SVM-Based Blood Exam Classification for Predicting Defining Factors in Metabolic Syndrome Diagnosis
Abstract
:1. Introduction
2. Metabolic Syndrome
2.1. Metabolic Syndrome Markers
2.1.1. A Large Waist
2.1.2. A High Triglyceride Level
2.1.3. Reduced HDL (“Good” Cholesterol)
2.1.4. Elevated Fasting Blood Sugar (Fasting Glucose)
2.1.5. Increased Blood Pressure
2.2. Genetics—Cardiac Effects of Obesity and MetS
3. Related Work and Methodology Comparison
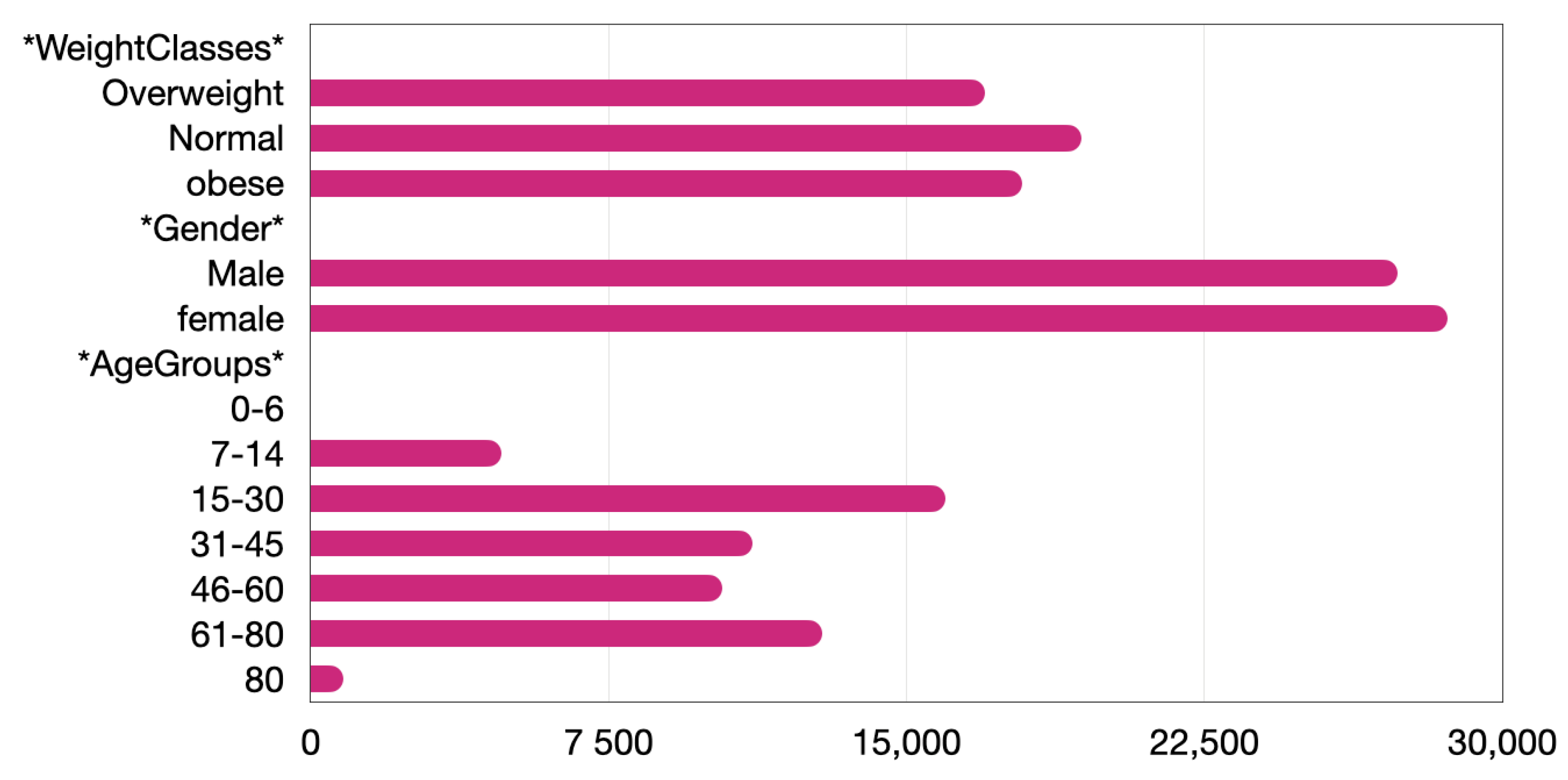
4. Exploratory Data Analysis and Bias Evaluation
5. Predicting BMI Based on Blood Exams
5.1. Neural Networks vs. SVCs, Challenges and Objectives
5.2. One-Class SVM Identifiers
5.3. One-vs.-One SVC
5.4. Cascaded SVC
6. Implementing a More Precise System
6.1. MetS Prediction Model
- While classifying for BMI, blood exams did not include triglycerides, glucose or cholesterol levels.
- Total cholesterol is computed using both LDL (bad cholesterol) and HDL (good cholesterol) and 20% of triglycerides. The out-of-bounds values of total cholesterol when, at the same time, triglycerides are out of range suggests that LDL is more prominent than HDL and that HDL has a greater probability of being low. To test this hypothesis, results need to be compared for total cholesterol or only HDL used as features.
- Since it was previously shown that BMI can be predicted quite accurately by using blood exams and BMI is a more common factor of MetS, there is good reason to believe that the classifier can identify patterns related to metabolism and, as such, be used in more refined calculations.
- The tests will not be gender specific. Any gender specificities will probably be identified by the model. Data will be balanced, but the road is always open for further experiments to be carried out in the future.
6.1.1. Total Cholesterol as a Feature
6.1.2. HDL as a Feature
6.2. Blood Pressure Predictor
7. Summary, Discussion, Itemization of Key Findings and Contribution
8. Conclusions and Future Research Avenues
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SBP | Systolic blood pressure |
| METS | Metabolic Syndrome |
| BMI | Body mass index |
| ASCVD | Atherosclerotic cardiovascular disease |
| HDL | High-density lipoprotein |
| LDL | Low-density lipoprotein |
| NCD | Noncommunable disease |
| SVM | Support vector machine |
| SVC | Support vector classifier |
| Trig | Triglycerides |
| Cholestrl | Cholesterol |
References
- Thomas, D.; Ravi, K. National health and nutrition examination survey: Sample design. Future Healthc. J. 2019, 6, 94. [Google Scholar]
- Strimbu, K.; Tavel, J.A. What are biomarkers? Curr. Opin. HIV AIDS 2010, 5, 463. [Google Scholar] [CrossRef] [PubMed]
- Panagoulias, D.P.; Sotiropoulos, D.N.; Tsihrintzis, G.A. Towards Personalized Nutrition Applications with Nutritional Biomarkers and Machine Learning. In Advances in Assistive Technologies: Selected Papers in Honour of Professor Nikolaos G. Bourbakis—Vol. 3; Tsihrintzis, G.A., Virvou, M., Esposito, A., Jain, L.C., Eds.; Springer: Cham, Switzerland, 2022; Volume 28, pp. 73–122. [Google Scholar]
- Panagoulias, D.P.; Sotiropoulos, D.N.; Tsihrintzis, G.A. Nutritional Biomarkers and Machine Learning for Personalized Nutrition Applications and Health Optimization. Intell. Decis. Technol. 2021, 15, 645–653. [Google Scholar] [CrossRef]
- Panagoulias, D.P.; Sotiropoulos, D.N.; Tsihrintzis, G.A. Nutritional Biomarkers and Machine Learning for Personalized Nutrition Applications and Health Optimization. In Proceedings of the Twelfth IEEE International Conference on Information, Intelligence, Systems and Applications, Chania, Greece, 12–14 July 2021; pp. 731–733. [Google Scholar]
- Panagoulias, D.P.; Sotiropoulos, D.N.; Tsihrintzis, G.A. Biomarker-based Deep Learning for Personalized Nutrition. In Proceedings of the 33rd IEEE International Conference on Tools with Artificial Intelligence (ICTAI), Virtually, 1–3 November 2021; pp. 73–122. [Google Scholar]
- Massaro, A.; Ricci, G.; Selicato, S.; Raminelli, S.; Galiano, A. Decisional Support System with Artificial Intelligence oriented on Health Prediction using a Wearable Device and Big Data. In Proceedings of the 2020 IEEE International Workshop on Metrology for Industry 4.0 IoT, Roma, Italy, 3–5 June 2020; pp. 718–723. [Google Scholar]
- Usman, A.; Won, L.J.; Syed, M.B.H.; Taqdir, A.; Ali, K.W.; Sungyoung, L. The Impact of Big Data in Healthcare Analytics. In Proceedings of the 2020 International Conference on Information Networking (ICOIN), Barcelona, Spain, 7–10 January 2020; pp. 61–63. [Google Scholar]
- Ralf, H.; Michael, G. Artificial Intelligence Applications in Human Pathology; World Scientific Pub Co Inc.: Singapore, 2022; 216p. [Google Scholar]
- WHO. Obesity and Overweight. 2021. Available online: https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight (accessed on 9 June 2021).
- Kakudi, H.A.; Kiong, L.C.; Moy, F.M.; Kau, L.C.; Pasupa, K. Diagnosis of metabolic syndrome using machine learning, statistical and risk quantifiaction techniques: A systematic literature review. Malays. J. Comput. Sci. 2021, 34, 221–241. [Google Scholar]
- Huiling, C.; Bo, Y.; Dayou, L.; Wenbin, L.; Yanlong, L.; Xiuhua, Z.; Lufeng, H. Using Blood Indexes to Predict Overweight Statuses: An Extreme Learning Machine-Based Approach. PLoS ONE 2015, 10, e0143003. [Google Scholar]
- Khatiwada, S.; Sah, S.K.; Kc, R.; Baral, N.; Lamsal, M. Thyroid dysfunction in metabolic syndrome patients and its relationship with components of metabolic syndrome. Clin. Diabetes Endocrinol. 2016, 2, 3. [Google Scholar] [CrossRef] [Green Version]
- Sanisoglu, S.Y.; Oktenli, C.; Hasimi, A.; Yokusoglu, M.; Ugurlu, M. Prevalence of metabolic syndrome-related disorders in a large adult population in Turkey. Clin. Diabetes Endocrinol. 2016, 6, 92. [Google Scholar] [CrossRef] [Green Version]
- Denson, J.L.; Gillet, A.S.; Zu, Y.; Brown, M.; Pham, T.; Yoshida, Y.; Mauvais-Jarvis, F.; Douglas, I.S.; Moore, M.; Tea, K.; et al. Metabolic Syndrome and Acute Respiratory Distress Syndrome in Hospitalized Patients With COVID-19. JAMA Netw. Open 2021, 4, e2140568. [Google Scholar] [CrossRef]
- Dagan, S.S.; Segev, S.; Novikov, I.; Dankner, R. Waist, circumference vs. body mass index in association with cardiorespiratory fitness in healthy men and women: A cross sectional analysis of 403 subjects. Nutr. J. 2013, 12, 12. [Google Scholar] [CrossRef] [Green Version]
- Grundy, S.M. Metabolic syndrome: A multiplex cardiovascular risk factor. J. Clin. Endocrinol. Metab. 2007, 92, 399–404. [Google Scholar] [CrossRef]
- Cui, R.; Iso, H.; Yamagishi, K.; Saito, I.; Kokubo, Y.; Inoue, M.; Tsugane, S.; JPHC Study Group. High serum total cholesterol levels is a risk factor of ischemic stroke for general Japanese population: The JPHC study. Atherosclerosis 2012, 221, 565–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nathan, D.M.; Davidson, M.B.; DeFronzo, R.A.; Heine, R.J.; Henry, R.R.; Pratley, R.; Zinman, B. Impaired fasting glucose and impaired glucose tolerance: Implications for care. Diabetes Care 2007, 30, 753–759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- American Heart Association. What Is High Blood Pressure? South Carolina State Documents Depository; South Carolina State Library: Columbia, SC, USA, 2017; pp. 565–5698. [Google Scholar]
- Nilsson, P.M.; Tuomilehto, J.; Ryden, L. The metabolic syndrome—What is it and how should it be managed? Eur. J. Prev. Cardiol. 2019, 26, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.; Kim, J.Y.; Nam, Y. ECG Signal Analysis for Patient with Metabolic Syndrome based on 1D-Convolution Neural Network. In Proceedings of the 2020 International Conference on Computational Science and Computational Intelligence (CSCI), Las Vegas, NV, USA, 16–18 December 2020; pp. 731–733. [Google Scholar]
- Artham, S.M.; Lavie, C.J.; Milani, R.V.; Ventura, H.O. Obesity and hypertension, heart failure, and coronary heart disease—Risk factor, paradox, and recommendations for weight loss. Ochsner J. 2009, 9, 124–132. [Google Scholar]
- Lemieux, I.; Lamarche, B.; Couillard, C.; Pascot, A.; Cantin, B.; Bergeron, J.; Dagenais, G.R.; Després, J.P. Total cholesterol/HDL cholesterol ratio vs. LDL cholesterol/HDL cholesterol ratio as indices of ischemic heart disease risk in men: The Quebec Cardiovascular Study. Arch. Intern. Med. 2001, 161, 2685–2692. [Google Scholar] [CrossRef]
- Lampropoulos, A.S.; Sotiropoulos, D.N.; Tsihrintzis, G.A. Cascade hybrid recommendation as a combination of one-class classification and collaborative filtering. Int. J. Artif. Intell. Tools 2014, 23, 2685–2692. [Google Scholar] [CrossRef]
- Sotiropoulos, D.N.; Tsihrintzis, G.A. Machine Learning Paradigms—Artificial Immune Systems and Their Applications in Software Personalization; Springer: Berlin/Heidelberg, Germany, 2017; Volume 118, p. 330. [Google Scholar]
- Morteza, S.; Yadollah, M.; Anoshirvan, K.; Farzad, H. Comparison of artificial neural network, logistic regression and discriminant analysis methods in prediction of metabolic syndrome. Iran. J. Endocrinol. Metab. 2010, 11, 645–653. [Google Scholar]
- Hiroshi, H.; Tetsuro, T.; Shigenari, H.; Toshifumi, H.; Ikuo, S. Prediction of metabolic syndrome using artificial neural network system based on clinical data including insulin resistance index and serum adiponectin. Comput. Biol. Med. 2011, 41, 1051–1056. [Google Scholar] [CrossRef]
- Xia, S.-J.; Gao, B.-Z.; Wang, S.-H.; Guttery, D.S.; Li, C.-D.; Zhang, Y.-D. Metabolic syndrome, Machine learning, Diagnosis model, Symptoms, Traditional Chinese medicine, Physicochemical indexes. Biomed. Pharmacother. 2021, 137, 111–367. [Google Scholar] [CrossRef]
- Worachartcheewan, A.; Nantasenamat, C.; Isarankura-Na-Ayudhya, C.; Prachayasittikul, V. Quantitative population-health relationship (QPHR) for assessing metabolic syndrome. EXCLI J. 2013, 12, 569. [Google Scholar]
- Dietterich, T.G. Ensemble methods in machine learning. In Archives of Internal Medicine; Springer: Berlin/Heidelberg, Germany, 2000. [Google Scholar]
- Huang, P.L. A comprehensive definition for metabolic syndrome. Dis. Model. Mech. 2009, 2, 231–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cooney, M.T.; Dudina, A.; De Bacquer, D.; Wilhelmsen, L.; Sans, S.; Menotti, A.; De Backer, G.; Jousilahti, P.; Keil, U.; Thomsen, T.; et al. HDL cholesterol protects against cardiovascular disease in both genders, at all ages and at all levels of risk. Atherosclerosis 2009, 206, 611–616. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.L.; Dohrmann, S.M.; Burt, V.L.; Mohadjer, L.K. National Health and Nutrition Examination Survey: Sample Design, 2011–2014; Current Opinion in HIV and AIDS; US Department of Health and Human Services, Centers for Disease Control: Oxfordshire, UK, 2014. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]

















| Name | Description | Health Results |
|---|---|---|
| Waist to hip ratio | Abdominal obesity index | Hypertension, CHD, insulin-dependent diabetes and stroke |
| Total cholesterol | Helps in the synthesis of bile acids and steroid hormones | At middle age: Coronary heart disease (CHD) and mortality of all causes at an older age: u-shaped relation to death |
| Systolic blood pressure (SBP) | Cardiovascular activity index: maximum pressure in an artery when the heart supplies the body with blood | Cardiovascular death (CVD), stroke, coronary heart disease (CHD) |
| Fasting glucose | Measures the amount of sugar in the diabetes index | Diabetes, CHD, mortality, poor cognitive function |
| Metrics | Cascaded SVC | QPHR1 [30] | ANN [28] |
|---|---|---|---|
| Accuracy (%) | 84 | >95 | >95 |
| Size of sample | 5000 | 15,000 | 410 |
| Balanced sample | yes | no | no |
| Features | Standard Biochemistry profile. Mets defining factors not included | Uses defining factors to predict MetS (triglycerides, Bmi) | Uses defining factors to predict MetS (BMI, diastolic blood pressure, HDL-cholesterol, LDL-cholesterol) |
| Metrics | 4 Class Neural Network [5] | 4 Class SVC | 3 Class Neural Network [6] | 3 Class SVC | Cascaded SVM | Extreme LM [12] |
|---|---|---|---|---|---|---|
| Accuracy (%) | 56 | 55 | 58 | 62 | 85 | 90.54 |
| Size of sample | 75,000 | 15,000 | 33,000 | 15,000 | 10,000 | 500 |
| Balanced sample | no | no | yes | yes | yes | no |
| All Classes? | yes | yes | no | no | yes | no (Overweight class) |
| Features | Full Biochemistry Profile | Full Biochemistry Profile | Full Biochemistry Profile | Full Biochemistry Profile | Without defining factors of MetS (15 features) | Blood Indexes (39 features) + age |
| Set/Kernels | rbf | Linear | Poly | Sigmoid |
|---|---|---|---|---|
| training | 0.83 | 0.804 | 0.8 | 0.7 |
| testing | 0.81 | 0.803 | 0.785 | 0.7 |
| Set/Classifiers | Gaussian Naive Bayes | Random Forest Classifier | AdaBooster |
|---|---|---|---|
| training | 0.7511 | 0.83 | 0.805 |
| testing | 0.732 | 0.72 | 0.781 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panagoulias, D.P.; Sotiropoulos, D.N.; Tsihrintzis, G.A. SVM-Based Blood Exam Classification for Predicting Defining Factors in Metabolic Syndrome Diagnosis. Electronics 2022, 11, 857. https://doi.org/10.3390/electronics11060857
Panagoulias DP, Sotiropoulos DN, Tsihrintzis GA. SVM-Based Blood Exam Classification for Predicting Defining Factors in Metabolic Syndrome Diagnosis. Electronics. 2022; 11(6):857. https://doi.org/10.3390/electronics11060857
Chicago/Turabian StylePanagoulias, Dimitrios P., Dionisios N. Sotiropoulos, and George A. Tsihrintzis. 2022. "SVM-Based Blood Exam Classification for Predicting Defining Factors in Metabolic Syndrome Diagnosis" Electronics 11, no. 6: 857. https://doi.org/10.3390/electronics11060857
APA StylePanagoulias, D. P., Sotiropoulos, D. N., & Tsihrintzis, G. A. (2022). SVM-Based Blood Exam Classification for Predicting Defining Factors in Metabolic Syndrome Diagnosis. Electronics, 11(6), 857. https://doi.org/10.3390/electronics11060857








