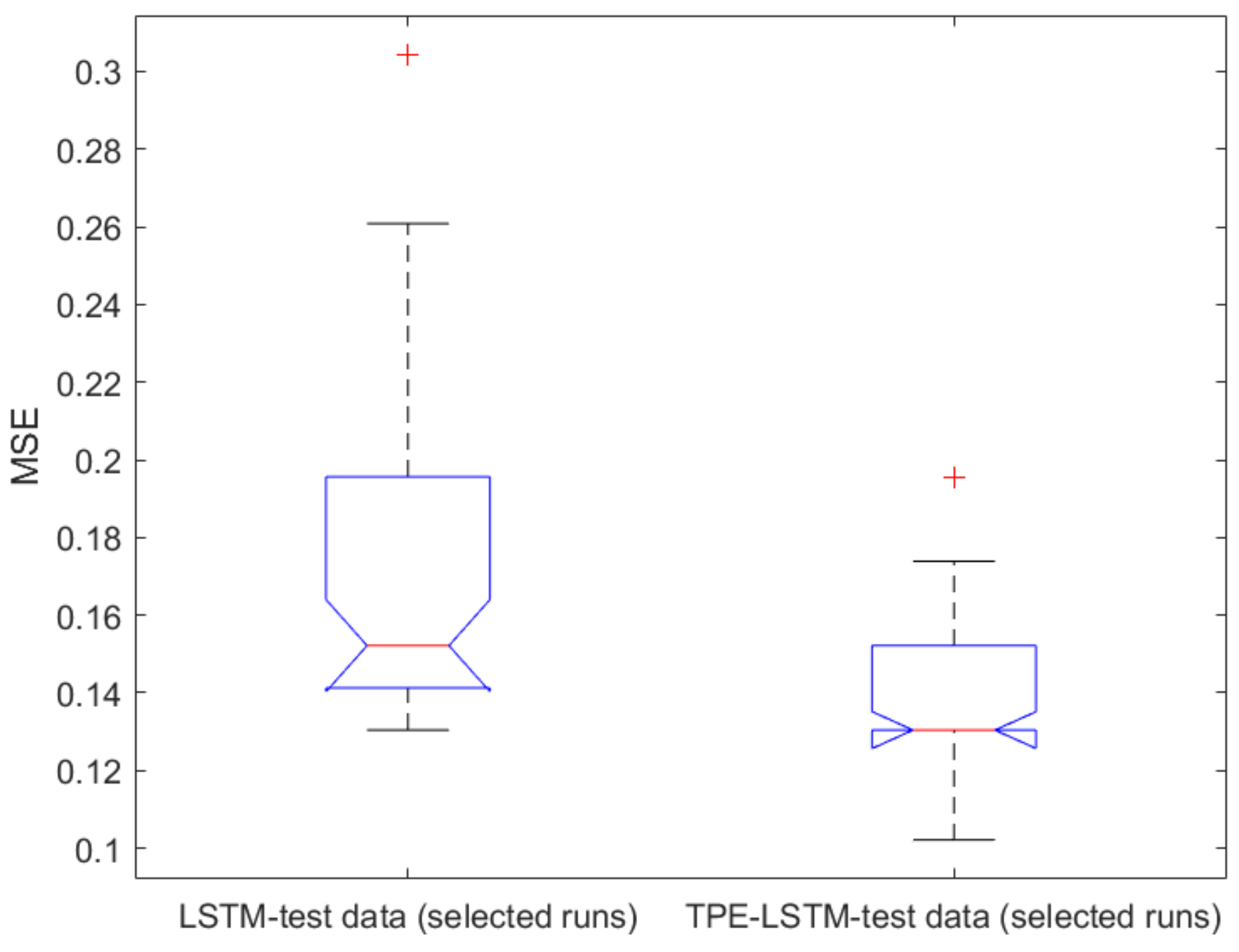
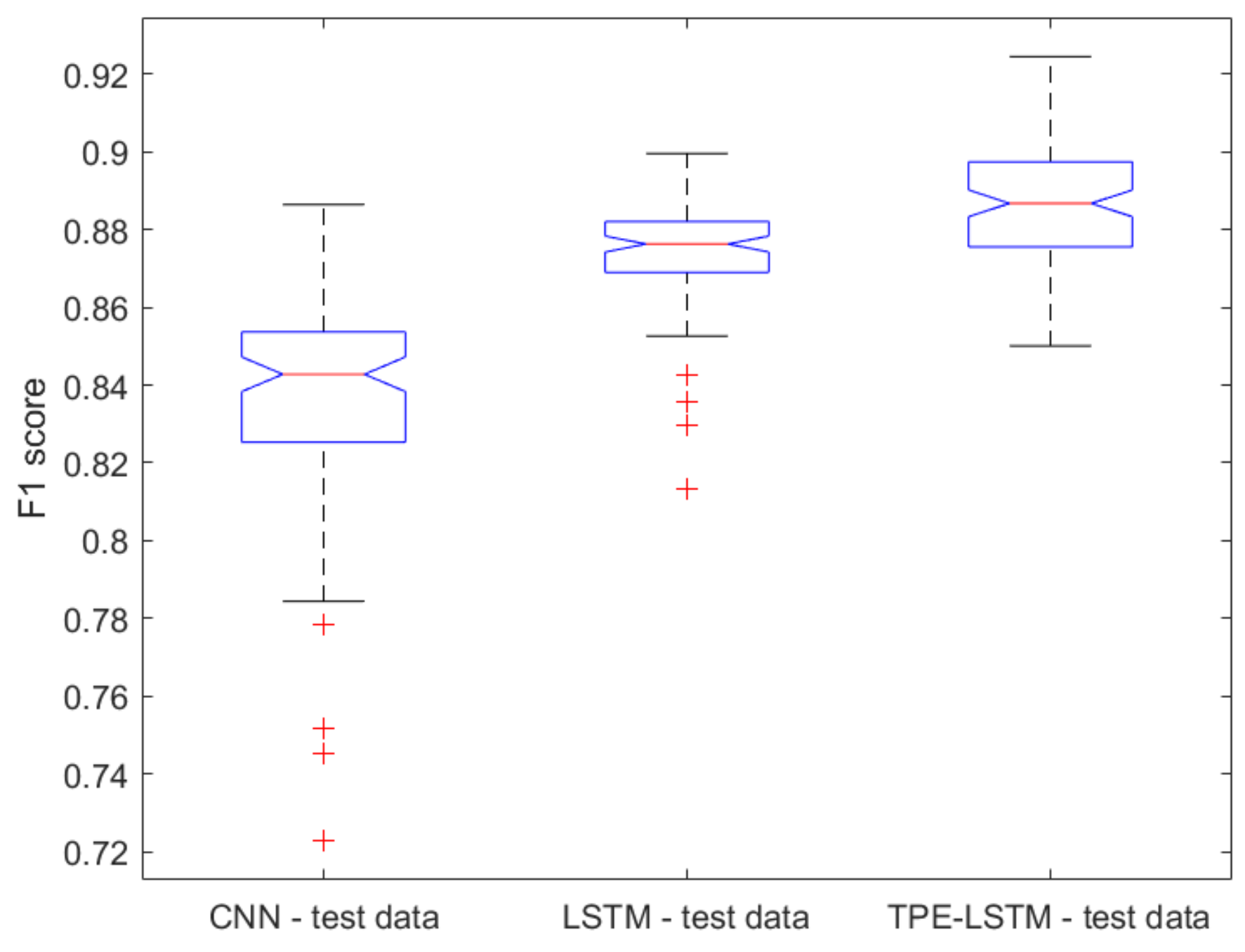
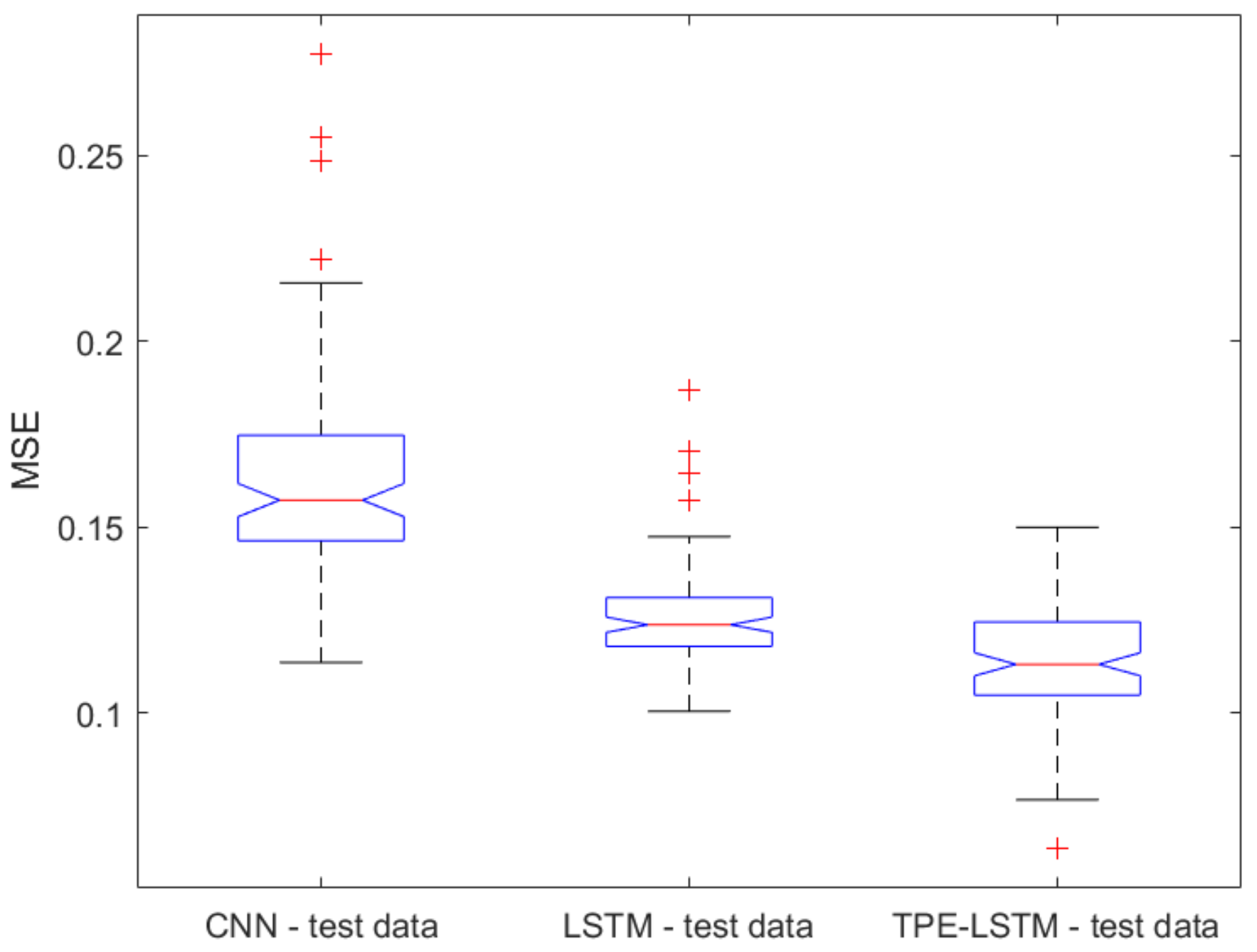
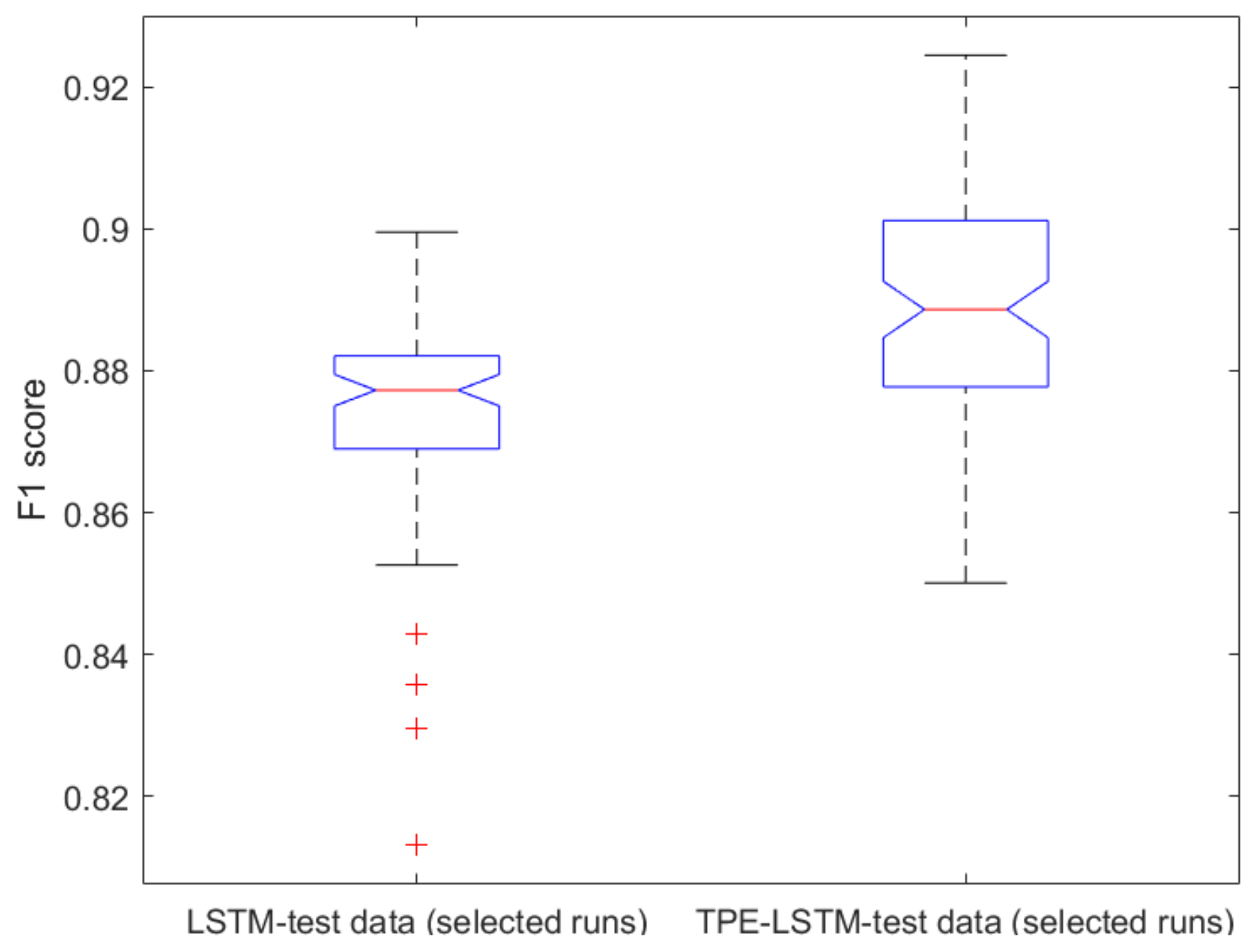
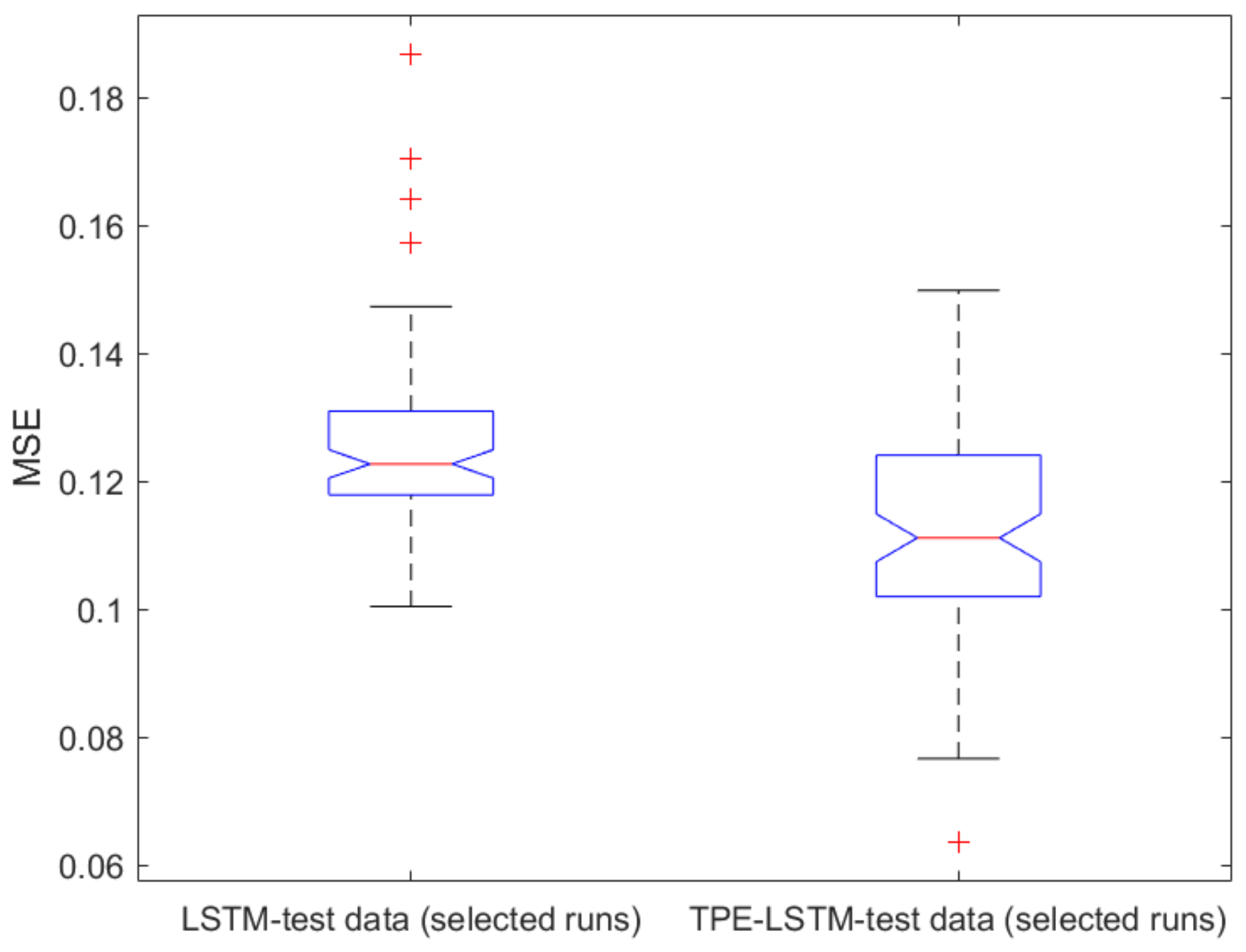
2.1. Classical ML Techniques
In [
5], three different approaches based on classical statistical methods were used to identify CVDs. The first approach is based on the usage of various machine learning (ML) algorithms, such as random forest, logistic regression, K-Nearest Neighbors (KNN), support vector machine (SVM), decision tree, and XGBoost, using the UCI Heart Disease dataset. Note that feature selection and outliers detection were not considered. The second approach used only the feature selection mechanism, while the third approach brought both feature selection and outliers detection into practice. The most accurate algorithm was obtained in the last approach by KNN, with the percentage of correct classification being 84.86%.
The research reported in [
6] also uses statistical models such as SVM, Gaussian Naïve Bayes (GNB), logistic regression, LightGBM, CGBoost, and random forest (RF) to create a classifier that is tested and trained on the Cleveland Clinic Foundation for Heart Disease dataset. The accuracy of the models is measured using performance matrices and confusion matrices. The experimentally established results pointed out that the random forest classifier achieved the best accuracy, followed by SVM and logistic regression.
Various ML techniques, including SVMs, decision trees (DTs), and Naïve Bayes (NB), were used in [
7] to generate diagnosis results on the South African Heart Disease dataset. The performance of these models was compared according to accuracy, sensitivity, specificity, true positives (TPs), true negatives (TNs), false positives (FPs), and false negatives (FNs). Results indicated that NB had the highest accuracy rate; however, it was not satisfactory in terms of specificity and sensitivity. On the contrary, SVMs and DTs provided higher specificity ratings but displayed inadequate sensitivity. Thus, it was concluded that further research is necessary to elevate the performance and increase sensitivity and specificity scores.
Additionally, the research work reported in [
8] has demonstrated the efficacy of using a cost-sensitive ensemble method for the diagnosis of heart diseases. To assess the performance of this approach, the Statlog, Cleveland, and Hungarian heart disease datasets were selected for analysis. Furthermore, various metrics, such as E, MC, G-mean, precision, recall, specificity, and AUC, were used to measure the effectiveness of the classifiers. Relief algorithms were employed to identify the most pertinent features and eliminate any effects of irrelevant features. This study has provided promising results and is a step forward in the development of more sophisticated classifiers with improved accuracy when used in combination with new algorithms.
Recent research has demonstrated the utility of ML techniques in forecasting the 90-day prognosis of individuals diagnosed with transient ischemic attack and minor stroke. The study, conducted in [
9], utilized data from the CNSR-III prospective registry study, which included demographic, physiological, and medical history information of patients with the medical condition. The authors found that models constructed using logistic regression and machine learning exhibited superior performance, as evidenced by their Area Under the Curve (AUC) measure exceeding 0.8. Of the models employed, the Catboost model demonstrated the highest AUC score at 0.839.
2.2. Deep Learning Techniques
With the development of computing power appeared the chance to use more resource demanding algorithms, which are a subcategory of machine learning models named deep learning (DL). Those algorithms have evolved rapidly in recent years and have proven useful, with robust results for various projects, in different areas of interest.
Diagnosing diseases is a good example of showcasing the abilities of deep learning models and the different areas they excel in classification problems. In [
10], the possibility of categorizing and understanding MRI scans to diagnose brain tumors is studied. By comparing two models, the convolutional neural networks (CNN) and a deep neural network (DNN), which is typically a feed-forward network and makes it a perfect fit for these types of problems, it is concluded that the latter provides the most accurate results.
The authors of [
11] proposed the usage of multiple data mining and deep learning techniques, using a dataset containing information selected by taking into account the history of patients’ heart problems in correlation with other medical aspects. Experimentally established results showed that the best classification scores of the proposed ML approach were obtained by the random forest classifier: accuracy (90.21%), precision (90.22%), recall (90.21%), and F1 score (90.21%).
In [
12], a CNN-based diagnosis system was proposed. This model was found to be comparably effective to traditional machine learning models, such as SVM and random forests, particularly in predicting negative cases, meaning those cases without coronary heart disease. The architecture of the proposed model was a sequential feedforward one-input–one-output network, which begins with the application of LASSO regression, which adds a penalty and eliminates coefficients that help control true negatives in the dataset.
There has been research that used both machine and deep learning algorithmic approaches to classify and create predictions for heart diseases. It has been proven that taking into account the medical history of the patients allows deep learning models to outperform machine learning algorithms and yield high accuracy [
11].
2.3. Hybrid Methods
A hybrid method aiming to extract significant features using ML techniques for CVDs prediction was reported in [
13]. The classification model was developed with various feature combinations and is based on the aggregation of random forest and linear classification models. The proposed algorithm, hybrid random forest with a linear model (HRFLM), was assessed to have an 88.7% accuracy. The study also points to the idea that new feature selection methods and new combinations of ML techniques can be used to achieve highly accurate classification algorithms.
Another method to classify patients suffering from CVDs is reported in [
14]. The method is based on the usage of ML techniques and ontology to build an efficient model capable of accurately predicting the presence of cardiac disease and facilitating early diagnosis. The main purpose is to extract the relevant rules from the DT algorithm, then to implement these rules in an ontology using the Semantic Web Rule Language (SWRL). The model reached a level of accuracy of 75% and an F1 score of 80.5%, outperforming the standard DT model (73.1% accuracy, 73.8% F1 score).
In the case study presented in [
15], the usage of evolutionary algorithms (EAs), such as genetic algorithms (GAs) and particle swarm optimization (PSO), was proved to raise the overall accuracy of ML algorithms. The reported research combines EAs with Naïve Bayes and support vector machine for feature selection. The most successful algorithm in terms of classification accuracy uses GA as a feature extraction strategy.
Various techniques can be used to enhance deep learning algorithms to increase their accuracy. An example is the combination of a Multilayer Perceptron (MLP) algorithm with the use of the Back Propagation of Errors Algorithm, which fine tunes the weights based on the error rate acquired from the previous iteration [
16]. It was experimentally proved that the new model has improved performance compared to similar approaches, thus creating a better tuned MLP model for the use of classification problems.
Another way to optimize the algorithms is to use swarm intelligence optimization techniques. Such a technique is particle swarm optimization (PSO) which helps the model to determine the optimal weight and bias values. Combining this technique with a MLP model and testing on the heart disease dataset gives a model that outperformed the initial model [
17].
In [
18], various techniques for recognizing cardiovascular diseases (CVDs) were examined, including Data Mining (DM), DL, ML, and Soft Computing. The authors reviewed the literature on CVD recognition and presented the findings in terms of advantages, limitations, and accuracy levels. The results revealed that certain approaches, such as the utilization of DM and GAs, yielded high accuracy scores of around 96%. However, other methods produced lower accuracy levels, with accuracy scores of around 45%. The best accuracy was observed in the use of Neural Networks (NN), which achieved a score of 99%.
The capabilities of different methods in predicting heart disease can be further enhanced by analyzing the data obtained from other sources such as electrocardiograms (ECGs). ECG waves are widely used to diagnose cardiovascular illnesses. The work reported in [
19] aims to develop a non-linear vector decomposed neural network (NVDN) to classify ECG data. The proposed method was tested using well-known datasets from UCI and Physio net. After denoising the images with the use of frequency wavelet decay strategies, a subset of common features is identified. The NVDN model is then used to predict CVDs. The model produces decent results in terms of the F1 score, accuracy, sensitivity, and specificity; however, the forecasts are inaccurate. It is thus proposed to minimize time complexity and improve categorization.
In [
20], neuro-fuzzy systems were used to learn predictive models from training data to create decision rules meant to support the decision-making process in cardiovascular risk assessment. The reported accuracy has reached 0.91, proving that artificial intelligence models are a valuable help for clinicians.
The literature review shows that there are many possibilities and opportunities for optimization techniques and different approaches to improve the accuracy of the deep learning models, which makes them even better candidates for classification problems.
Recent studies aiming to increase the accuracy of the diagnosis systems indicate that hyperparameter optimization techniques proved powerful tools. In [
21], a radial basis function neural network (RBFNN) designed to identify and diagnose non-linear systems is proposed. The hyperparameters of the RBFNN are computed using PSO-based techniques. The resulting algorithm, which incorporates a spiral search mechanism, proves to improve prediction accuracy and can be extended to various types of neural networks. A PSO algorithm is also used for parameter optimization in [
22]. The proposed diagnostic system is based on CNN and recognizes malignant and benign people in the attempt of identifying early-stage breast cancer.
A novel ensemble technique combining NB, DT, and SVM is introduced in [
23] to classify heart diseases. The proposed approach involves two layers of base learners and a final meta-learner used to optimize the prediction accuracy. Other approaches involving a more accurate representation of neuronal activity use spiking neural networks [
24]. To optimize the recognition rate, various heuristic algorithms including Cuckoo Search Algorithm, Grasshopper Optimization Algorithm, and Polar Bears Algorithm are used to compute the parameters of the spiking NN.