Computer Vision-Based Kidney’s (HK-2) Damaged Cells Classification with Reconfigurable Hardware Accelerator (FPGA)
Abstract
1. Introduction
- (1)
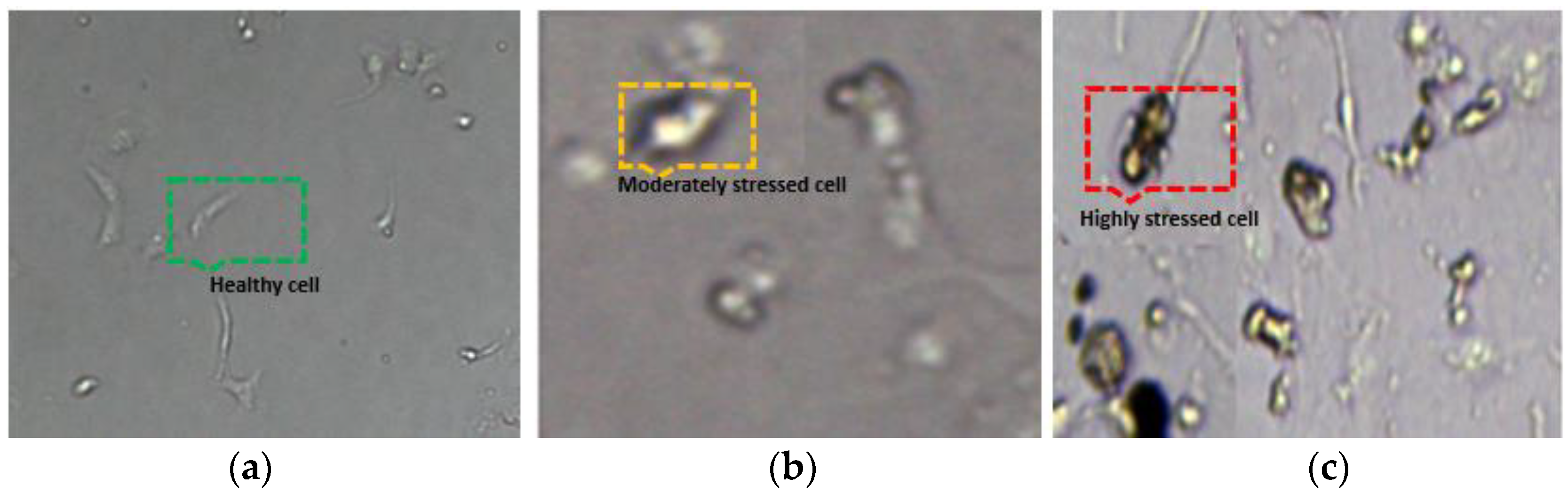
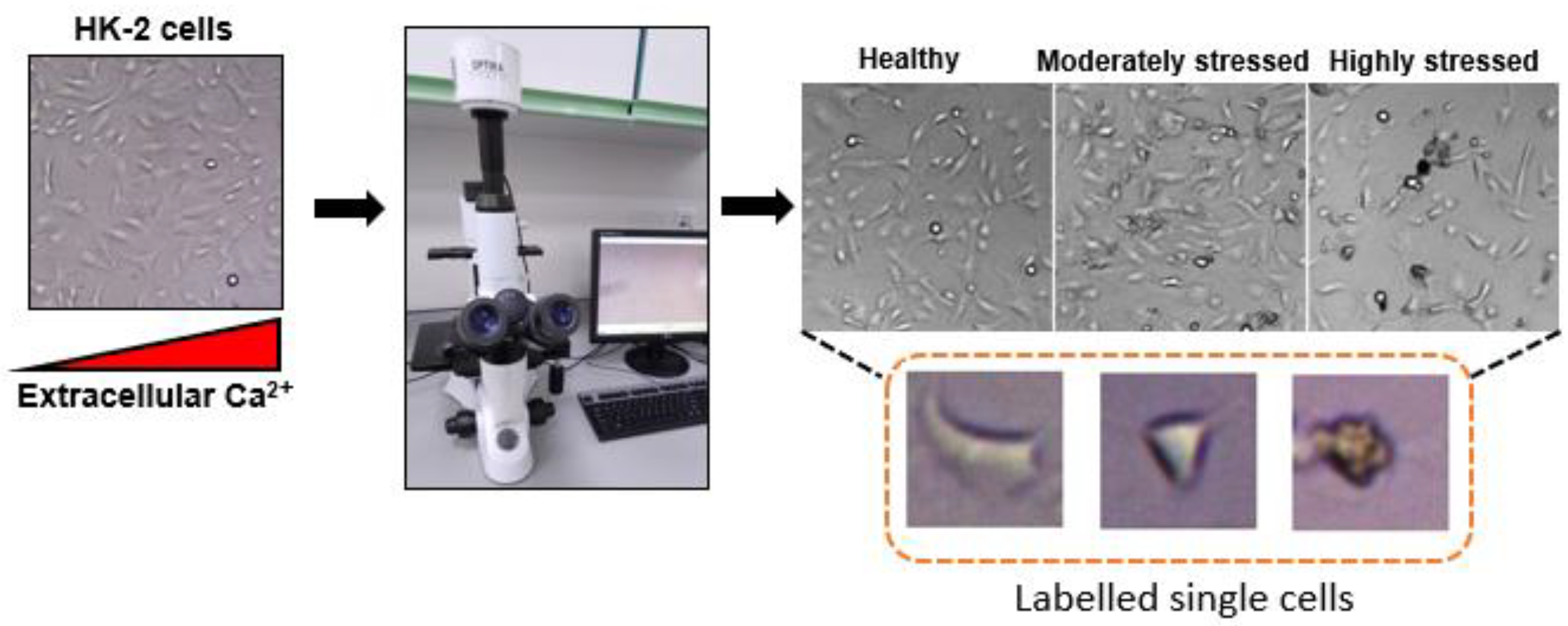
- Bespoke data collected from cell cultures in the biotechnology lab and cell labelling for HK-2 kidney cells (healthy, moderate and highly stressed).
- (2)
- Developing an integrated enhanced Canny edge detector to calculate pixel area for backend neural classifier implementation and retrospective comparison with histogram-based technique. This technique offers a simpler shallow network that is easier to implement on both software and reconfigurable hardware platforms with almost 100% accuracy.
- (3)
- A small number of training samples are required, which is demonstrated as a technological solution for the real-time classification of HK-2 damaged kidney cells.
2. Materials and Methods
2.1. Cell Preparation and Data Collection
2.2. Image Pre-Processing
2.3. Backend Classification Using Artificial Neural Network (ANN)
3. Results
3.1. Software
3.2. Hardware Accelerator (FPGA)
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Himavathi, S.; Anitha, D.; Muthuramalingam, A. Feedforward Neural Network Implementation in FPGA Using Layer Multiplexing for Effective Resource Utilization. IEEE Trans. Neural Netw. 2007, 18, 880–888. [Google Scholar] [CrossRef] [PubMed]
- Medus, L.D.; Iakymchuk, T.; Frances-Villora, J.V.; Bataller-Mompean, M.; Rosado-Munoz, A. A Novel Systolic Parallel Hardware Architecture for the FPGA Acceleration of Feedforward Neural Networks. IEEE Access 2019, 7, 76084–76103. [Google Scholar] [CrossRef]
- Zhang, C.; Wu, D.; Sun, J.; Sun, G.; Luo, G.; Cong, J. Energy-efficient CNN implementation on a deeply pipelined FPGA cluster. In Proceedings of the ISLPED’16: International Symposium on Low Power Electronics and Design, San Francisco, CA, USA, 8–10 August 2016; pp. 326–331. [Google Scholar]
- Ghani, A.; McGinnity, T.M.; Maguire, L.P.; Harkin, J. Area Efficient Architecture for Large Scale Implementation of Biologically Plausible Spiking Neural Networks on Reconfigurable Hardware. In Proceedings of the 2006 International Conference on Field Programmable Logic and Applications, Madrid, Spain, 28–30 August 2006; pp. 1–2. [Google Scholar] [CrossRef]
- Bataller-Mompeán, M.; Martínez-Villena, J.M.; Rosado-Muñoz, A.; Francés-Víllora, J.V.; Guerrero-Martínez, J.F.; Wegrzyn, M.; Adamski, M. Support tool for the combined software/hardware design of on-chip ELM training for SLFF neural networks. IEEE Trans. Ind. Informat. 2016, 12, 1114–1123. [Google Scholar] [CrossRef]
- Nikitakis, A.; Makantasis, K.; Tampouratzis, N.; Papaefstathiou, I. A Unified Novel Neural Network Approach and a Prototype Hardware Implementation for Ultra-Low Power EEG Classification. IEEE Trans. Biomed. Circuits Syst. 2019, 13, 670–681. [Google Scholar] [CrossRef] [PubMed]
- Ghani, A.; Aina, A.; See, C.H.; Yu, H.; Keates, S. Accelerated Diagnosis of Novel Coronavirus (COVID-19)—Computer Vision with Convolutional Neural Networks (CNNs). Electronics 2022, 11, 1148. [Google Scholar] [CrossRef]
- Ghani, A.; See, C.H.; Sudhakaran, V.; Ahmad, J.; Abd-Alhameed, R. Accelerating Retinal Fundus Image Classification Using Artificial Neural Networks (ANNs) and Reconfigurable Hardware (FPGA). Electronics 2019, 8, 1522. [Google Scholar] [CrossRef]
- Chen, X.; Zhou, X.; Wong, S.T.C. Automated Segmentation, Classification, and Tracking of Cancer Cell Nuclei in Time-Lapse Microscopy. IEEE Trans. Biomed. Eng. 2006, 53, 762–766. [Google Scholar] [CrossRef]
- Moen, E.; Bannon, D.; Kudo, T.; Graf, W.; Covert, M.; Van Valen, D. Deep learning for cellular image analysis. Nat. Methods 2019, 16, 1233–1246. [Google Scholar] [CrossRef]
- Grimm, J.B.; Muthusamy, A.K.; Liang, Y.; Brown, T.A.; Lemon, W.C.; Patel, R.; Lu, R.; Macklin, J.J.; Keller, P.J.; Ji, N.; et al. A general method to fne-tune fuorophores for live-cell and in vivo imaging. Nat. Methods 2017, 14, 987–994. [Google Scholar] [CrossRef]
- Megason, S.G. In toto imaging of embryogenesis with confocal time-lapse microscopy. Methods Mol. Biol. 2009, 546, 317–332. [Google Scholar]
- Chen, B.-C.; Legant, W.R.; Wang, K.; Shao, L.; Milkie, D.E.; Davidson, M.W.; Janetopoulos, C.; Wu, X.S.; Hammer, J.A., 3rd; Liu, Z.; et al. Lattice light-sheet microscopy: Imaging molecules to embryos at high spatiotemporal resolution. Science 2014, 346, 1257998. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.; Piccinini, F.; Balassa, T.; Koos, K.; Danka, T.; Azizpour, H.; Horvath, P. Phenotypic Image Analysis Software Tools for Exploring and Understanding Big Image Data from Cell-Based Assays. Cell Syst. 2018, 6, 636–653. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Greka, A. Calcium-permeable ion channels in the kidney. Am. J. Physiol. Renal. Physiol. 2016, 310, F1157–F1167. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rouse, D.; Suki, W.N. Renal control of extracellular calcium. Kidney Int. 1990, 38, 700–708. [Google Scholar] [CrossRef] [PubMed]
- Hodeify, R.; Ghani, A.; Matar, R.; Vazhappilly, C.G.; Merheb, M.; Zouabi, H.A.; Marton, J. Adenosine Triphosphate Protects from Elevated Extracellular Calcium-Induced Damage in Human Proximal Kidney Cells: Using Deep Learning to Predict Cytotoxicity. Cell. Physiol. Biochem. 2022, 56, 484–499. [Google Scholar] [CrossRef]
- Huang, Q.; Li, W.; Zhang, B.; Li, Q.; Tao, R.; Lovell, N.H. Blood Cell Classification Based on Hyperspectral Imaging With Modulated Gabor and CNN. IEEE J. Biomed. Health Inform. 2019, 24, 160–170. [Google Scholar] [CrossRef]
- Saraswat, M.; Arya, K. Automated microscopic image analysis for leukocytes identification: A survey. Micron 2014, 65, 20–33. [Google Scholar] [CrossRef]
- Wang, Q.; Chang, L.; Zhou, M.; Li, Q.; Liu, H.; Guo, F. A spectral and morphologic method for white blood cell classification. Opt. Laser Technol. 2016, 84, 144–148. [Google Scholar] [CrossRef]
- Froom, P.; Havis, R.; Barak, M. The rate of manual peripheral blood smear reviews in outpatients. Clin. Chem. Lab. Med. 2009, 47, 1401–1405. [Google Scholar] [CrossRef]
- Li, W.; Du, Q. Gabor-filtering based nearest regularized subspace for hyperspectral image classification. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 1012–1022. [Google Scholar] [CrossRef]
- Kang, X.; Li, C.; Li, S.; Lin, H. Classification of hyperspectral images by Gabor filtering based deep network. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2017, 11, 1166–1178. [Google Scholar] [CrossRef]
- Hinton, G.E.; Osindero, S.; Teh, Y.-W. A fast learning algorithm for deep belief nets. Neural Comput. 2006, 18, 1527–1554. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. In Proceedings of the Advances in Neural Information, Processing Systems, Lake Tahoe, NV, USA, 3–6 December 2012; pp. 1097–1105. [Google Scholar]
- Kumar, A.; Kim, J.; Lyndon, D.; Fulham, M.; Feng, D. An ensemble of fine-tuned convolutional neural networks for medical image classification. IEEE J. Biomed. Health Inform. 2017, 21, 31–40. [Google Scholar] [CrossRef]
- Gao, Z.; Wang, L.; Zhou, L.; Zhang, J. Hep-2 cell image classification with deep convolutional neural networks. IEEE J. Biomed. Health Inform. 2017, 21, 416–428. [Google Scholar] [CrossRef]
- Wang, Q.; Zheng, Y.; Yang, G.; Jin, W.; Chen, X.; Yin, Y. Multiscale rotation-invariant convolutional neural networks for lung texture classification. IEEE J. Biomed. Health Inform. 2018, 22, 184–195. [Google Scholar] [CrossRef]
- Canny, J. A computational approach to edge detection. IEEE Trans. Pattern Anal. Mach. Intell. 1986, PAMI-8, 679–698. [Google Scholar] [CrossRef]
- Marr, D.; Hildreth, E. Theory of edge detection. Proc. R. Soc. London. Ser. B. Biol. Sci. 1980, 207, 187–217. [Google Scholar]
- Torre, V.; Poggio, T.A. On edge detection. IEEE Trans. Pattern Anal. Mach. Intell. 1986, PAMI-8, 147–163. [Google Scholar] [CrossRef]
- Gunn, S.R. On the discrete representation of the Laplacian of Gaussian. Pattern Recognit. 1999, 32, 1463–1472. [Google Scholar] [CrossRef]
- Mlsna, P.A.; Rodriguez, J.J. Gradient and Laplacian edge detection. In The Essential Guide to Image Processing; Academic Press: Cambridge, MA, USA, 2009; pp. 495–524. [Google Scholar]
- Parker, J.R. Algorithms for Image Processing and Computer Vision; John Wiley & Sons, Inc.: New York, NY, USA, 1997; pp. 23–29. [Google Scholar]
- Sadanandan, S.K.; Ranefall, P.; Le Guyader, S.; Wählby, C. Automated training of deep convolutional neural networks for cell segmentation. Sci. Rep. 2017, 7, 7860. [Google Scholar] [CrossRef]
- Stringer, C.; Wang, T.; Michaelos, M.; Pachitariu, M. Cellpose: A generalist algorithm for cellular segmentation. Nat. Methods 2020, 18, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Hertz, K. A Simple Weight Decay Can Improve Generalization. In Advances in Neural Information Processing Systems 4; Morgan Kaufmann: San Francisco, CA, USA, 1992. [Google Scholar]
- MacKay, D.J.C. Bayesian interpolation. Neural Comput. 1992, 4, 415–447. [Google Scholar] [CrossRef]
- Oei, R.W.; Hou, G.; Liu, F.; Zhong, J.; Zhang, J.; An, Z.; Xu, L.; Yang, Y.-H. Convolutional neural network for cell classification using microscope images of intracellular actin networks. PLoS ONE 2019, 14, e0213626. [Google Scholar] [CrossRef]
- Shin, H.C.; Roth, H.R.; Gao, M.; Lu, L.; Xu, Z.; Nogues, I.; Yao, J.; Mollura, D.; Summers, R.M. Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning. IEEE Trans. Med. Imaging 2016, 35, 1285. [Google Scholar] [CrossRef]














| Static Power Consumption | 0.53 W (1%) | |
|---|---|---|
| Dynamic Power Consumption | 18.47 W (99%) | Logic (14.5 W) |
| Signals (3.79 W) | ||
| I/O (0.067 W) | ||
| NN Output (MATLAB) | NN Output FPGA | NN Output (MATLAB) | NN Output FPGA |
|---|---|---|---|
| 0.1425 (Healthy) | 0 (Healthy) | 0.942 (Moderately stressed | 1 (Moderately stressed) |
| 0.0058 (Healthy) | 0 (Healthy) | 0.9213 (Moderately stressed) | 1 (Moderately stressed) |
| 0.0418 (Healthy) | 0 (Healthy) | 0.942 (Moderately stressed | 1 (Moderately stressed) |
| 0.0056 (Healthy) | 0 (Healthy) | 0.942 (Moderately stressed | 1 (Moderately stressed) |
| 0.0086 (Healthy) | 0 (Healthy) | 0.9213 (Moderately stressed) | 1 (Moderately stressed) |
| 0.0425 (Healthy) | 0 (Healthy) | 0.942 (Moderately stressed | 1 (Moderately stressed) |
| 0.0058 (Healthy) | 0 (Healthy) | 0.9213 (Moderately stressed) | 1 (Moderately stressed) |
| 0.0418 (Healthy) | 0 (Healthy) | 0.942 (Moderately stressed | 1 (Moderately stressed) |
| 0.0056 (Healthy) | 0 (Healthy) | 0.942 (Moderately stressed | 1 (Moderately stressed) |
| 0.0086 (Healthy) | 0 (Healthy) | 0.9213 (Moderately stressed) | 1 (Moderately stressed) |
| NN Output (MATLAB) | NN Output FPGA | NN Output (MATLAB) | NN Output FPGA |
|---|---|---|---|
| 0.0016 (moderately stressed) | 0 (moderately stressed) | 1.0044 (highly stressed) | 1 (highly stressed) |
| 0.0015 (moderately stressed) | 0 (moderately stressed) | 1.0059(highly stressed) | 1 (highly stressed) |
| 0.0017 (moderately stressed) | 0 (moderately stressed) | 1.0047(highly stressed) | 1 (highly stressed) |
| 0.0014 (moderately stressed) | 0 (moderately stressed) | 0.9302(highly stressed) | 1 (highly stressed) |
| 0.0015 (moderately stressed) | 0 (moderately stressed) | 1.0044 (highly stressed) | 1 (highly stressed) |
| 0.0314 (moderately stressed | 0 (moderately stressed) | 0.0035 (moderately stressed) | 0 (moderately stressed) |
| 0.0215 (moderately stressed) | 0 (moderately stressed) | 1.0047(highly stressed) | 1 (highly stressed) |
| 0.0051 (moderately stressed) | 0 (moderately stressed) | 0.9302(highly stressed) | 1 (highly stressed) |
| 0.0745 (moderately stressed) | 0 (moderately stressed) | 1.0044 (highly stressed) | 1 (highly stressed) |
| 0.0562 (moderately stressed) | 0 (moderately stressed) | 0.0016 (moderately stressed) | 0 (moderately stressed) |
| FPGA Resources | Available | Utilised |
|---|---|---|
| Slice (LUTs) | 63,400 | 2408 (<4%) |
| Slice registers (FFs) | 126,800 | 35 (<1%) |
| Bonded IO blocks | 210 | 3 (<2%) |
| BUFGCTRL | 31 | 1 (<4%) |
| FPGA Logic Blocks | Utilisation |
| Adder | 10 |
| Multiplier | 490 |
| Sigmoid | 1890 |
| UART | 119 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghani, A.; Hodeify, R.; See, C.H.; Keates, S.; Lee, D.-J.; Bouridane, A. Computer Vision-Based Kidney’s (HK-2) Damaged Cells Classification with Reconfigurable Hardware Accelerator (FPGA). Electronics 2022, 11, 4234. https://doi.org/10.3390/electronics11244234
Ghani A, Hodeify R, See CH, Keates S, Lee D-J, Bouridane A. Computer Vision-Based Kidney’s (HK-2) Damaged Cells Classification with Reconfigurable Hardware Accelerator (FPGA). Electronics. 2022; 11(24):4234. https://doi.org/10.3390/electronics11244234
Chicago/Turabian StyleGhani, Arfan, Rawad Hodeify, Chan H. See, Simeon Keates, Dah-Jye Lee, and Ahmed Bouridane. 2022. "Computer Vision-Based Kidney’s (HK-2) Damaged Cells Classification with Reconfigurable Hardware Accelerator (FPGA)" Electronics 11, no. 24: 4234. https://doi.org/10.3390/electronics11244234
APA StyleGhani, A., Hodeify, R., See, C. H., Keates, S., Lee, D.-J., & Bouridane, A. (2022). Computer Vision-Based Kidney’s (HK-2) Damaged Cells Classification with Reconfigurable Hardware Accelerator (FPGA). Electronics, 11(24), 4234. https://doi.org/10.3390/electronics11244234








