Image Formation Algorithms for Low-Cost Freehand Ultrasound Scanner Based on Ego-Motion Estimation and Unsupervised Clustering
Abstract
1. Introduction
- Philips Lumify—200 USD/month per probe + 75 USD/month warranty.
- Sonosite IVIZ—USD 10,000.
- GE VSAN Extend—starts at USD 2995.
- Clarius—starts at USD 6900.
- Butterfly IQ—USD 1999 + 420 USD/year for cloud user license.
- Optimisation of the decorrelation-based velocity estimation technique for linear scans and application of the unscented Kalman filter (UKF) to filter out the effects of noisy velocity estimates to improve trajectory estimates.
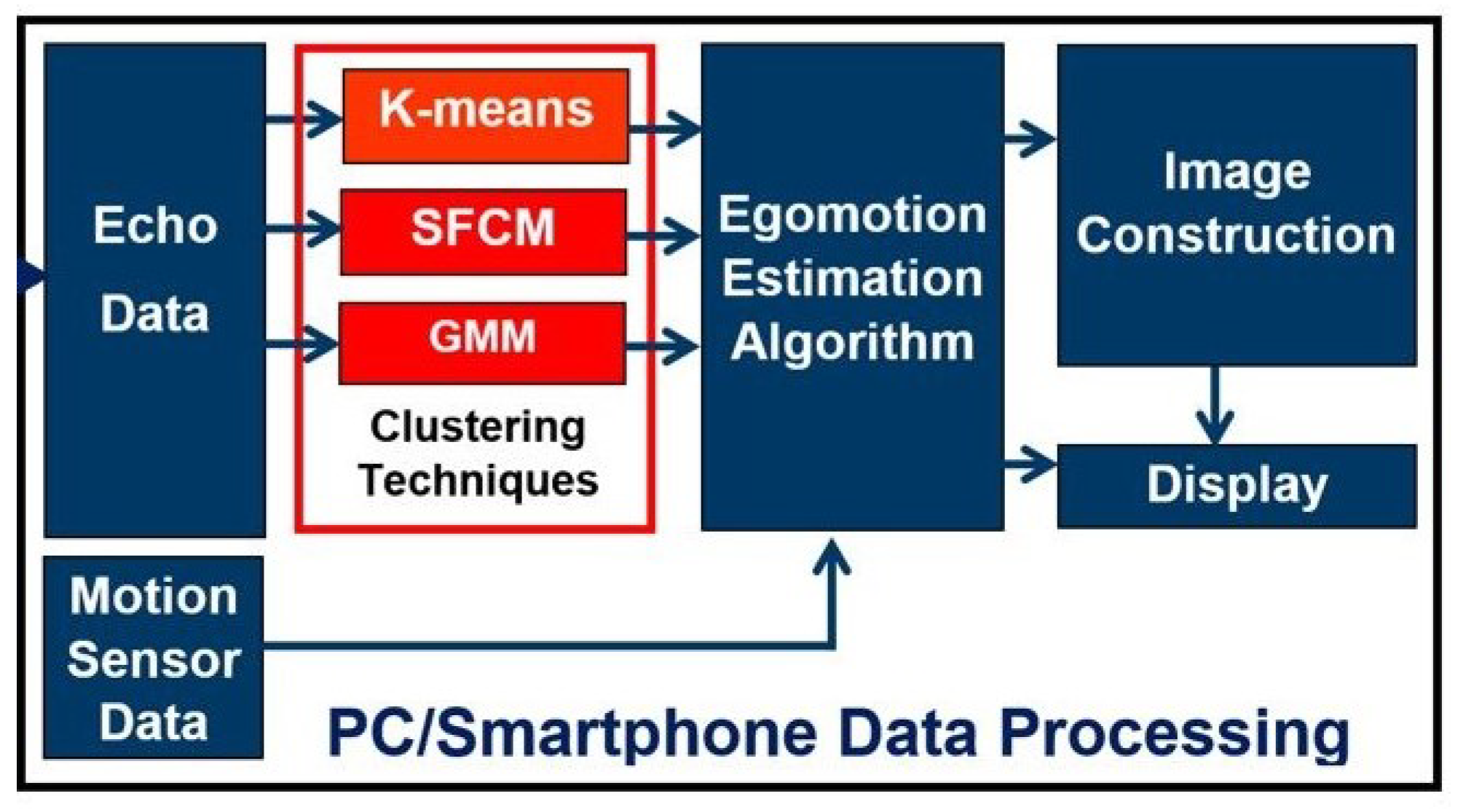
- First-time exploitation of unsupervised clustering (K-means, SFCM and GMM) on the 1D raw ultrasound imaging data.
- Reconstruction of geometrically correct 2D ultrasound images of phantom and in vivo data from the single-element transducer.
2. Related Research
3. Material and Methods
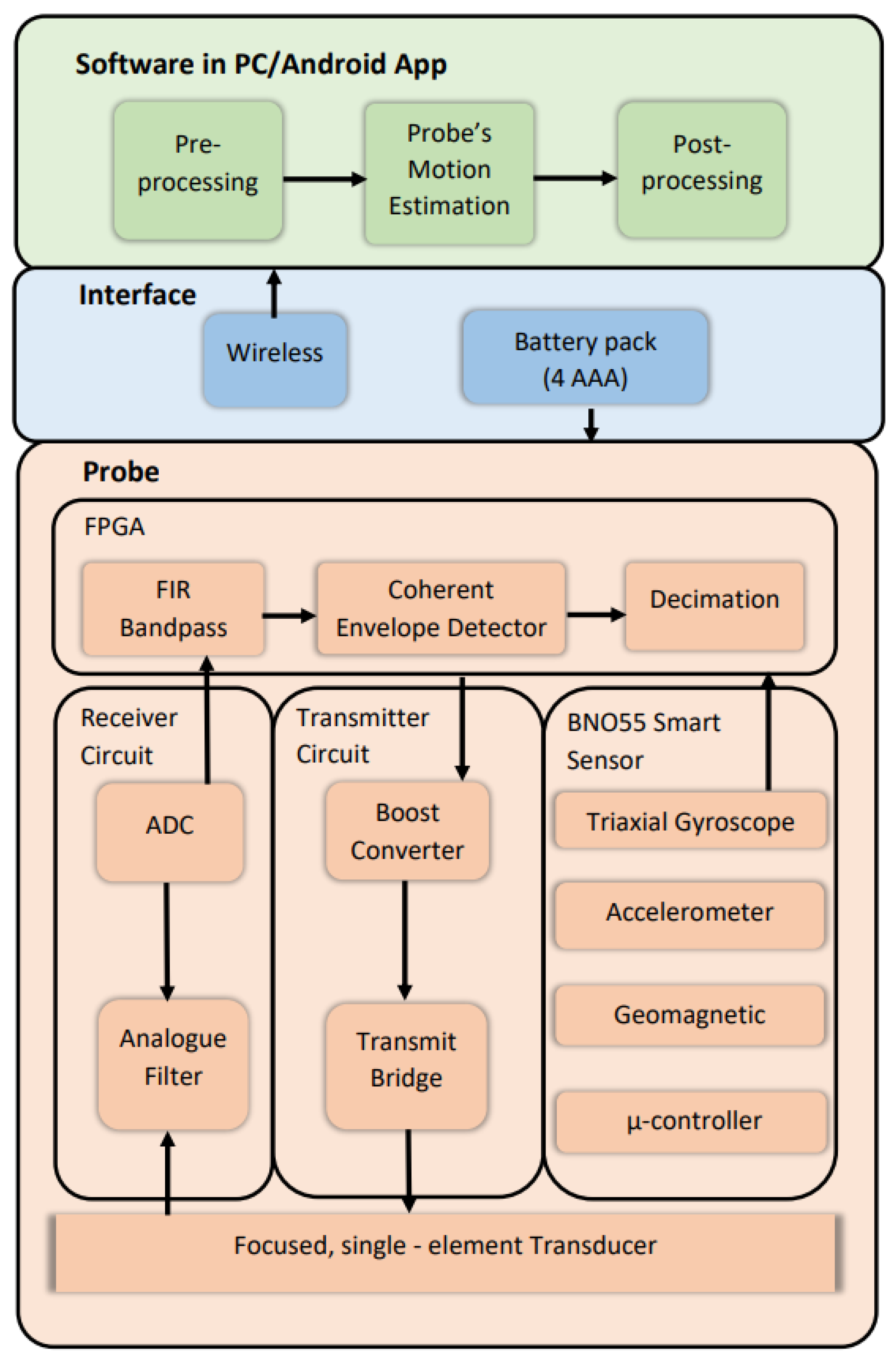
3.1. Proposed Ultrasound Scanner Design
3.2. Pre-Processing the Echo Data
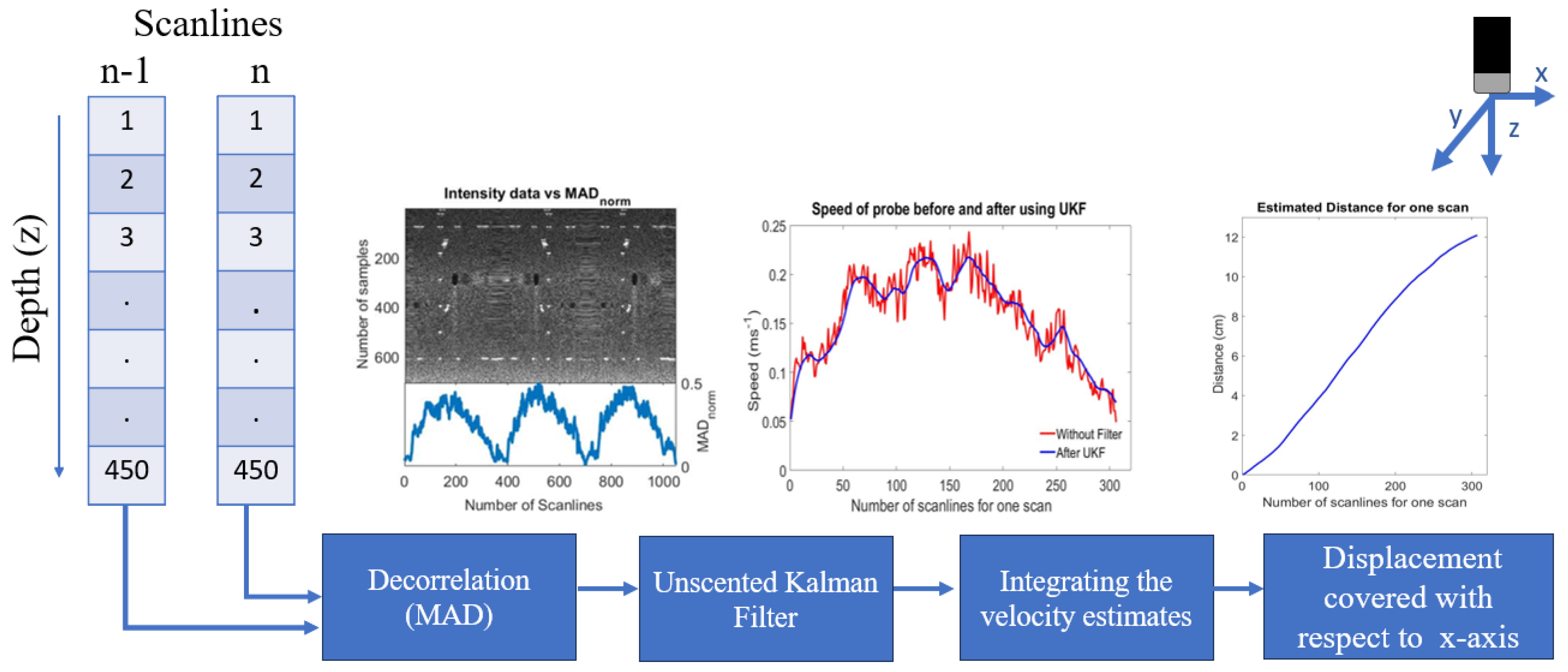
3.3. Ultrasound Decorrelation Measurements
- (1)
- The absolute differences between the echo signal intensities of each consecutive scan line were calculated.
- (2)
- The mean of the absolute differences was calculated.
- (3)
- Steps 1 and 2 were repeated for all the other scan lines by moving successively through all the data.
3.3.1. Velocity Calculation for the Ultrasound Probe
3.3.2. Unscented Kalman Filter
3.4. K-Means Clustering
- The number of clusters, K, is given as a priori and it has been chosen to be equal to 3 in this project. This is because echo data needs to be clustered in three clusters, i.e., hyperechoic, anechoic and hypoechoic.
- Cluster centres are chosen randomly by the algorithm.
- The distance d between echo data, , and the cluster centre, , is calculated using the Euclidean distance formula as follows:
- The membership function, , is computed in which denote the membership degree of the th data point to the th cluster, ∈ [0,1].where, k = 3 because echo data need to be clustered in three clusters, i.e., hyperechoic, anechoic and hypoechoic.
| Algorithm 1: Kmeans clustering pseudocode. |
 |
- 5.
- Then, the k-means objective function is calculated as follows:is minimized by iterating the k-means algorithm.
- 6.
- The cluster centres, , and membership function, , are updated using the equations shown below:
3.5. SFCM Clustering
- Set values for K, l and . Where,K = number of clusters = 3.l = is the weighting exponent (>1) on each fuzzy membership that controls the fuzziness of resultant segmentation.= termination criterion between [0,1].
- Initialise the fuzzy membership function matrix whereas, the membership functions are subject to the following constraints:
- Calculate the cluster centres as follows:
| Algorithm 2: SFCM clustering pseudocode. |
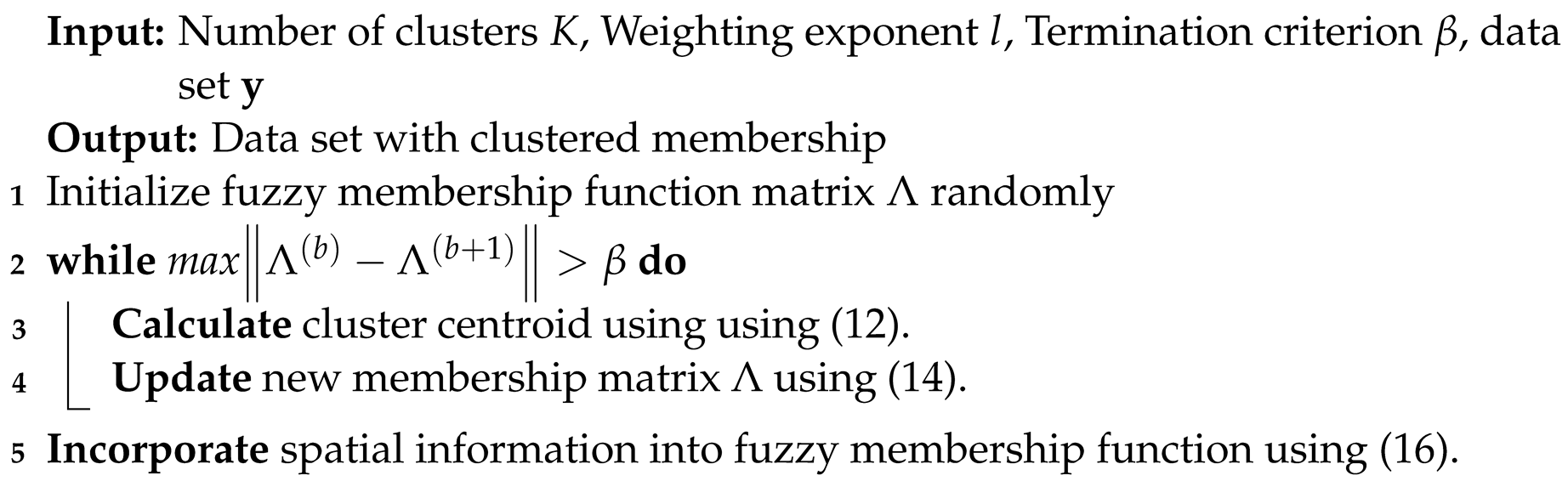 |
- 4.
- Compute the membership function . For (), calculate the following:for the th column of the matrix, compute new membership values:
- if = ø then
- Otherwise, = 0 for all I and ;
- 5.
- If , stop; otherwise, keep incrementing the loop and repeat step 3, 4 and 5. Where ( number of iterations = 100).
3.6. GMM Clustering
- K-means clustering for a reduced number of iterations using a random parameter initialization.
- GMM-EM clustering using the parameter initialization is given by the results of the previous K-means clustering phase. Assuming expectation maximization for a Gaussian mixture model (GMM-EM), the goal is to maximize the likelihood function with respect to the parameters (comprising the means and covariances of the components and the mixing coefficients). The steps of the EM clustering are further presented:
- (a)
- Compute the means , covariance matrices and mixing coefficients (where ) as a result of the previous phase of K-means clustering, by considering them as initialization parameters for the present GMM-EM phase and evaluate the initial value of the log-likelihood.
- (b)
- E step. Evaluate the responsibilities using the current parameter values.
- (c)
- M step. Re-estimate the parameters using the current responsibilities.where,
- (d)
- Evaluate the log-likelihood.
- (e)
- Check for convergence of either the parameters or the log-likelihood. If the convergence criterion is not satisfied, return to Step B.
| Algorithm 3: GMM clustering pseudocode. |
 |
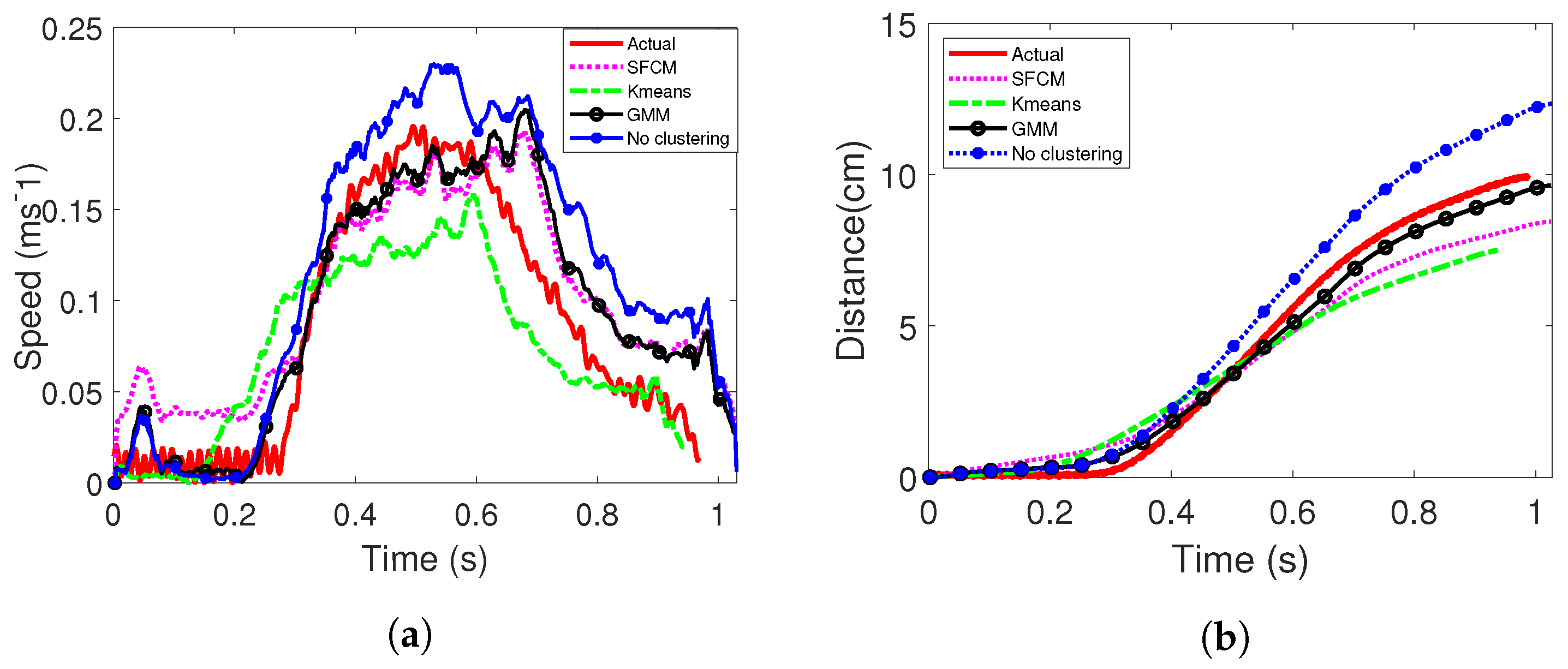
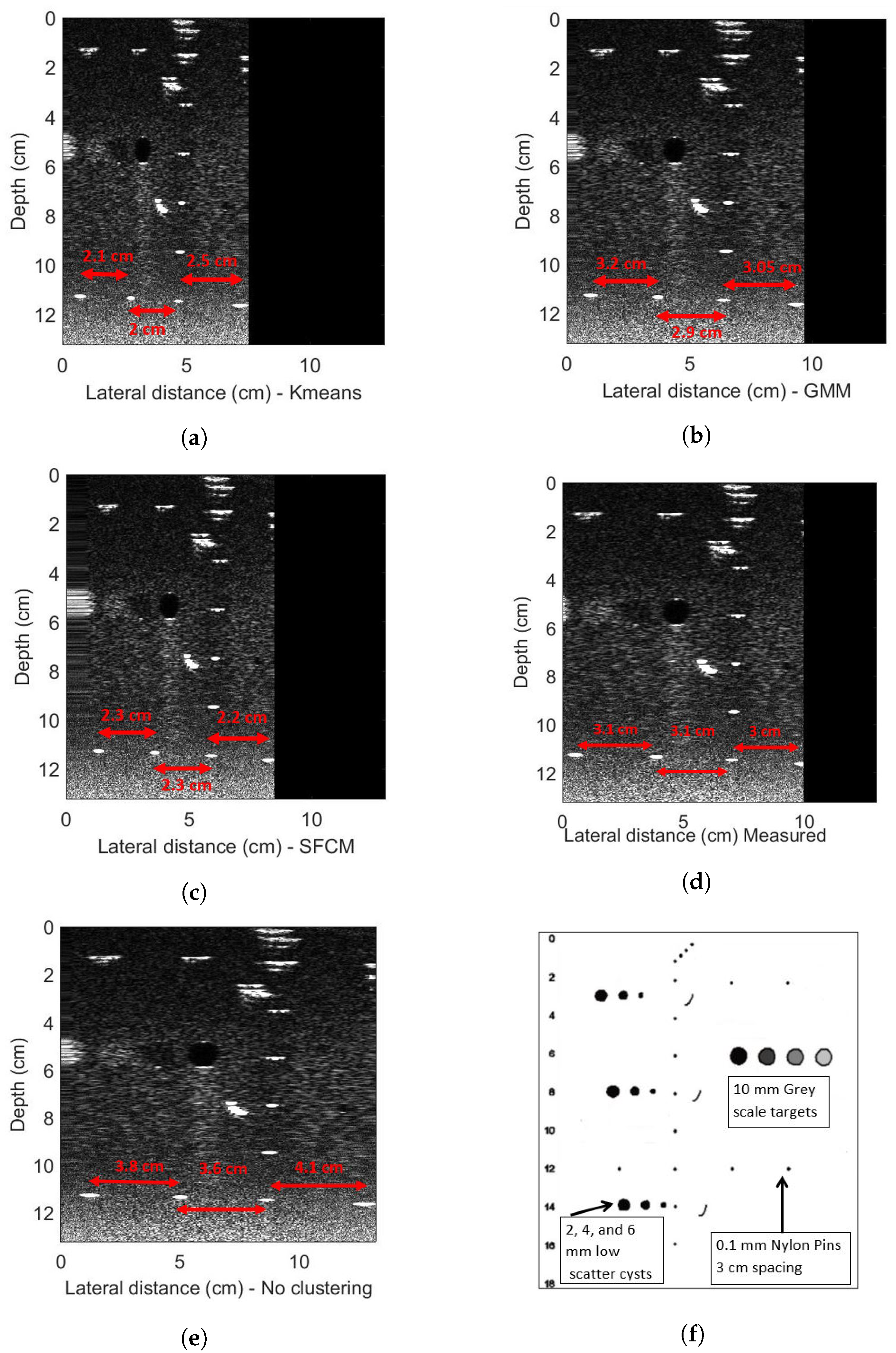
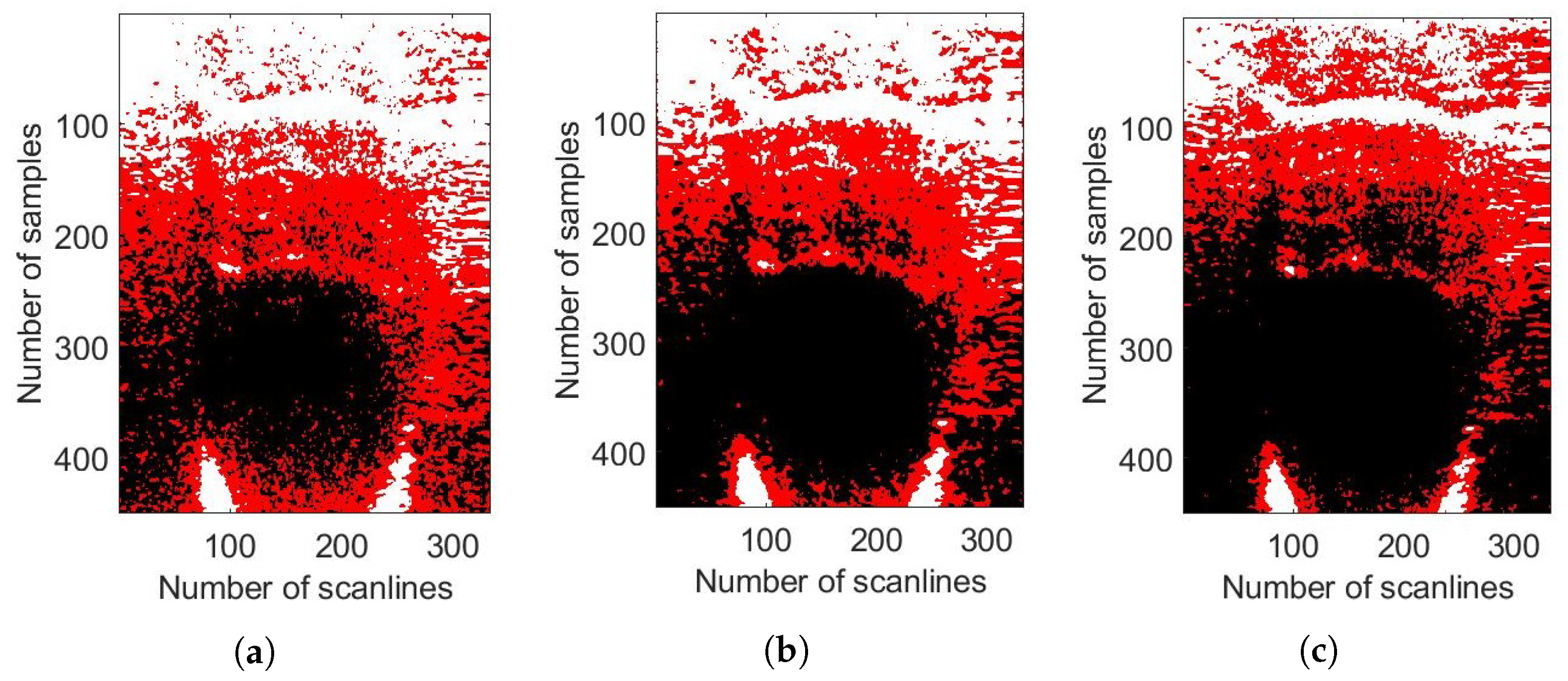
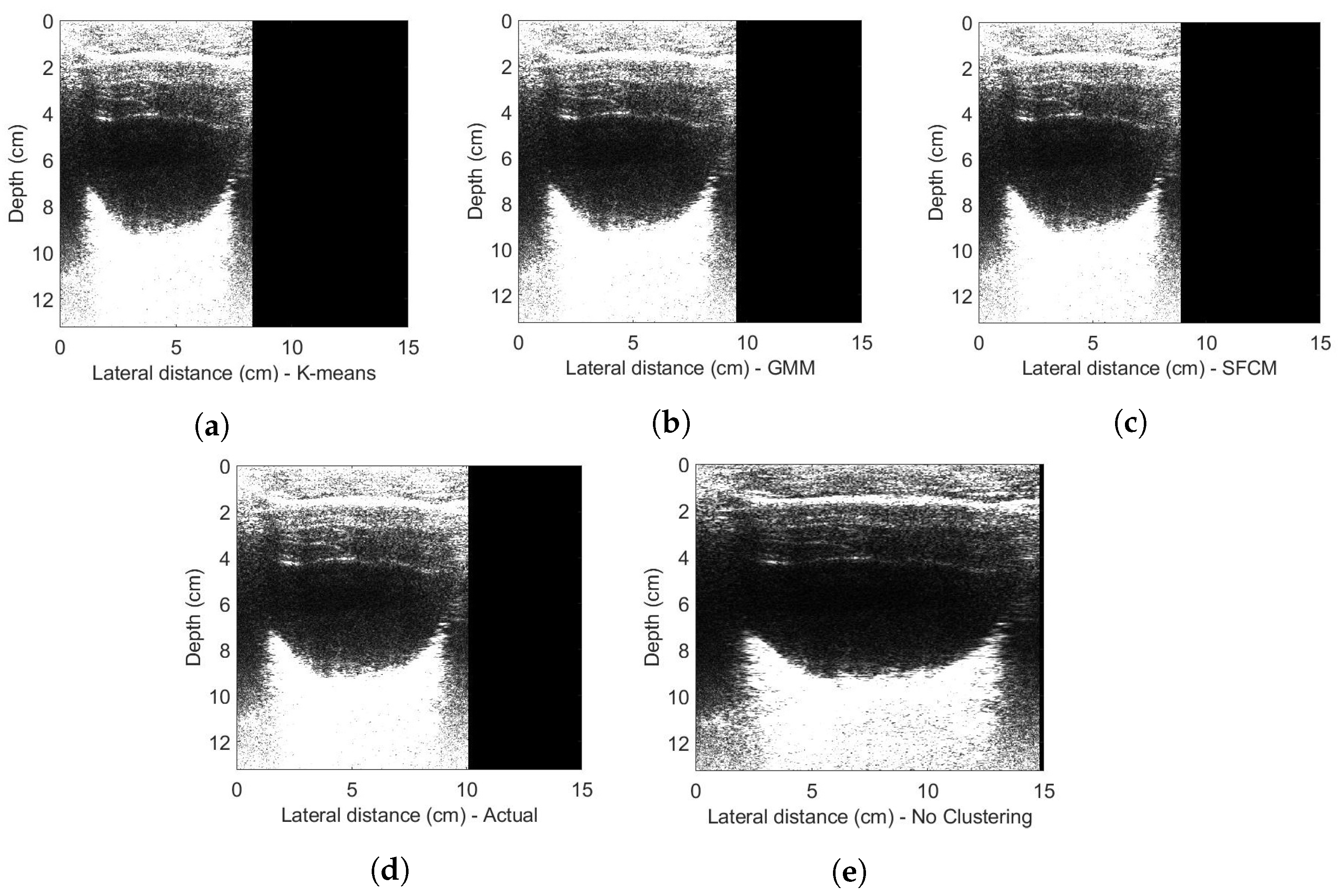
4. Experimental Evaluations
4.1. Set-Up for Scanning
4.2. Phantom Experiments
4.3. In Vivo Experiments
5. Conclusions and Future Work
Future Work
- Physical beam shape—in a fixed focus ultrasonic design, it is clear that the beam width and depth of focus have a strong influence on the backscattered signals and hence both the image resolution and the performance of correlation-based ego-motion estimation. More investigation is required to understand this relationship and to determine the optimum parameters for different applications.
- Synthetic focusing—a fixed focus design places constraints on lateral resolution vs. depth. It is interesting to consider whether monostatic synthetic aperture concepts as in [9] could be applied to the freehand scanner. This is very challenging, however, due to the requirement for precise tracking/control of the transducer trajectory and orientation.
- Frequency band—the frequency band considered in this paper (fc = 4.2 MHz, B = 2 MHz) was chosen for abdominal imaging with penetration up to 15 cm. Other possible applications aimed at imaging more superficial structures (e.g., muscles, vascular, breast) would favour a higher frequency design and the scalability of the proposed scanner design and the algorithms needs to be investigated.
- Front end—the ultrasound front end design in these experiments is sub-optimal in terms of noise performance and this limits both the image SNR and the depth range, which can be effectively utilised for correlation. A review of the front-end amplifier and impedance matching arrangements has significant potential to improve on this.
Author Contributions
Funding
Conflicts of Interest
References
- WHO|Maternal Mortality: Levels and Trends. Available online: https://www.who.int/reproductivehealth/publications/maternal-mortality-2000-2017/en/?fbclid=IwAR0AHXsRyLH9llPPSVGrRB3DgJOzkhEnl9bey4PUKI8no3QFe2H-nIHkBUA (accessed on 3 February 2022).
- UN Sustainable Development Goals—Five Talents UK. Available online: https://www.fivetalents.org.uk/un-sustainable-development-goals/?gclid=Cj0KCQiA09eQBhCxARIsAAYRiynwWr91YomGUIeahKPnRGOPn48udtwar1pDvmQRAJ8AXqmTvI0K6YUaAj6cEALw_wcB&fbclid=IwAR1m-SK9ERouQpz3eAPx2C0e-Wn6xDl0yJDPzjnb7Zk1hwad9efss9HBhlE (accessed on 3 February 2022).
- Martín-Arguedas, C.J.; Romero-Laorden, D.; Martinez-Graullera, O.; Perez-Lopez, M.; Gomez-Ullate, L. An Ultrasonic Imaging SystemBbased on a New SAFT Approach and a GPU Beamformer. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2012, 59, 1402–1412. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.A.; Karaman, M.; Khuri-Yakub, B.T. Coherent-array imaging using phased subarrays. Part I: Basic principles. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2005, 52, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Karaman, M.; Li, P.C.; O’Donnell, M. Synthetic aperture imaging for small scale systems. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 1995, 42, 429–442. [Google Scholar] [CrossRef]
- Vrijburg, K.; Hernández-Peña, P. Global spending on health: Weathering the storm 2020. In World Health Organization Working Paper; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Medical Imaging: Develop New Technologies in the UK. Available online: https://www.gov.uk/government/publications/medical-imaging-develop-new-technologies-in-the-uk/medical-imaging-develop-new-technologies-in-the-uk (accessed on 19 July 2023).
- Ultrasound Equipment Market Size, Share: Global Report [2028]. Available online: https://www.alliedmarketresearch.com/ultrasound-devices-market (accessed on 19 July 2023).
- van den Heuvel, T.L.; Graham, D.J.; Smith, K.J.; de Korte, C.L.; Neasham, J.A. Development of a Low-Cost Medical Ultrasound Scanner Using a Monostatic Synthetic Aperture. IEEE Trans. Biomed. Circuits Syst. 2017, 11, 849–857. [Google Scholar] [CrossRef] [PubMed][Green Version]
- The Global Ultrasound Devices Market Size to Surpass USD 10 Billion by 2027: DelveInsight. Available online: https://www.globenewswire.com/en/news-release/2022/07/26/2486260/0/en/The-Global-Ultrasound-Devices-Market-Size-to-Surpass-USD-10-Billion-by-2027-DelveInsight.html (accessed on 19 July 2023).
- Abbas, A.; Neasham, J.; Naqvi, S.M. Ego-motion Estimation for Low-cost Freehand Ultrasound Scanner. In Proceedings of the ICASSP 2019—2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; pp. 1050–1054. [Google Scholar] [CrossRef]
- B, L.; Thittai, A.K. Design of a low cost ultrasound system using diverging beams and synthetic aperture approach: Preliminary study. In Proceedings of the 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), Melbourne, Australia, 18–21 April 2017; pp. 1108–1111. [Google Scholar] [CrossRef]
- Fuller, M.I.; Ranganathan, K.; Zhou, S.; Blalock, T.N.; Hossack, J.A.; Walker, W.F. Experimental System Prototype of a Portable, Low-Cost, C-Scan Ultrasound Imaging Device. IEEE Trans. Biomed. Eng. 2008, 55, 519–530. [Google Scholar] [CrossRef]
- Fuller, M.; Ranganathan, K.; Zhou, S.; Blalock, T.; Hossack, J.; Walker, W. Portable, low-cost medical ultrasound device prototype. In Proceedings of the IEEE Ultrasonics Symposium, Montreal, QC, Canada, 23–27 August 2004; Volume 1, pp. 106–109. [Google Scholar] [CrossRef]
- Goldsmith, A.; Pedersen, P.; Szabo, T. An inertial-optical tracking system for portable, quantitative, 3D ultrasound. In Proceedings of the 2008 IEEE Ultrasonics Symposium, Beijing, China, 2–5 November 2008; pp. 45–49. [Google Scholar]
- Owen, K.; Mauldin, F.W.; Hossack, J.A. Transducer motion estimation using combined ultrasound signal decorrelation and optical sensor data for low-cost ultrasound systems with increased field of view. In Proceedings of the 2011 IEEE International Ultrasonics Symposium, Orlando, FL, USA, 18–21 October 2011; pp. 1431–1434. [Google Scholar] [CrossRef]
- Cai, Q.; Peng, C.; Prieto, J.C.; Rosenbaum, A.J.; Stringer, J.S.; Jiang, X. A low-cost camera-based ultrasound probe tracking system: Design and prototype. In Proceedings of the 2019 IEEE International Ultrasonics Symposium (IUS), Glasgow, UK, 6–9 October 2019; pp. 997–999. [Google Scholar]
- Chen, J.F.; Fowlkes, J.B.; Carson, P.L.; Rubin, J.M. Determination of scan-plane motion using speckle decorrelation: Theoretical considerations and initial test. Int. J. Imaging Syst. Technol. 1997, 8, 38–44. [Google Scholar] [CrossRef]
- Krupa, A.; Fichtinger, G.; Hager, G.D. Full Motion Tracking in Ultrasound Using Image Speckle Information and Visual Servoing. In Proceedings of the 2007 IEEE International Conference on Robotics and Automation, Roma, Italy, 10–14 April 2007; pp. 2458–2464. [Google Scholar] [CrossRef]
- Gee, A.H.; James Housden, R.; Hassenpflug, P.; Treece, G.M.; Prager, R.W. Sensorless freehand 3D ultrasound in real tissue: Speckle decorrelation without fully developed speckle. Med. Image Anal. 2006, 10, 137–149. [Google Scholar] [CrossRef]
- Chang, R.F.; Wu, W.J.; Chen, D.R.; Chen, W.M.; Shu, W.; Lee, J.H.; Jeng, L.B. 3-D US frame positioning using speckle decorrelation and image registration. Ultrasound Med. Biol. 2003, 29, 801–812. [Google Scholar] [CrossRef]
- Erickson, B.; Korfiatis, P.; Akkus, Z.; Kline, T. Machine learning for medical imaging. Radio Graph. 2017, 37, 505–515. [Google Scholar] [CrossRef]
- Chan, H.P.; Lo, S.C.; Sahiner, B.; Lam, K.; Helvie, M. Computer-aided detection of mammographic microcalcifications: Pattern recognition with an artificial neural network. Med. Phys. 1995, 22, 1555–1567. [Google Scholar] [CrossRef]
- Summers, R.M. Improving the Accuracy of CTC Interpretation: Computer-Aided Detection. Gastrointest. Endosc. Clin. N. Am. 2010, 20, 245–257. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yoshida, H.; Näppi, J. CAD in CT colonography without and with oral contrast agents: Progress and challenges. Comput. Med. Imaging Graph. 2007, 31, 267–284. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, T.M.; Shinkareva, S.V.; Carlson, A.; Chang, K.M.; Malave, V.L.; Mason, R.A.; Just, M.A. Predicting human brain activity associated with the meanings of nouns. Science 2008, 320, 1191–1195. [Google Scholar] [CrossRef] [PubMed]
- Davatzikos, C.; Fan, Y.; Wu, X.; Shen, D.; Resnick, S.M. Detection of prodromal Alzheimer’s disease via pattern classification of magnetic resonance imaging. Neurobiol. Aging 2008, 29, 514–523. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Burge, J.; Lane, T.; Pearlson, G.; Kiehl, K.; Calhoun, V. Hybrid ICA–Bayesian network approach reveals distinct effective connectivity differences in schizophrenia. NeuroImage 2008, 42, 1560–1568. [Google Scholar] [CrossRef][Green Version]
- Schoepf, U.J.; Schneider, A.; Das, M.; Wood, S.A.; Cheema, J.I.; Costello, P. Pulmonary Embolism: Computer-aided Detection at Multidetector Row Spiral Computed Tomography. J. Thorac. Imaging 2007, 22, 319–323. [Google Scholar] [CrossRef]
- Dundar, M.M.; Fung, G.; Krishnapuram, B.; Rao, R.B. Multiple-Instance Learning Algorithms for Computer-Aided Detection. IEEE Trans. Biomed. Eng. 2008, 55, 1015–1021. [Google Scholar] [CrossRef]
- Ueno, Y.; Forghani, B.; Forghani, R.; Dohan, A.; Zeng, X.Z.; Chamming’s, F.; Arseneau, J.; Fu, L.; Gilbert, L.; Gallix, B.; et al. Endometrial carcinoma: MR imaging–based texture model for preoperative risk stratification—A preliminary analysis. Radiology 2017, 284, 748–757. [Google Scholar] [CrossRef]
- Sherin, L.; Sohail, A.; Shujaat, S. Time-dependent AI-modeling of the anticancer efficacy of synthesized gallic acid analogues. Comput. Biol. Chem. 2019, 79, 137–146. [Google Scholar] [CrossRef]
- Ogasawara, J.; Ikenoue, S.; Yamamoto, H.; Sato, M.; Kasuga, Y.; Mitsukura, Y.; Ikegaya, Y.; Yasui, M.; Tanaka, M.; Ochiai, D. Deep neural network-based classification of cardiotocograms outperformed conventional algorithms. Sci. Rep. 2021, 11, 13367. [Google Scholar] [CrossRef]
- Abrantes, J.; Silva, M.J.; Meneses, J.; Oliveira, C.; Calisto, F.M.; Filice, R. External Validation of a Deep Learning Model for Breast Density Classification; ESR—European Society of Radiology: Vienna, Austria, 2023. [Google Scholar] [CrossRef]
- Calisto, F.M.; Nunes, N.; Nascimento, J.C. Modeling adoption of intelligent agents in medical imaging. Int. J. Hum.-Comput. Stud. 2022, 168, 102922. [Google Scholar] [CrossRef]
- Calisto, F.M.; Fernandes, J.; Morais, M.; Santiago, C.; Abrantes, J.M.; Nunes, N.; Nascimento, J.C. Assertiveness-based Agent Communication for a Personalized Medicine on Medical Imaging Diagnosis. In Proceedings of the 2023 CHI Conference on Human Factors in Computing Systems, Hamburg, Germany, 23–28 April 2023; pp. 1–20. [Google Scholar]
- Ray, M.; Mahata, N.; Sing, J.K. Uncertainty parameter weighted entropy-based fuzzy c-means algorithm using complemented membership functions for noisy volumetric brain MR image segmentation. Biomed. Signal Process. Control 2023, 85, 104925. [Google Scholar] [CrossRef]
- Pregitha, R.E.; Kumar, R.; Selvakumar, C.E. Segmentation of ultrasound fetal image using spatial fuzzy C-Mean clustering method. AIP Conference Proceedings; AIP Publishing: College Park, MA, USA, 2022; Volume 2519. [Google Scholar]
- Meenakshi, S.; Suganthi, M.; Sureshkumar, P. Segmentation and boundary detection of fetal kidney images in second and third trimesters using kernel-based fuzzy clustering. J. Med. Syst. 2019, 43, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kissara, W.A.; Hassan, B.F. Determination of Fetal sex by Fetal anatomy parameters using a Fuzzy C-Mean Cluster. Al-Kitab J. Pure Sci. 2021, 5, 9–25. [Google Scholar] [CrossRef]
- Li, B.N.; Chui, C.K.; Chang, S.; Ong, S. Integrating spatial fuzzy clustering with level set methods for automated medical image segmentation. Comput. Biol. Med. 2011, 41, 1–10. [Google Scholar] [CrossRef]
- Zhang, Q.; Huang, C.; Li, C.; Yang, L.; Wang, W. Ultrasound image segmentation based on multi-scale fuzzy c-means and particle swarm optimization. In Proceedings of the IET International Conference on Information Science and Control Engineering 2012 (ICISCE 2012), Shenzhen, China, 7–9 December 2012; pp. 1–5. [Google Scholar] [CrossRef]
- Yang, H.; Christopher, L.A.; Duric, N.; West, E.; Bakic, P. Performance analysis of EM-MPM and K-means clustering in 3D ultrasound image segmentation. In Proceedings of the 2012 IEEE International Conference on Electro/Information Technology, Indianapolis, IN, USA, 6–8 May 2012; pp. 1–4. [Google Scholar] [CrossRef]
- Sombutkaew, R.; Kumsang, Y.; Chitsobuk, O. Adaptive quantization with Fuzzy C-mean clustering for liver ultrasound compression. In Proceedings of the 2014 14th International Conference on Control Automation and Systems (ICCAS 2014), Gyeonggi-do, Republic of Korea, 22–25 October 2014; pp. 521–524. [Google Scholar] [CrossRef]
- Çiklaçandir, F.G.Y.; Ertaylan, A.; Bınzat, U.; Kut, A. Lesion Detection from the Ultrasound Images Using K-Means Algorithm. In Proceedings of the 2019 Medical Technologies Congress (TIPTEKNO), Izmir, Turkey, 3–5 October 2019; pp. 1–4. [Google Scholar] [CrossRef]
- Upadhyay, P.; Sharma, A.; Chandra, S. A Novel Approach of Intuitive K-means Clustering for Renal Calculi Detection in Ultrasound Images. Int. J. Electr. Eng. Inform. 2018, 10, 126–139. [Google Scholar] [CrossRef]
- Xu, Y. Image decomposition based ultrasound image segmentation by using fuzzy clustering. In Proceedings of the 2009 IEEE Symposium on Industrial Electronics Applications, Kuala Lumpur, Malaysia, 4–6 October 2009; Volume 1, pp. 6–10. [Google Scholar] [CrossRef]
- Mandwe, A.A.; Anjum, A. Detection of Brain Tumor Using K-Means Clustering. Int. J. Sci. Res. (IJSR) 2016, 5, 420–423. [Google Scholar]
- Agrawal, R.; Jajulwar, K.; Agrawal, U. A Design Approach for Performance Analysis of Infants Abnormality Using K Means Clustering. In Proceedings of the 2021 5th International Conference on Trends in Electronics and Informatics (ICOEI), Tirunelveli, India, 3–5 June 2021; pp. 992–997. [Google Scholar]
- Hussain, A.A.; Kanger, E.F.; Hassan, W.A. Isolation and Identify Biometrics of Fetuses using Segmentation Techniques. Indian J. Ecol. 2022, 49, 462–468. [Google Scholar]
- Khanna, A.; Sood, M.; Devi, S. US Image Segmentation Based on Expectation Maximization and Gabor Filter. Int. J. Model. Optim. 2012, 2, 230–233. [Google Scholar] [CrossRef]
- Huang, K.; Xu, M.; Qi, X. NGMMs: Neutrosophic Gaussian Mixture Models for Breast Ultrasound Image Classification. In Proceedings of the 2021 43rd Annual International Conference of the IEEE Engineering in Medicine Biology Society (EMBC), Mexico City, Mexico, 1–5 November 2021; pp. 3943–3947. [Google Scholar] [CrossRef]
- Belciug, S.; Iliescu, D.G. Deep learning and Gaussian Mixture Modelling clustering mix. A new approach for fetal morphology view plane differentiation. J. Biomed. Inform. 2023, 143, 104402. [Google Scholar] [CrossRef]
- Ravindraiah, R.; Tejaswini, K. IVUS Image Segmentation By Using Expectation-Maximization Approach. Int. J. Adv. Res. Comput. Commun. Eng. 2014, 3, 5662–5664. [Google Scholar]
- Nsugbe, E.; Reyes-Lagos, J.J.; Adams, D.; Samuel, O.W. On the prediction of premature births in Hispanic labour patients using uterine contractions, heart beat signals and prediction machines. Healthc. Technol. Lett. 2023, 10, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Gammex. 403 GSLE Precision Multi-Purpose Phantom8. Available online: https://www.sunnuclear.com/solutions/diagnostic/ultrasoundqa (accessed on 1 October 2018).
- Sinaga, K.P.; Yang, M.S. Unsupervised K-Means Clustering Algorithm. IEEE Access 2020, 8, 80716–80727. [Google Scholar] [CrossRef]
- Li, B.; Jiang, Q.; Chen, L. A Heuristically Weighting K-Means algorithm for subspace clustering. In Proceedings of the 2008 2nd International Conference on Anti-Counterfeiting, Security and Identification, Guiyang, China, 20–23 August 2008; pp. 268–271. [Google Scholar] [CrossRef]
- Jain, A.K. Data clustering: 50 years beyond K-means. Pattern Recognit. Lett. 2010, 31, 651–666. [Google Scholar] [CrossRef]
- Yao, J.; Summers, R. Chapter 20—Polyp Segmentation on CT Colonography. In Medical Image Recognition, Segmentation and Parsing; Zhou, S.K., Ed.; The Elsevier and MICCAI Society Book Series; Academic Press: Cambridge, MA, USA, 2016; pp. 451–484. [Google Scholar] [CrossRef]
- Yang, Y.; Huang, S. Image Segmentation by Fuzzy C-Means Clustering Algorithm with a Novel Penalty Term. Comput. Artif. Intell. 2007, 26, 17–31. [Google Scholar]
- Chuang, K.S.; Tzeng, H.L.; Chen, S.; Wu, J.; Chen, T.J. Fuzzy c-means clustering with spatial information for image segmentation. Comput. Med. Imaging Graph. 2006, 30, 9–15. [Google Scholar] [CrossRef]
- Virupakshappa, K.; Oruklu, E. Unsupervised Machine Learning for Ultrasonic Flaw Detection using Gaussian Mixture Modeling, K-Means Clustering and Mean Shift Clustering. In Proceedings of the 2019 IEEE International Ultrasonics Symposium (IUS), Glasgow, UK, 6–9 October 2019; pp. 647–649. [Google Scholar] [CrossRef]
- Neagoe, V.E.; Chirila-Berbentea, V. Improved Gaussian mixture model with expectation-maximization for clustering of remote sensing imagery. In Proceedings of the 2016 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Beijing, China, 10–15 July 2016; pp. 3063–3065. [Google Scholar] [CrossRef]
- Patel, E.; Kushwaha, D.S. Clustering Cloud Workloads: K-Means vs Gaussian Mixture Model. Procedia Comput. Sci. 2020, 171, 158–167. [Google Scholar] [CrossRef]
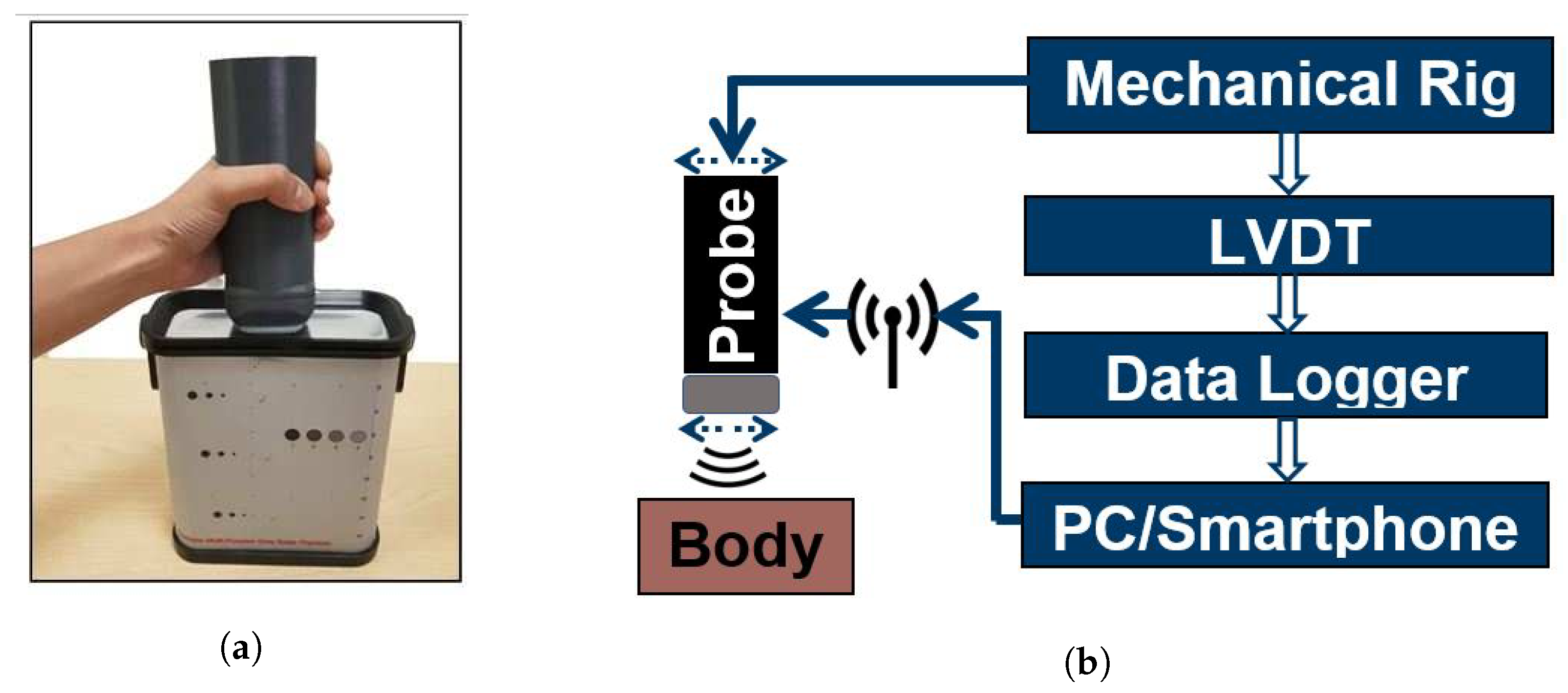

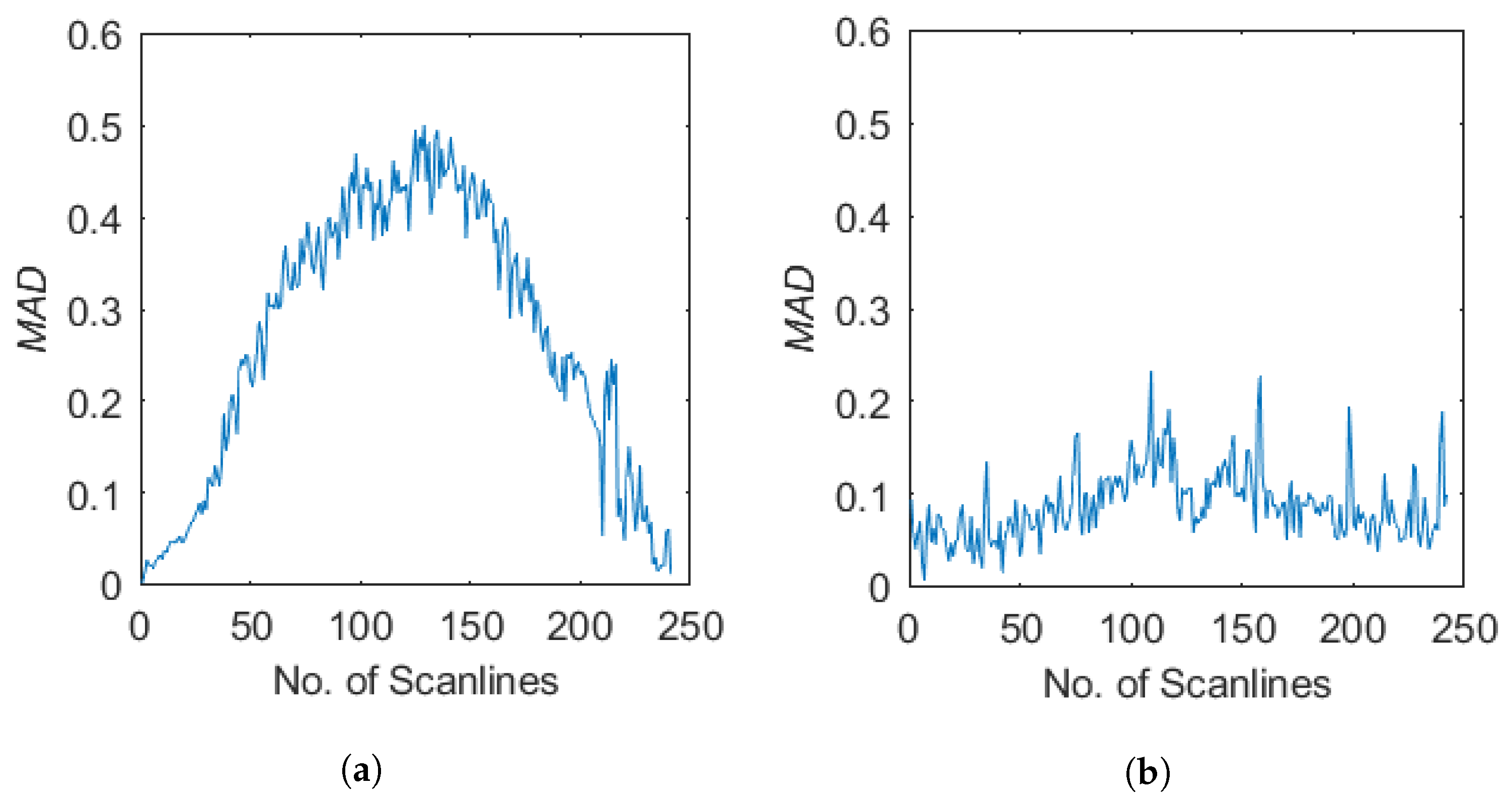
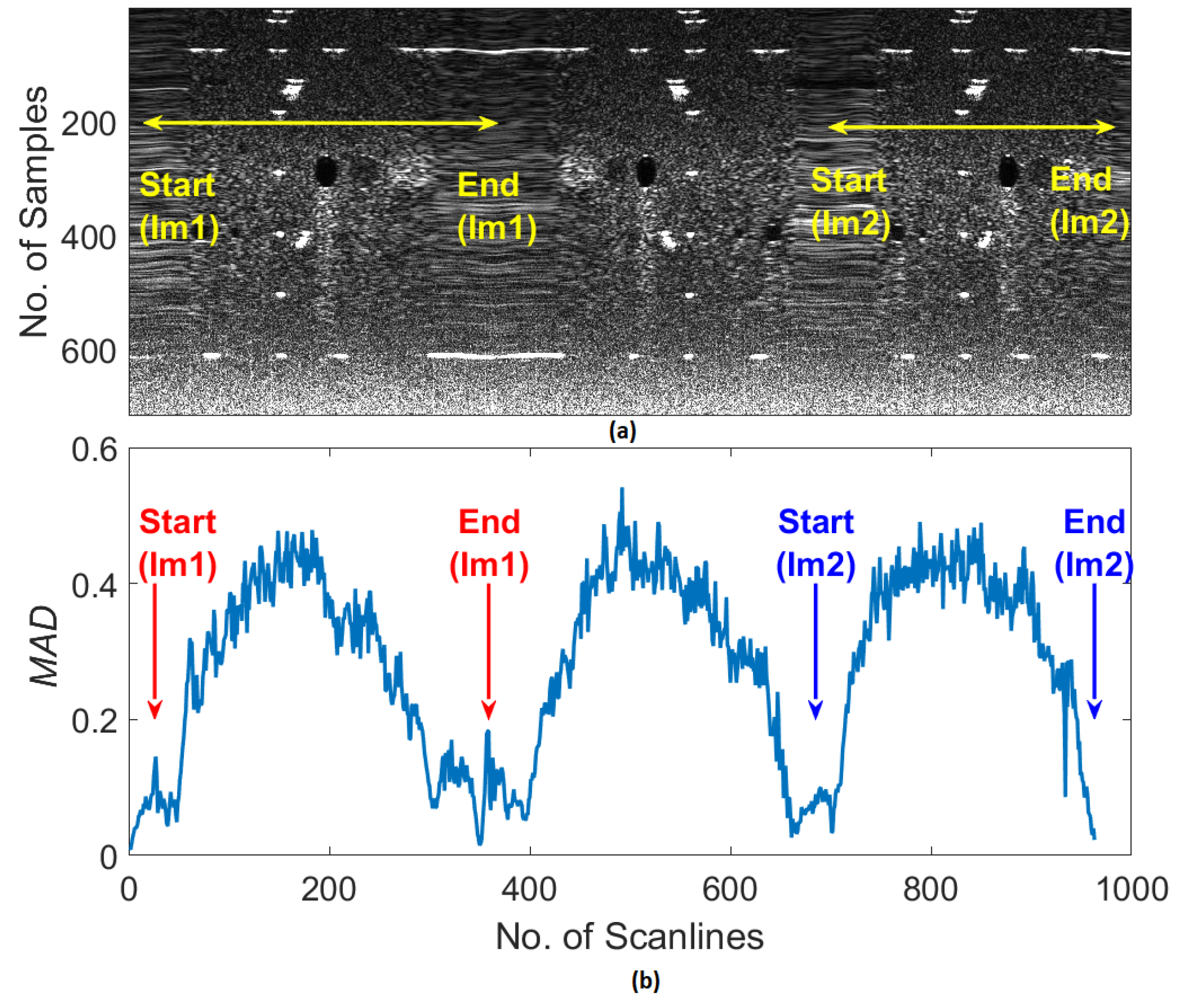
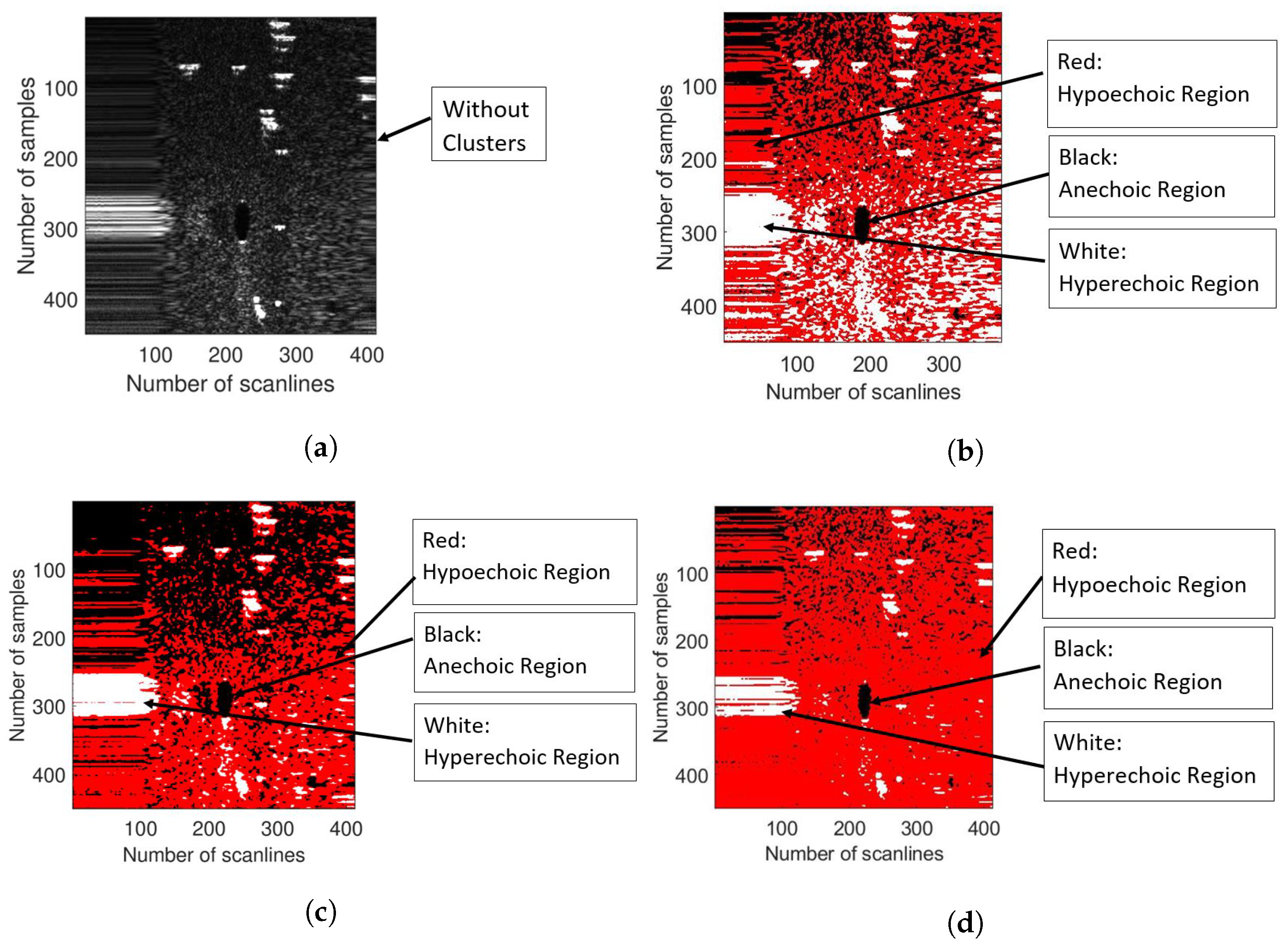
| Parameter | Symbol | Value |
|---|---|---|
| Centre Frequency | 4.2 MHz | |
| Bandwidth | 2 MHz | |
| Range | R | 0.15 m |
| Sampling Frequency | 25 MHz | |
| Speed of Sound in tissues | c | 1540 ms−1 |
| Element Width | 2 mm | |
| Element Length | 7 mm | |
| Element Thickness | 0.5 mm | |
| Frame/s | 400 |
| Parameter | Value |
|---|---|
| Elevation Focus (mm) | 50.5 |
| Slice thickness at elevation focus | 1.2 |
| Axial Resolution in focus (mm) | 0.37 ± 0.05 |
| Axial Resolution averaged over depth (mm) | 0.55 ± 0.13 |
| Lateral Resolution in focus (mm) | 1.25 ± 0.06 |
| Lateral Resolution averaged over depth (mm) | 2.71 ± 1.40 |
| Axial spatial conformity (%) | 2 ± 0.06 |
| Dynamic Range (dB) | 98 mm |
| Contrast sensitivity | 2.19 |
| Quantitative Metrics | Without Clustering | K-Means | SFCM | GMM |
|---|---|---|---|---|
| Mean PE (%) | 28.7 | 12.3 | 14.9 | 4.2 |
| Max PE (%) | 57.2 | 59.1 | 18.8 | 20.9 |
| Min PE (%) | 22.4 | 0.011 | 0.017 | 0.001 |
| MSE (10) | 37 | 5.5 | 15 | 2.1 |
| RMSE (10) | 1.94 | 0.74 | 1.22 | 0.46 |
| Time (s) | 0 | 2.72 | 5.7 | 291 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abbas, A.; Neasham, J.; Naqvi, M. Image Formation Algorithms for Low-Cost Freehand Ultrasound Scanner Based on Ego-Motion Estimation and Unsupervised Clustering. Electronics 2023, 12, 3634. https://doi.org/10.3390/electronics12173634
Abbas A, Neasham J, Naqvi M. Image Formation Algorithms for Low-Cost Freehand Ultrasound Scanner Based on Ego-Motion Estimation and Unsupervised Clustering. Electronics. 2023; 12(17):3634. https://doi.org/10.3390/electronics12173634
Chicago/Turabian StyleAbbas, Ayusha, Jeffrey Neasham, and Mohsen Naqvi. 2023. "Image Formation Algorithms for Low-Cost Freehand Ultrasound Scanner Based on Ego-Motion Estimation and Unsupervised Clustering" Electronics 12, no. 17: 3634. https://doi.org/10.3390/electronics12173634
APA StyleAbbas, A., Neasham, J., & Naqvi, M. (2023). Image Formation Algorithms for Low-Cost Freehand Ultrasound Scanner Based on Ego-Motion Estimation and Unsupervised Clustering. Electronics, 12(17), 3634. https://doi.org/10.3390/electronics12173634









