Chasing a Better Decision Margin for Discriminative Histopathological Breast Cancer Image Classification
Abstract
:1. Introduction
2. Related Work
3. Materials and Methods
3.1. Notations
- Matrices are represented using uppercase letters, whereas vectors are indicated by lowercase letters.
- corresponds to the features extracted from the i-th sample.
- The j-th column within the weight matrix is indicated as , where d represents the sample dimension, and C denotes the total number of classes.
- m serves as an additional angular margin, effectively employed to minimize fluctuations within class boundaries.
- denotes the angle formed between the weight vector and the feature vector , whereas represents the angle between the feature vector and the weight vector , with the stipulation that .
- s is a scaling factor applied to all logits, effectively altering their magnitude.
3.2. BreakHis Dataset
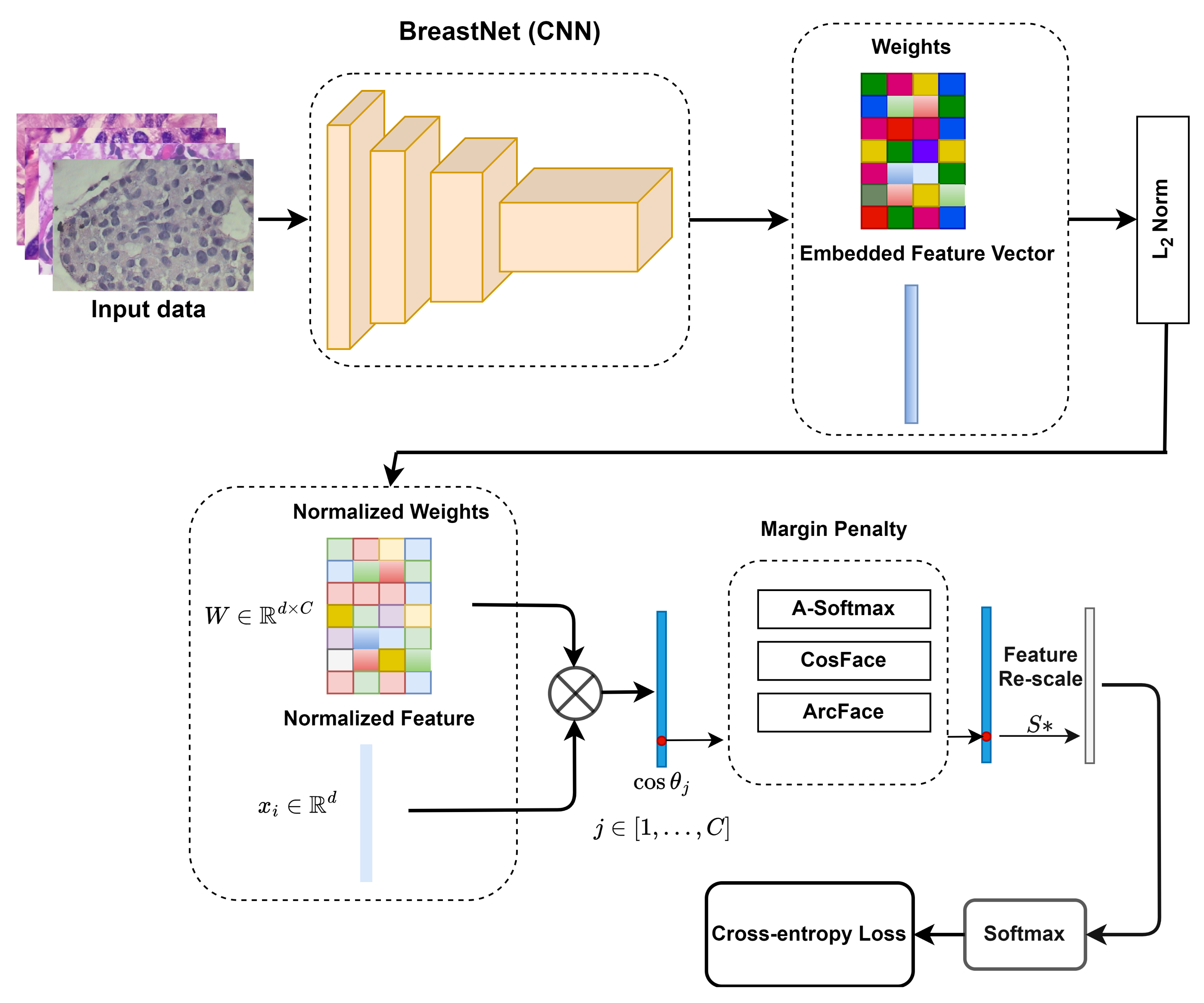
3.3. Pipeline of the Proposed System
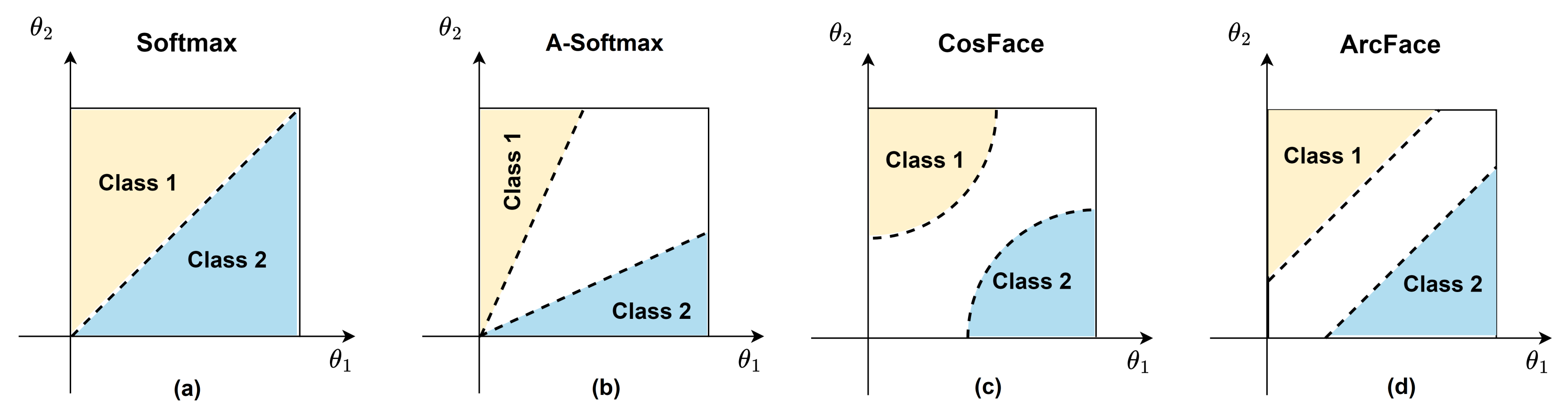
3.4. Margin Penalties on Angular Softmax Losses
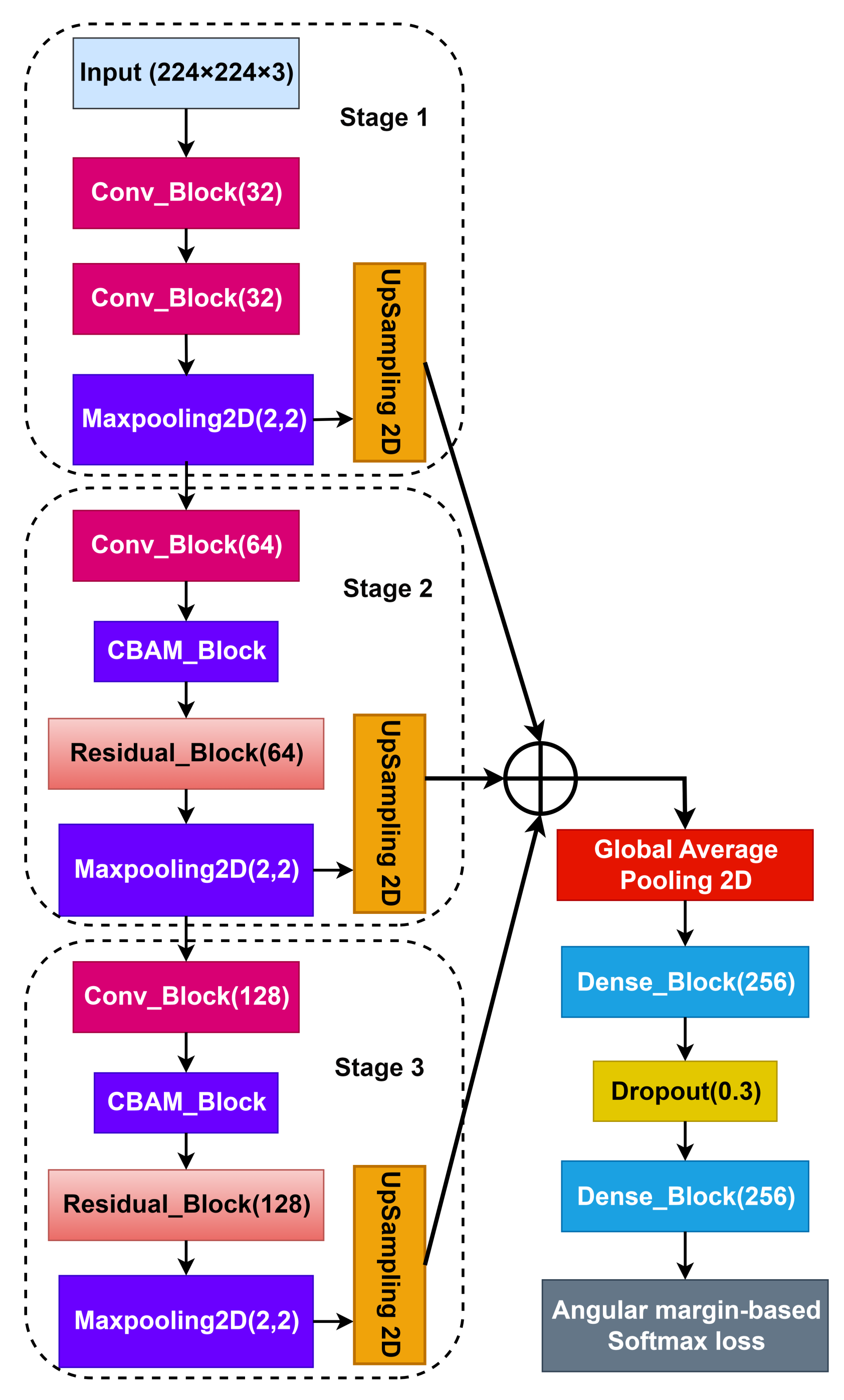
3.5. Convolutional Neural Networks
4. Experimental Setup
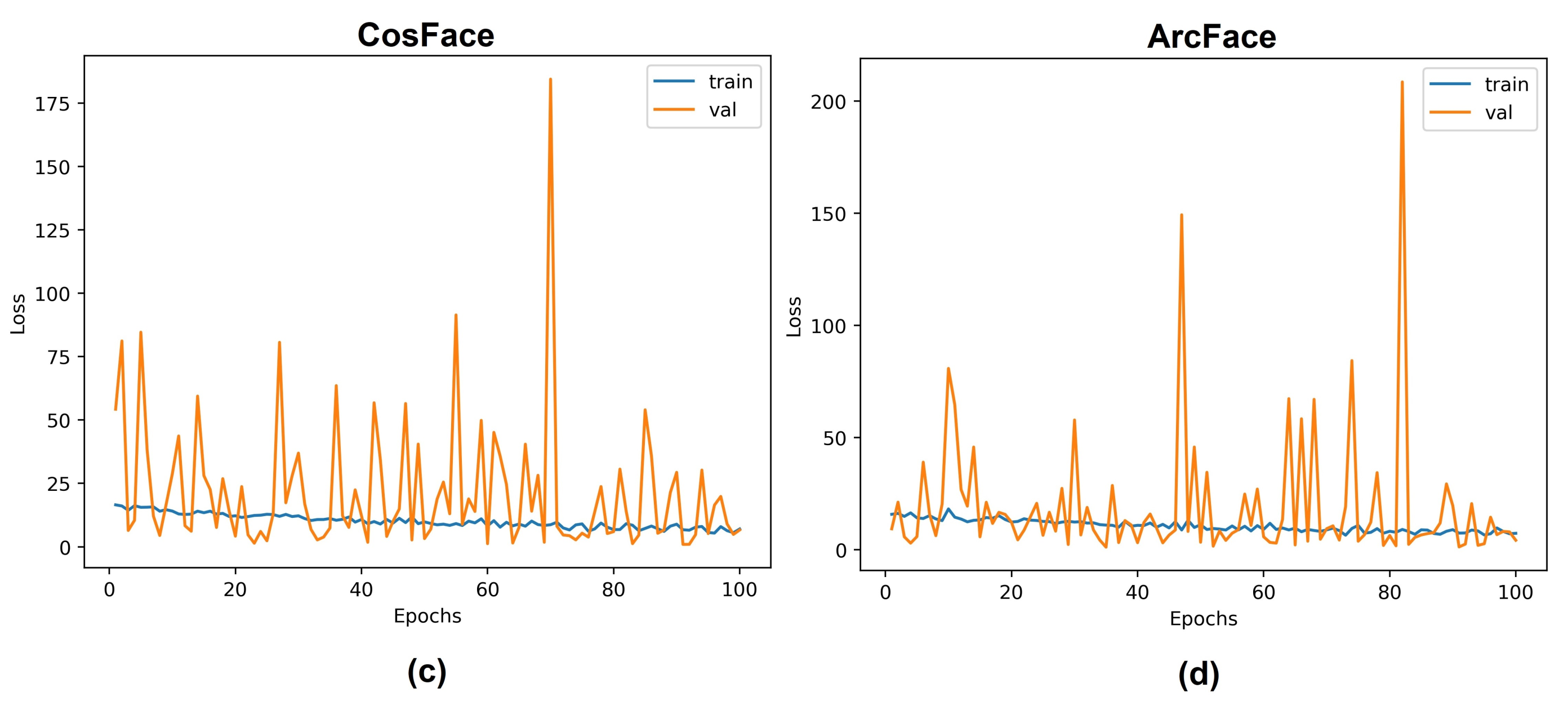
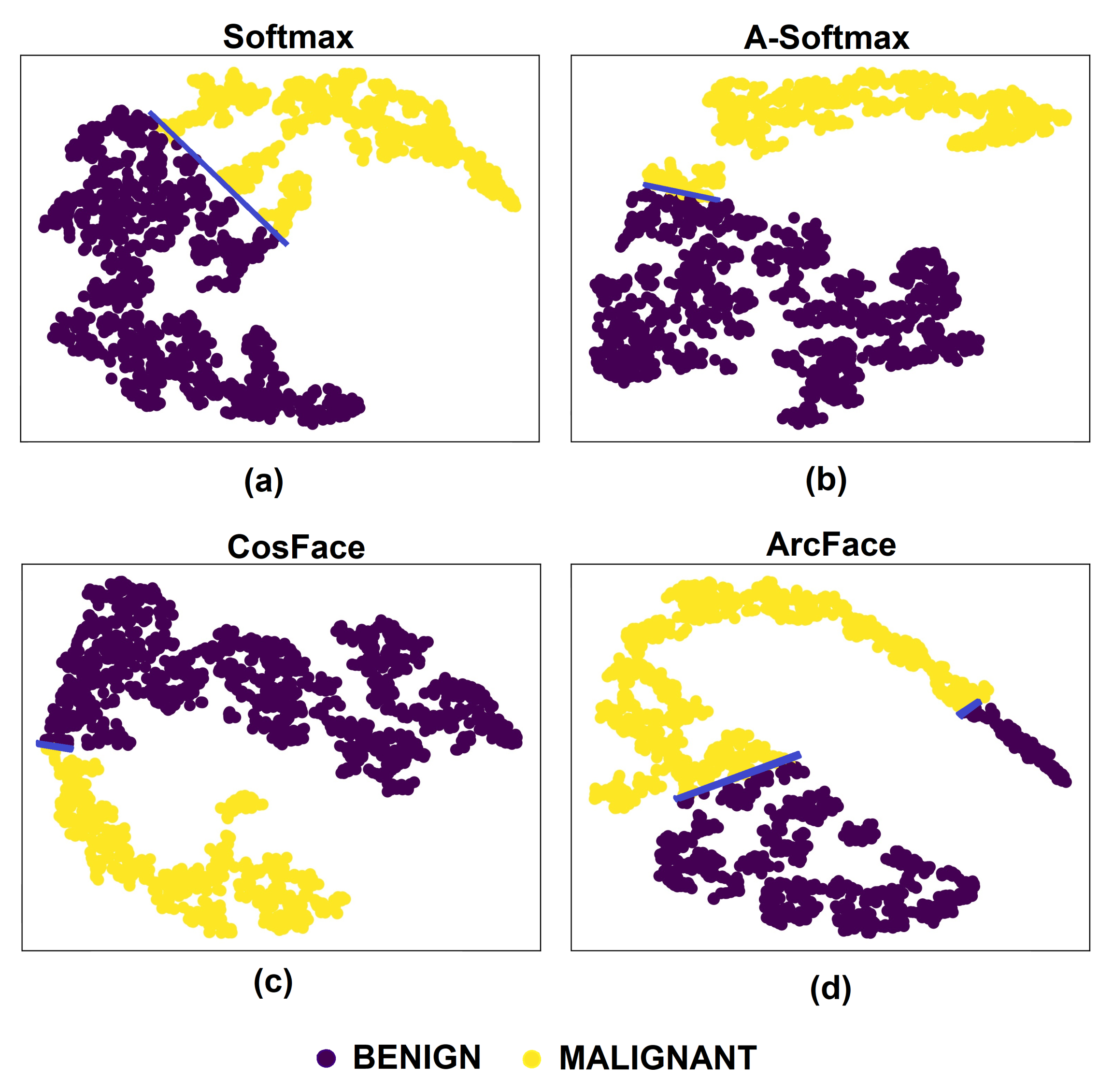
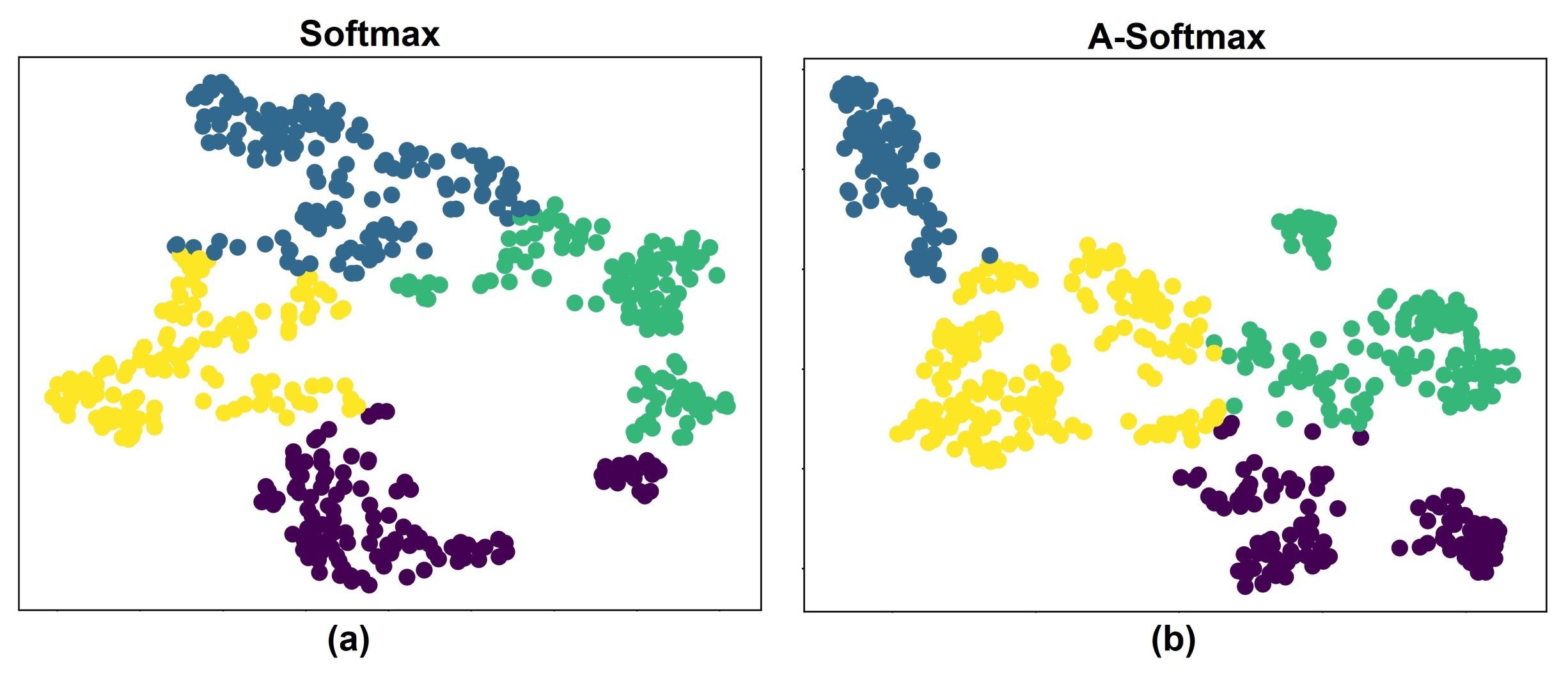
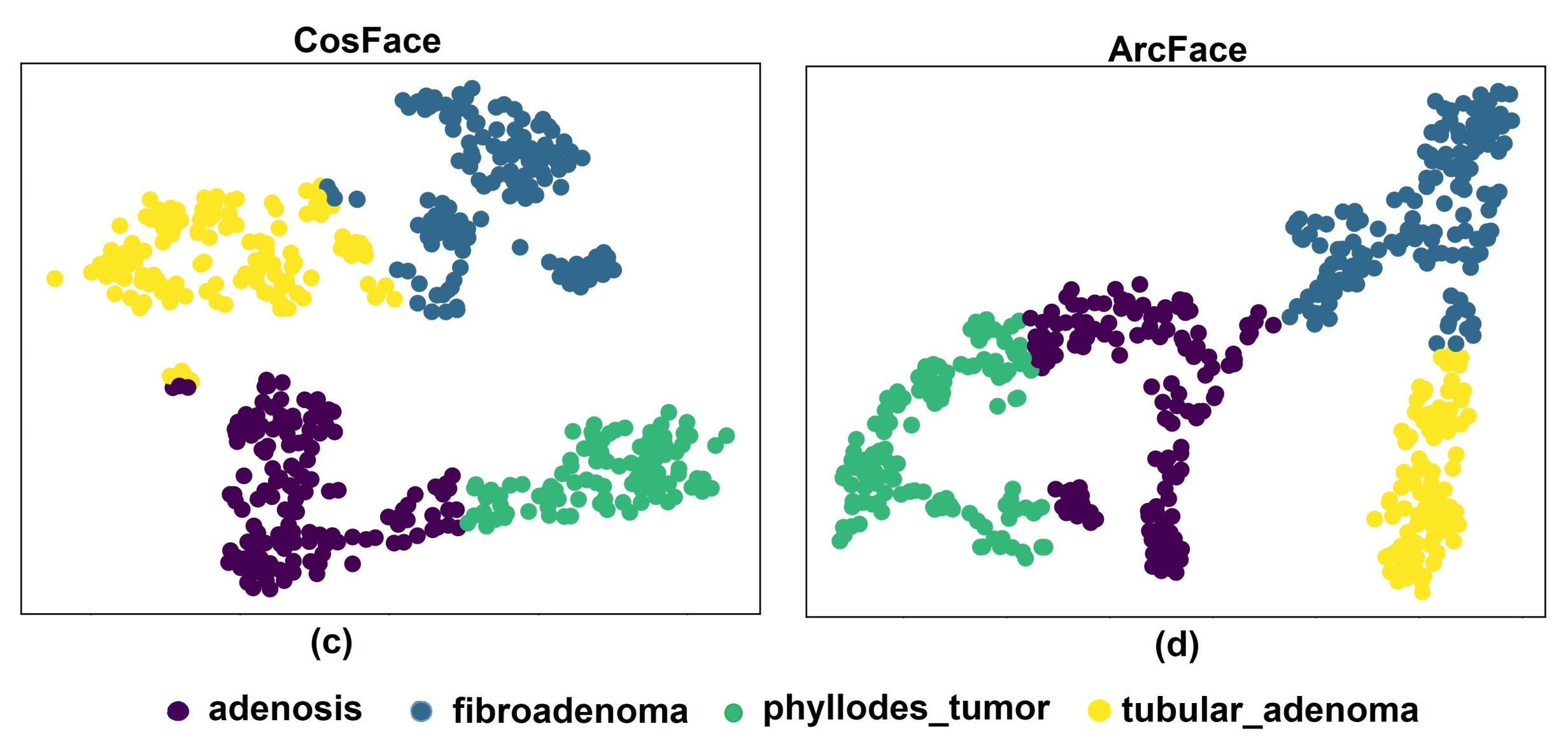
5. Results and Discussion
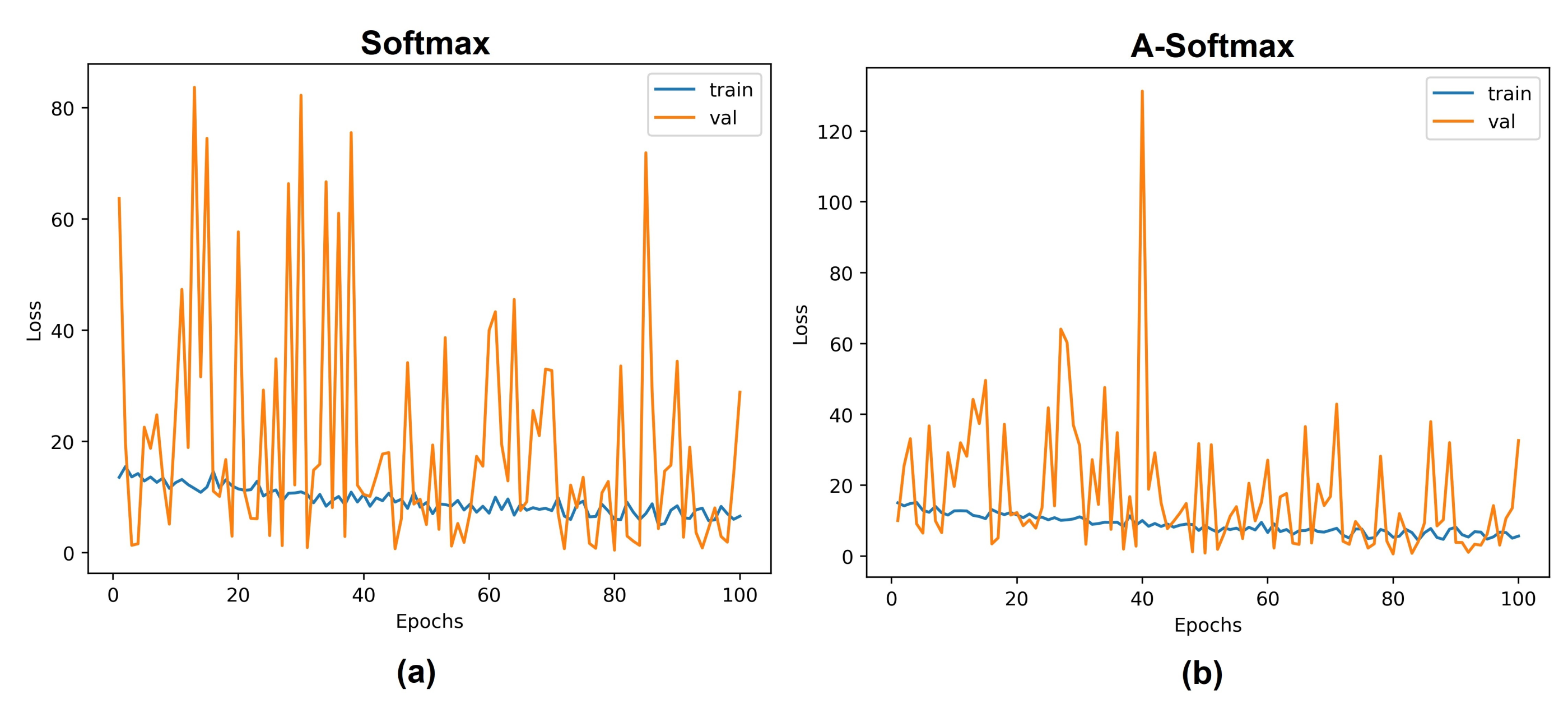
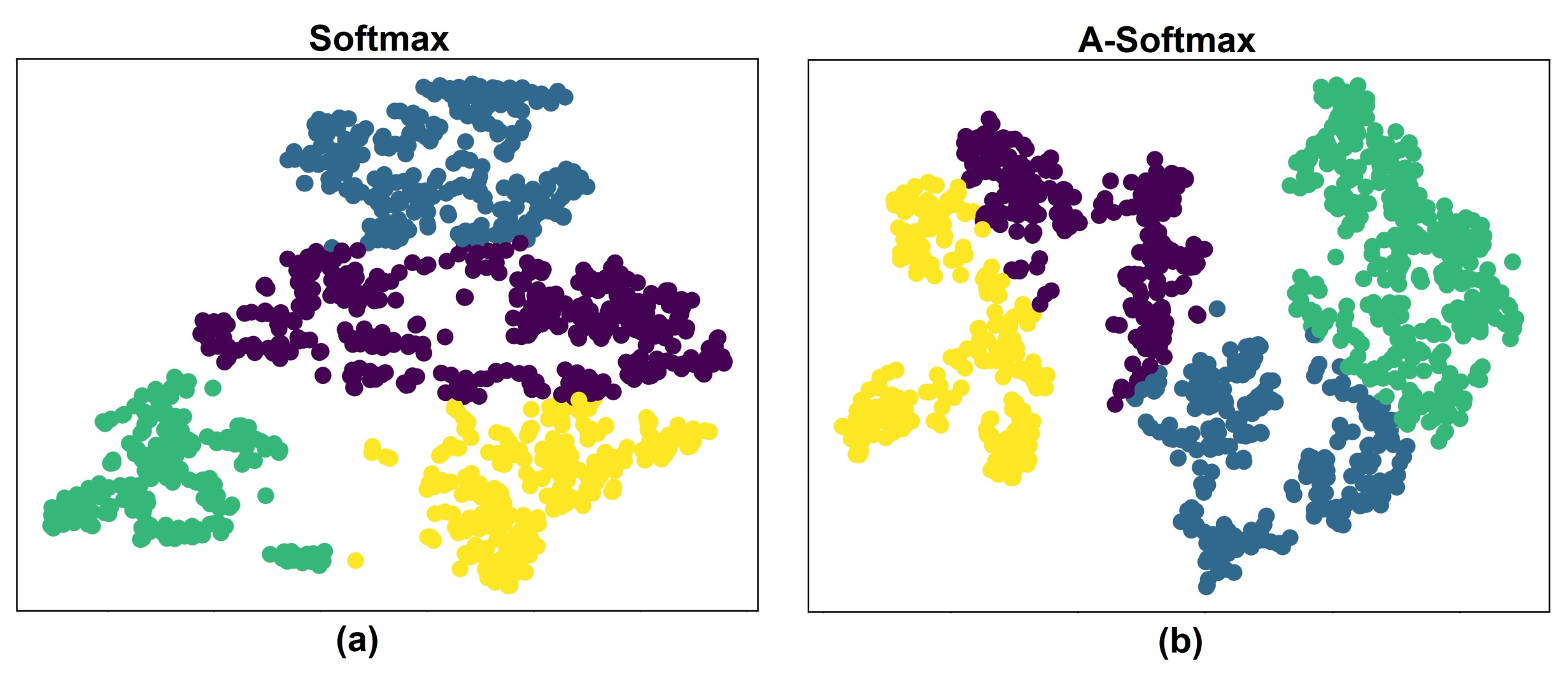
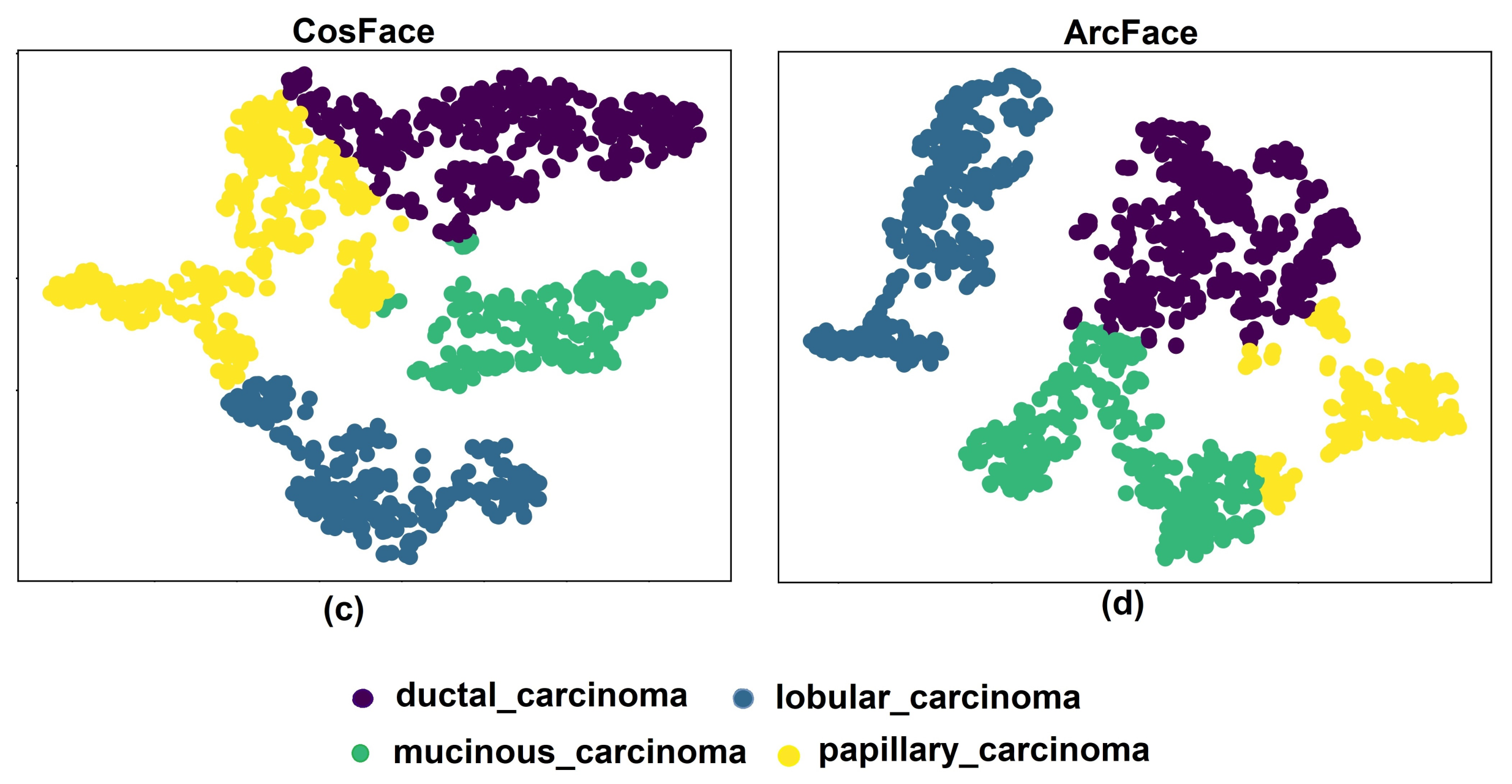
5.1. Experiments with Different Losses
5.2. Comparison with State-of-the-Art Methods
5.3. Discussion
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Veta, M.; Pluim, J.; Van Diest, P.; Viergever, M. Breast cancer histopathology image analysis: A review. IEEE Trans. Biomed. Eng. 2014, 61, 1400–1411. [Google Scholar] [CrossRef] [PubMed]
- Al-Hajj, M.; Wicha, M.; Benito-Hernandez, A.; Morrison, S.; Clarke, M. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, T.; Lee, Y.; Cho, M. Histopathological classification of breast cancer images using a multi-scale input and multi-feature network. Cancers 2020, 12, 2031. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Liu, R.; Luttrell, J., IV; Zhang, C. Deep learning based analysis of histopathological images of breast cancer. Front. Genet. 2019, 10, 80. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Jia, Z.; Wang, L.; Ai, Y.; Zhang, F.; Lai, M.; Chang, E. Large scale tissue histopathology image classification, segmentation, and visualization via deep convolutional activation features. BMC Bioinform. 2017, 18, 281. [Google Scholar] [CrossRef]
- Gandomkar, Z.; Brennan, P.; Mello-Thoms, C. MuDeRN: Multi-category classification of breast histopathological image using deep residual networks. Artif. Intell. Med. 2018, 88, 14–24. [Google Scholar] [CrossRef]
- Huang, Y.; Zheng, H.; Liu, C.; Ding, X.; Rohde, G. Epithelium-stroma classification via convolutional neural networks and unsupervised domain adaptation in histopathological images. IEEE J. Biomed. Health Inform. 2017, 21, 1625–1632. [Google Scholar] [CrossRef]
- Idlahcen, F.; Himmi, M.; Mahmoudi, A. Cnn-based approach for cervical cancer classification in whole-slide histopathology images. arXiv 2020, arXiv:2005.13924. [Google Scholar]
- Yu, X.; Zheng, H.; Liu, C.; Huang, Y.; Ding, X. Classify epithelium-stroma in histopathological images based on deep transferable network. J. Microsc. 2018, 271, 164–173. [Google Scholar] [CrossRef]
- Spanhol, F.; Oliveira, L.; Petitjean, C.; Heutte, L. Breast cancer histopathological image classification using convolutional neural networks. In Proceedings of the International Joint Conference On Neural Networks (IJCNN), Vancouver, BC, Canada, 24–29 July 2016; pp. 2560–2567. [Google Scholar]
- Büker, A.; Hanilçi, C. Angular Margin Softmax Loss and Its Variants for Double Compressed AMR Audio Detection. In Proceedings of the 2021 ACM Workshop On Information Hiding Furthermore, Multimedia Security, Virtual, 22–25 June 2021; pp. 45–50. [Google Scholar]
- Liu, W.; Wen, Y.; Yu, Z.; Li, M.; Raj, B.; Song, L. Sphereface: Deep hypersphere embedding for face recognition. In Proceedings of the IEEE Conference On Computer Vision Furthermore, Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 212–220. [Google Scholar]
- Wang, H.; Wang, Y.; Zhou, Z.; Ji, X.; Gong, D.; Zhou, J.; Li, Z.; Liu, W. Cosface: Large margin cosine loss for deep face recognition. In Proceedings of the IEEE Conference On Computer Vision Furthermore, Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 5265–5274. [Google Scholar]
- Wang, F.; Cheng, J.; Liu, W.; Liu, H. Additive margin softmax for face verification. IEEE Signal Process. Lett. 2018, 25, 926–930. [Google Scholar] [CrossRef]
- Deng, J.; Guo, J.; Xue, N.; Zafeiriou, S. Arcface: Additive angular margin loss for deep face recognition. In Proceedings of the IEEE/CVF Conference On Computer Vision Furthermore, Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019; pp. 4690–4699. [Google Scholar]
- Wang, P.; Wang, J.; Li, Y.; Li, P.; Li, L.; Jiang, M. Automatic classification of breast cancer histopathological images based on deep feature fusion and enhanced routing. Biomed. Signal Process. Control 2021, 65, 102341. [Google Scholar] [CrossRef]
- Zhang, C.; Bai, Y.; Yang, C.; Cheng, R.; Tan, X.; Zhang, W.; Zhang, G. Histopathological image recognition of breast cancer based on three-channel reconstructed color slice feature fusion. Biochem. Biophys. Res. Commun. 2022, 619, 159–165. [Google Scholar] [CrossRef]
- Zou, Y.; Chen, S.; Che, C.; Zhang, J.; Zhang, Q. Breast cancer histopathology image classification based on dual-stream high-order network. Biomed. Signal Process. Control 2022, 78, 104007. [Google Scholar] [CrossRef]
- Majumdar, S.; Pramanik, P.; Sarkar, R. Gamma function based ensemble of CNN models for breast cancer detection in histopathology images. Expert Syst. Appl. 2023, 213, 119022. [Google Scholar] [CrossRef]
- Toğaçar, M.; Özkurt, K.; Ergen, B.; Cömert, Z. BreastNet: A novel convolutional neural network model through histopathological images for the diagnosis of breast cancer. Phys. A Stat. Mech. Appl. 2020, 545, 123592. [Google Scholar] [CrossRef]
- Spanhol, F.; Oliveira, L.; Petitjean, C.; Heutte, L. A dataset for breast cancer histopathological image classification. IEEE Trans. Biomed. Eng. 2015, 63, 1455–1462. [Google Scholar] [CrossRef]
- Spanhol, F.; Oliveira, L.; Cavalin, P.; Petitjean, C.; Heutte, L. Deep features for breast cancer histopathological image classification. In Proceedings of the IEEE International Conference On Systems, Man, Furthermore, Cybernetics (SMC), Banff, AB, Canada, 5–8 October 2017; pp. 1868–1873. [Google Scholar]
- Alirezazadeh, P.; Schirrmann, M.; Stolzenburg, F. Improving Deep Learning-based Plant Disease Classification with Attention Mechanism. Gesunde Pflanz. 2023, 75, 49–59. [Google Scholar] [CrossRef]
- Alirezazadeh, P.; Rahimi-Ajdadi, F.; Abbaspour-Gilandeh, Y.; Landwehr, N.; Tavakoli, H. Improved digital image-based assessment of soil aggregate size by applying convolutional neural networks. Comput. Electron. Agric. 2021, 191, 106499. [Google Scholar] [CrossRef]
- Loshchilov, I.; Hutter, F. Sgdr: Stochastic gradient descent with warm restarts. arXiv 2016, arXiv:1608.03983. [Google Scholar]
- Buslaev, A.; Iglovikov, V.; Khvedchenya, E.; Parinov, A.; Druzhinin, M.; Kalinin, A. Albumentations: Fast and flexible image augmentations. Information 2020, 11, 125. [Google Scholar] [CrossRef]
- Zhu, C.; Song, F.; Wang, Y.; Dong, H.; Guo, Y.; Liu, J. Breast cancer histopathology image classification through assembling multiple compact CNNs. BMC Med. Inform. Decis. Mak. 2019, 19, 198. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Shen, X.; Zhou, Y.; Wang, X.; Li, T. Classification of breast cancer histopathological images using interleaved DenseNet with SENet (IDSNet). PLoS ONE 2020, 15, e0232127. [Google Scholar] [CrossRef] [PubMed]
- Budak, Ü.; Cömert, Z.; Rashid, Z.; Şengür, A.; Çıbuk, M. Computer-aided diagnosis system combining FCN and Bi-LSTM model for efficient breast cancer detection from histopathological images. Appl. Soft Comput. 2019, 85, 105765. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, S.; Saxena, S.; Lakshmanan, K.; Sangaiah, A.; Chauhan, H.; Shrivastava, S.; Singh, R. Deep feature learning for histopathological image classification of canine mammary tumors and human breast cancer. Inf. Sci. 2020, 508, 405–421. [Google Scholar] [CrossRef]
- Saini, M.; Susan, S. Deep transfer with minority data augmentation for imbalanced breast cancer dataset. Appl. Soft Comput. 2020, 97, 106759. [Google Scholar] [CrossRef]
- Sharma, S.; Kumar, S. The Xception model: A potential feature extractor in breast cancer histology images classification. ICT Express 2022, 8, 101–108. [Google Scholar] [CrossRef]
- Abbasniya, M.; Sheikholeslamzadeh, S.; Nasiri, H.; Emami, S. Classification of breast tumors based on histopathology images using deep features and ensemble of gradient boosting methods. Comput. Electr. Eng. 2022, 103, 108382. [Google Scholar] [CrossRef]
- Chattopadhyay, S.; Dey, A.; Singh, P.; Oliva, D.; Cuevas, E.; Sarkar, R. MTRRE-Net: A deep learning model for detection of breast cancer from histopathological images. Comput. Biol. Med. 2022, 150, 106155. [Google Scholar] [CrossRef]
| Key Symbol | Definition |
|---|---|
| N | Quantity of images within each batch |
| C | Total class count |
| The weight matrix , with each column corresponding to a specific class (j-th class). | |
| The ground truth weight vector associated with class | |
| The feature representation of the i-th sample. | |
| The angle between the feature vector and the corresponding weight vector . | |
| The angle formed between the feature vector and the weight vector for non-target classes (where ). | |
| m | Cosine and angular margin penalties for CosFace and ArcFace, respectively. |
| s | The scaling factor applied to all logit values. |
| Employed BreakHis Data | Method | Pr (%) | Re (%) | -Score (%) | Acc (%) |
|---|---|---|---|---|---|
| 40× | Softmax | 96.56 ± 3.85 | 95.30 ± 3.94 | 95.88 ± 3.88 | 96.49 ± 3.31 |
| A-Softmax | 97.05 ± 1.24 | 94.59 ± 3.68 | 95.58 ± 2.86 | 96.34 ± 2.22 | |
| CosFace | 97.47 ± 0.50 | 96.59 ± 0.97 | 97.01 ± 0.73 | 97.44 ± 0.62 | |
| ArcFace | 97.54 ± 1.22 | 96.31 ± 1.25 | 96.88 ± 1.22 | 97.34 ± 1.03 | |
| 100× | Softmax | 94.75 ± 5.08 | 94.98 ± 3.70 | 94.77 ± 4.54 | 95.35 ± 4.18 |
| A-Softmax | 95.87 ± 1.95 | 95.61 ± 1.88 | 95.74 ± 1.90 | 96.31 ± 1.65 | |
| CosFace | 96.72 ± 1.68 | 95.73 ± 2.54 | 96.18 ± 2.17 | 96.73 ± 1.81 | |
| ArcFace | 97.52 ± 0.54 | 96.42 ± 0.92 | 96.93 ± 0.59 | 97.36 ± 0.50 | |
| 200× | Softmax | 96.37 ± 1.43 | 96.92 ± 1.24 | 96.62 ± 1.27 | 97.07 ± 1.10 |
| A-Softmax | 97.32 ± 1.34 | 96.04 ± 3.54 | 96.54 ± 2.67 | 97.12 ± 2.21 | |
| CosFace | 97.01 ± 1.35 | 97.08 ± 2.04 | 96.99 ± 1.58 | 97.42 ± 1.32 | |
| ArcFace | 98.13 ± 1.20 | 97.26 ± 2.16 | 97.65 ± 1.69 | 98.01 ± 1.42 | |
| 400× | Softmax | 93.82 ± 3.59 | 94.88 ± 3.16 | 94.30 ± 3.41 | 94.94 ± 3.05 |
| A-Softmax | 94.03 ± 4.18 | 93.12 ± 6.54 | 93.39 ± 5.70 | 94.45 ± 4.52 | |
| CosFace | 95.64 ± 1.47 | 96.16 ± 0.89 | 95.88 ± 1.20 | 96.37 ± 1.09 | |
| ArcFace | 94.47 ± 1.37 | 94.50 ± 2.02 | 94.45 ± 1.65 | 95.16 ± 1.40 |
| Method | Number of Parameters | Training Time (s/epoch) | Classification Time (ms/image) |
|---|---|---|---|
| Softmax | 605,566 | 57.3 ± 2.64 | 13.3 ± 1.24 |
| A-Softmax | 605,582 | 58.8 ± 1.69 | 14.7 ± 1.44 |
| CosFace | 605,580 | 57.5 ± 1.76 | 15.7 ± 1.68 |
| ArcFace | 605,582 | 58.3 ± 2.32 | 15.1 ± 1.95 |
| Method | Employed BreakHis Data | Pr (%) | Re (%) | -Score (%) | Acc (%) | |
|---|---|---|---|---|---|---|
| Benign subset | Softmax | All magnifications | 92.82 ± 1.18 | 91.02 ± 0.86 | 91.70 ± 0.75 | 92.46 ± 0.61 |
| A-Softmax | All magnifications | 92.78 ± 1.87 | 92.26 ± 1.83 | 92.44 ± 1.85 | 92.90 ± 1.68 | |
| CosFace | All magnifications | 93.01 ± 1.56 | 92.99 ± 0.72 | 92.93 ± 1.11 | 93.55 ± 1.02 | |
| ArcFace | All magnifications | 93.36 ± 1.19 | 92.93 ± 1.82 | 93.09 ± 1.49 | 93.55 ± 1.33 | |
| Malignant subset | Softmax | All magnifications | 86.93 ± 1.74 | 86.09 ± 3.08 | 86.39 ± 2.28 | 90.04 ± 1.51 |
| A-Softmax | All magnifications | 87.83 ± 2.68 | 84.41 ± 5.92 | 85.89 ± 4.53 | 90.06 ± 2.76 | |
| CosFace | All magnifications | 89.41 ± 2.82 | 86.72 ± 6.30 | 87.73 ± 4.72 | 91.29 ± 3.05 | |
| ArcFace | All magnifications | 89.96 ± 2.39 | 86.28 ± 4.36 | 87.86 ± 3.60 | 91.42 ± 2.37 |
| Method | Acc (%) | Average | |||
|---|---|---|---|---|---|
| 40× | 100× | 200× | 400× | ||
| Hybrid CNN improved by SEP block [27] | 85.60 | 83.90 | 84.40 | 81.20 | 83.78 |
| DenseNet improved by SENet module [28] | 89.10 | 85.00 | 87.00 | 84.50 | 86.40 |
| FCN combined with BiLSTM network [29] | 95.69 | 93.60 | 96.30 | 94.29 | 92.47 |
| VGG16 feature extractor with SVM and RF classification [30] | 94.11 | 95.10 | 97.00 | 94.96 | 95.30 |
| Data augmentation by DCGAN for VGG16 training [31] | 96.40 | 94.00 | 95.50 | 93.00 | 94.73 |
| Xception feature extractor with SVM classification [32] | 96.25 | 96.25 | 95.74 | 94.11 | 95.59 |
| Inception-ResNet-v2 combined with CatBoost, XGBoost, and LightGBM [33] | 96.82 | 95.84 | 97.01 | 96.15 | 96.45 |
| Multi-scale dual residual recurrent network [34] | 97.12 | 95.20 | 96.80 | 97.81 | 96.73 |
| BreastNet architecture supervised by CosFace | 97.44 | 96.73 | 97.42 | 96.37 | 96.99 |
| BreastNet architecture supervised by ArcFace | 97.34 | 97.36 | 98.01 | 95.16 | 96.97 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alirezazadeh, P.; Dornaika, F.; Moujahid, A. Chasing a Better Decision Margin for Discriminative Histopathological Breast Cancer Image Classification. Electronics 2023, 12, 4356. https://doi.org/10.3390/electronics12204356
Alirezazadeh P, Dornaika F, Moujahid A. Chasing a Better Decision Margin for Discriminative Histopathological Breast Cancer Image Classification. Electronics. 2023; 12(20):4356. https://doi.org/10.3390/electronics12204356
Chicago/Turabian StyleAlirezazadeh, Pendar, Fadi Dornaika, and Abdelmalik Moujahid. 2023. "Chasing a Better Decision Margin for Discriminative Histopathological Breast Cancer Image Classification" Electronics 12, no. 20: 4356. https://doi.org/10.3390/electronics12204356
APA StyleAlirezazadeh, P., Dornaika, F., & Moujahid, A. (2023). Chasing a Better Decision Margin for Discriminative Histopathological Breast Cancer Image Classification. Electronics, 12(20), 4356. https://doi.org/10.3390/electronics12204356


_Kalra.png)




