Deep Learning Approach for Automatic Segmentation and Functional Assessment of LV in Cardiac MRI
Abstract
1. Introduction
- Variation in shape, intensity, and structure across the cardiac cycle, patients, and pathological conditions;
- Low contrast between myocardium and surrounding structure;
- High contrast between blood and the myocardium;
- Inherent noise due to motion artifacts; and
- Brightness heterogeneity due to blood flow.
- It provides context based localized segmentation;
- It does not require large samples for training and can produce acceptable levels of segmentation accuracy with just a few training samples;
- It provides more precise segmentation;
- It is faster to train than most other segmentation model due to its context-enabled segmentation approach.
2. Materials and Methods
2.1. Data
- Normal (N)A healthy group with ejection fraction (EF) > 55% and no hypertrophy.
- Hypertrophy (HYP)The left ventricle (LV) hypertrophy (HYP) group had normal EF (>55%), and the ratio of left ventricular (LV) mass over body surface area is >83 g/m2.
- Heart failure without infarction (HF)The group had EF <40% and no late Gadolinium (Gd) enhancement.
- Heart failure with infarction (HF-I)The group had ejection fraction (EF) <40% and evidence of late gadolinium (Gd) enhancement.
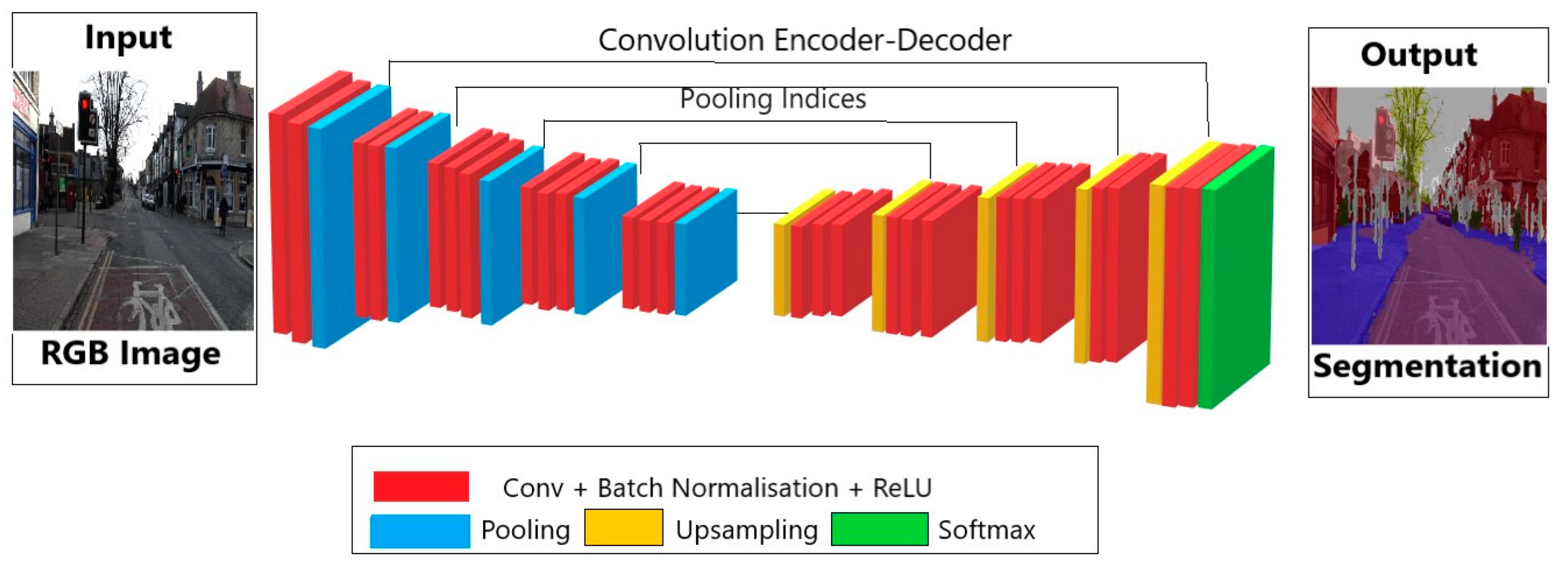
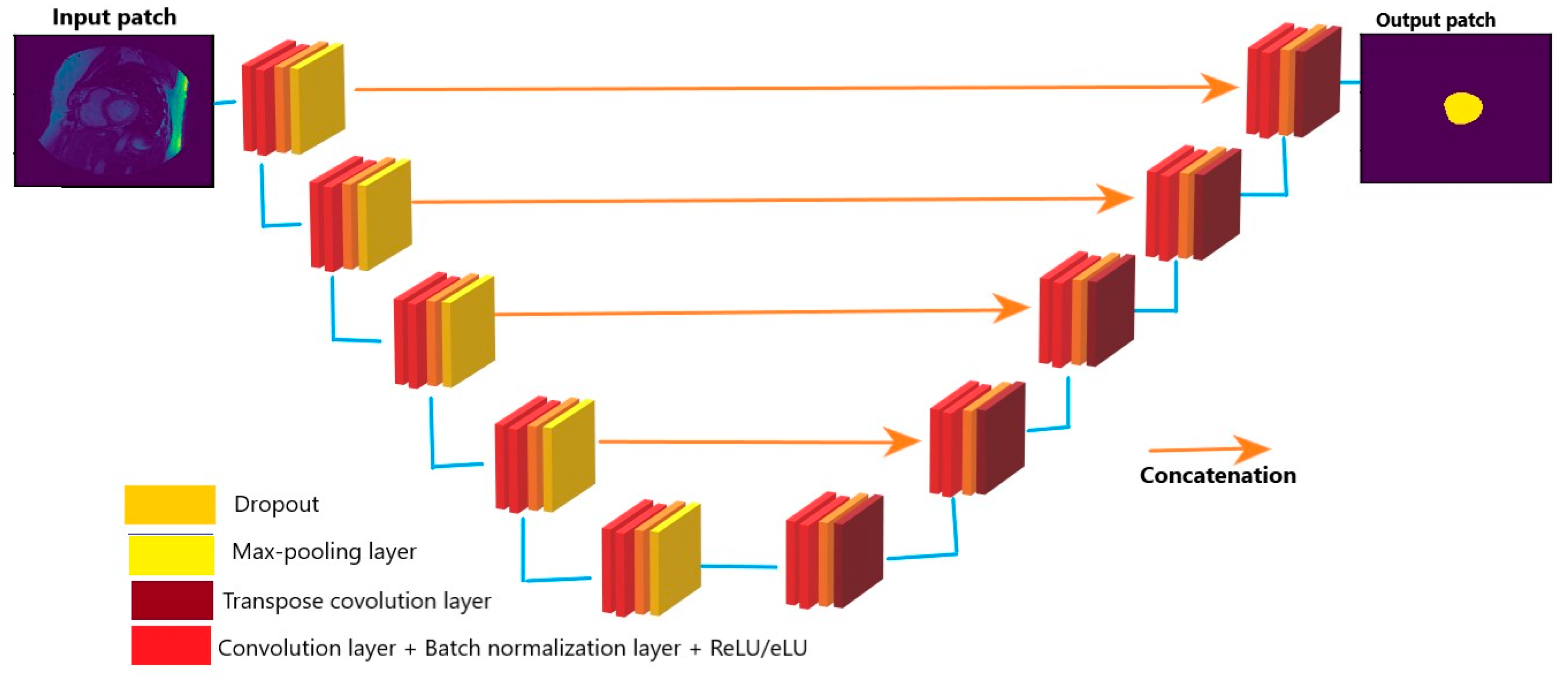
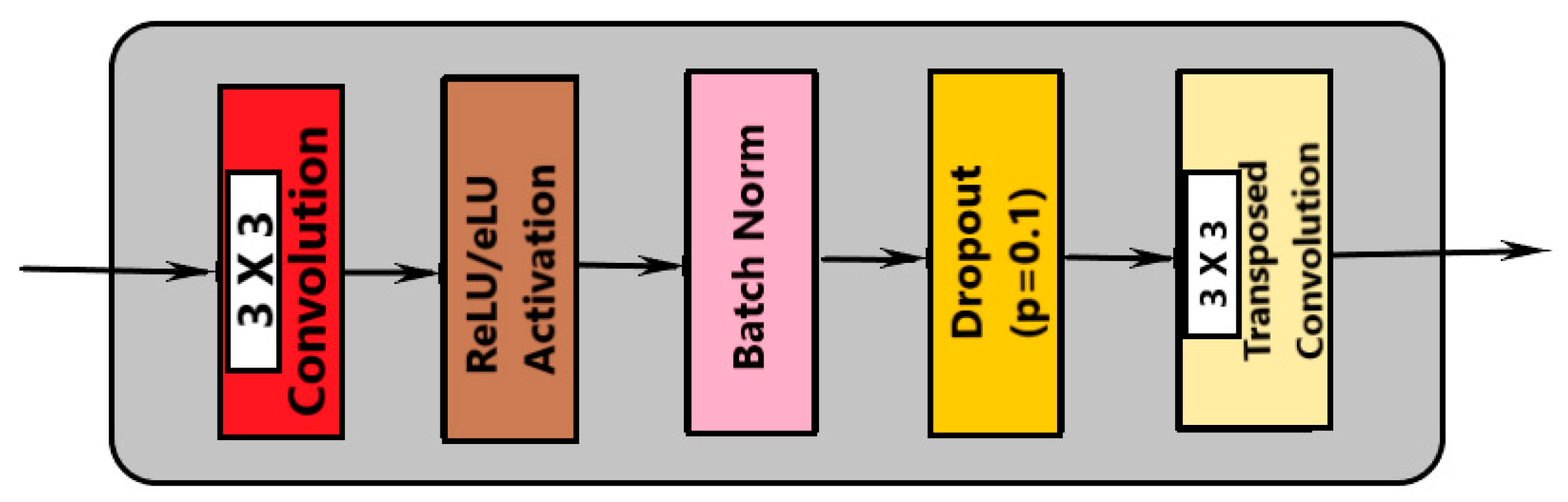
2.1.1. Automatic Segmentation of Cardiac MRI
- Batch size;
- Batch normalization;
- Activation function;
- Loss function; and
- Dropout.
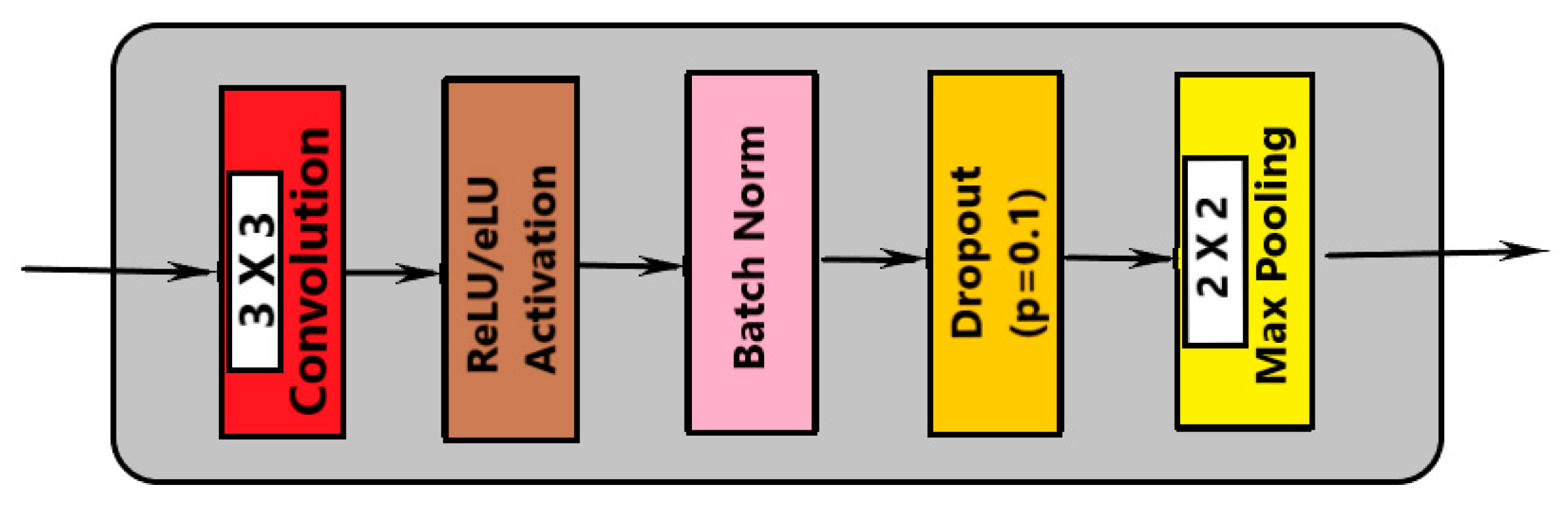
- Convolution with 3 × 3 kernel size;
- Batch normalization;
- Convolution with 3 × 3 kernel size;
- Batch normalization;
- Dropout;
- Max pooling (in contraction and transpose convolution in expansion).
2.1.2. Number of Convolution Layers
2.1.3. Number of Filters
2.1.4. Batch Normalization
2.1.5. Dropout Layers
2.1.6. Loss Functions
2.1.7. Optimization
2.2. Performance Evaluation
2.2.1. Segmented Accuracy Assessment Metrics
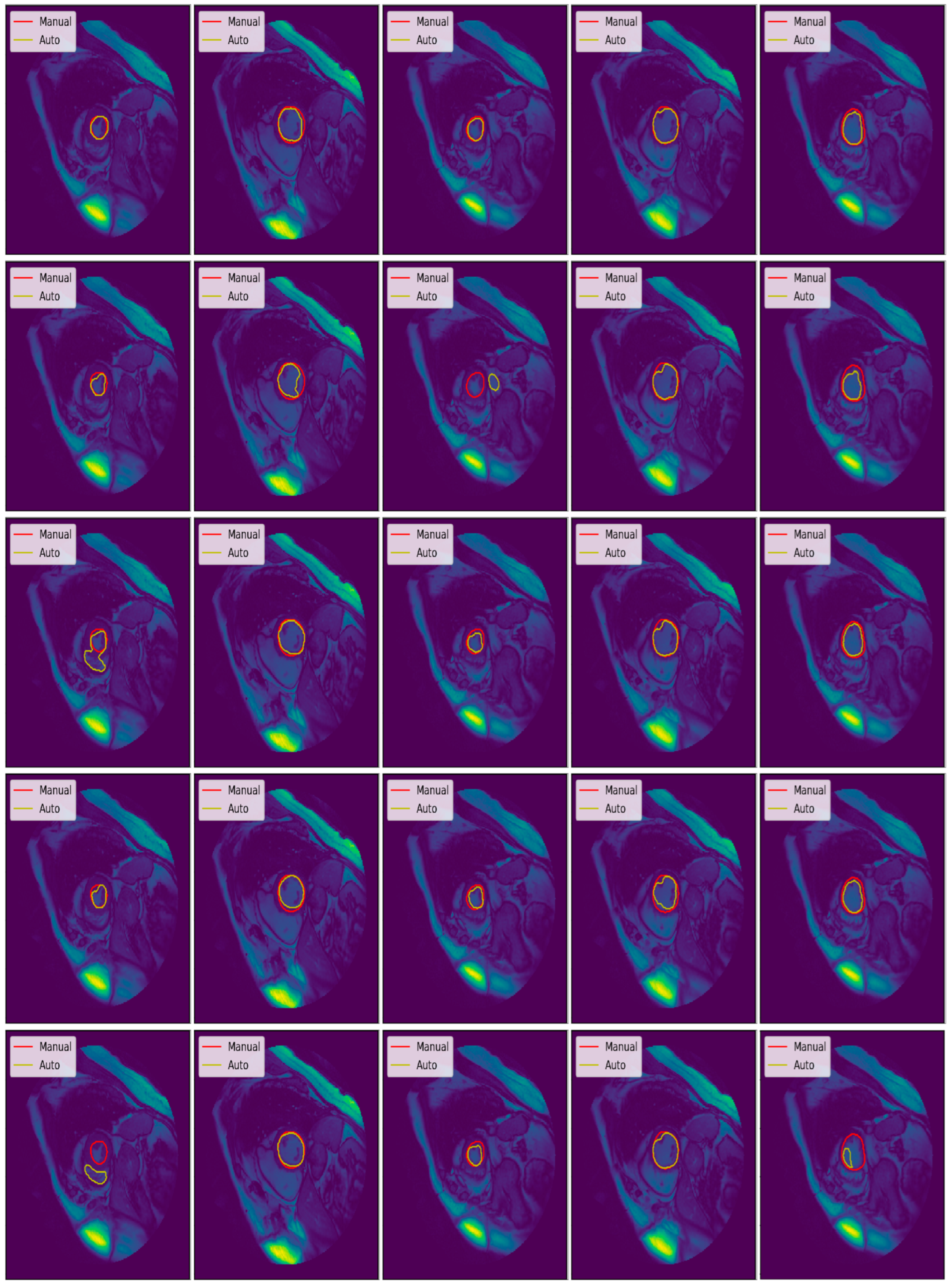
- Average perpendicular distance measures the distance between manual and auto contours, averaged over all contour points [31]. It was used to measure the closeness of the segmented boundaries; the smaller the value, the closer is the boundary. The value of APD measures in millimeters with the help of pixel spacing provided in the DICOM field named PixelSpacing.
- The dice metric was used to measure the overlapping or similarity between manual and auto contours. Its value lies between 0 to 1, where 0 means no overlap and 1 means perfect overlapping. The dice metric between auto and manual contours can be defined as:
- The percentage of good contours is evaluated in terms of APD. It is basically the fraction of contours out of total contours. The fraction of contours is selected if APD < 5 mm.
2.2.2. Clinical Metrics
3. Results and Discussion
3.1. Data Pre-processing
- Orientation: Orientation shift based on the DICOM InPlanePhaseEncoding metadata, which indicates the axis of phase encoding with respect to the image. A majority of the images were “Row” oriented; thus, if the image was “Col” oriented, it was flipped to be “Row” oriented.
- Rescale: The image is rescaled based on the image’s Pixel Spacing values.
- Rescale with the first Pixel Spacing value in both the x and y directions;
- Rescale to 1 mm × 1 mm.
- Crop: The image is cropped from the center to 256 × 256. In this project, 256 × 256 and 176 × 176 were used.
- ROI Location.
- The heart cavity containing the LV is near the center of the MRI Image;
- The frequency at which the LV moves is unique compared to the frequencies of other heart muscles;
- There is a degree of pixel variance around the LV muscle;
- The LV is circular in shape.
3.2. Data Preparation and Training
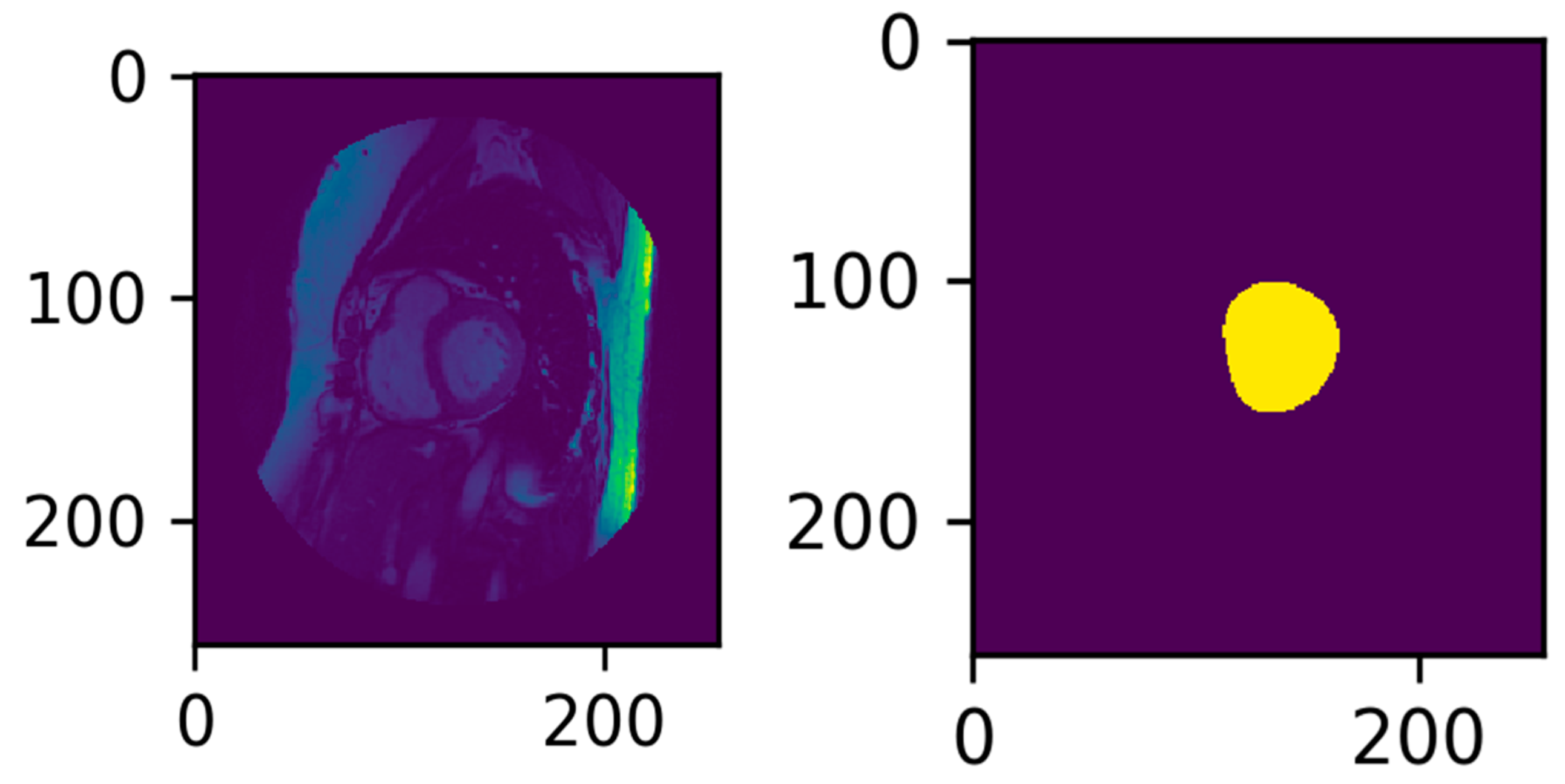
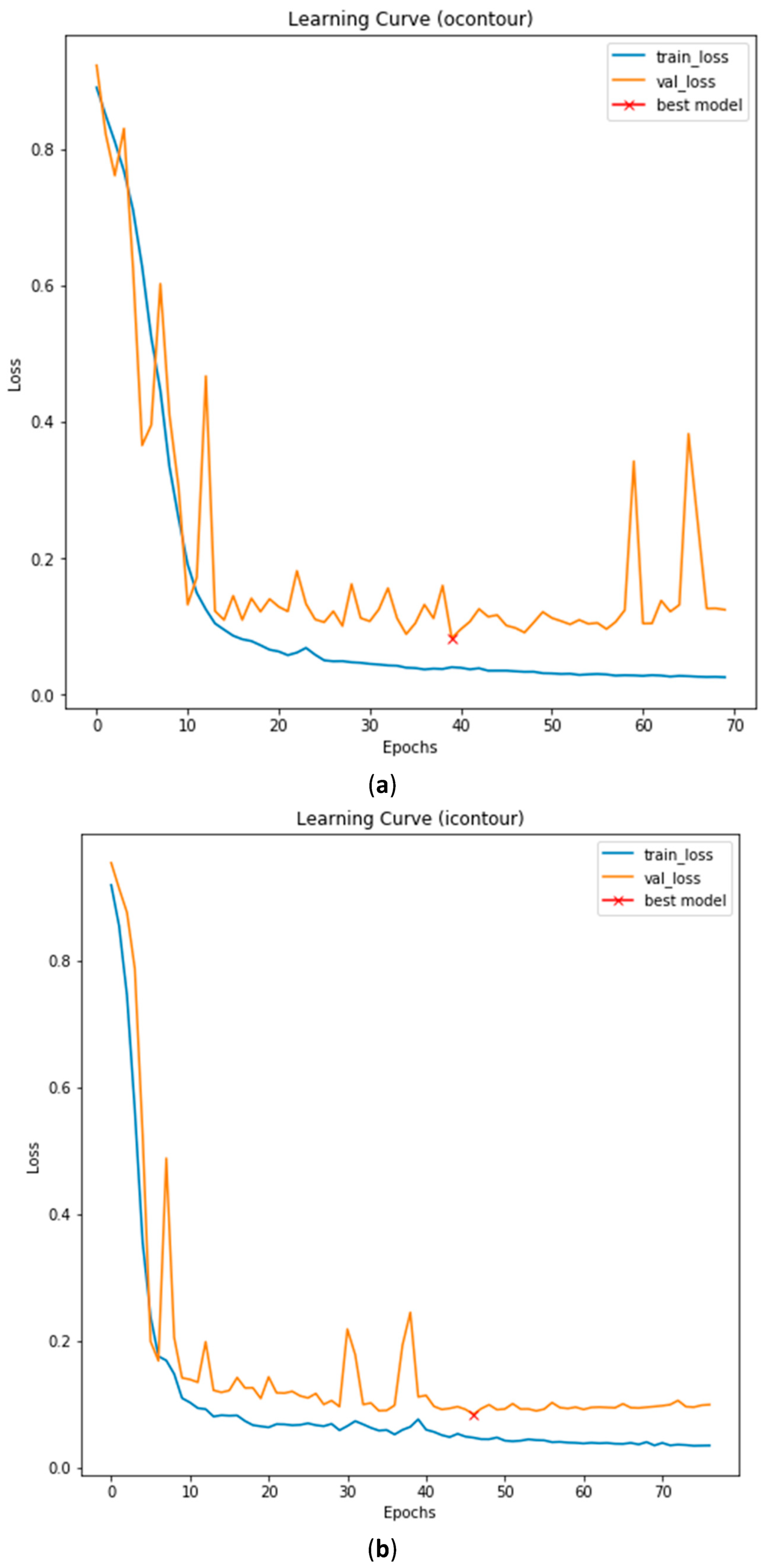
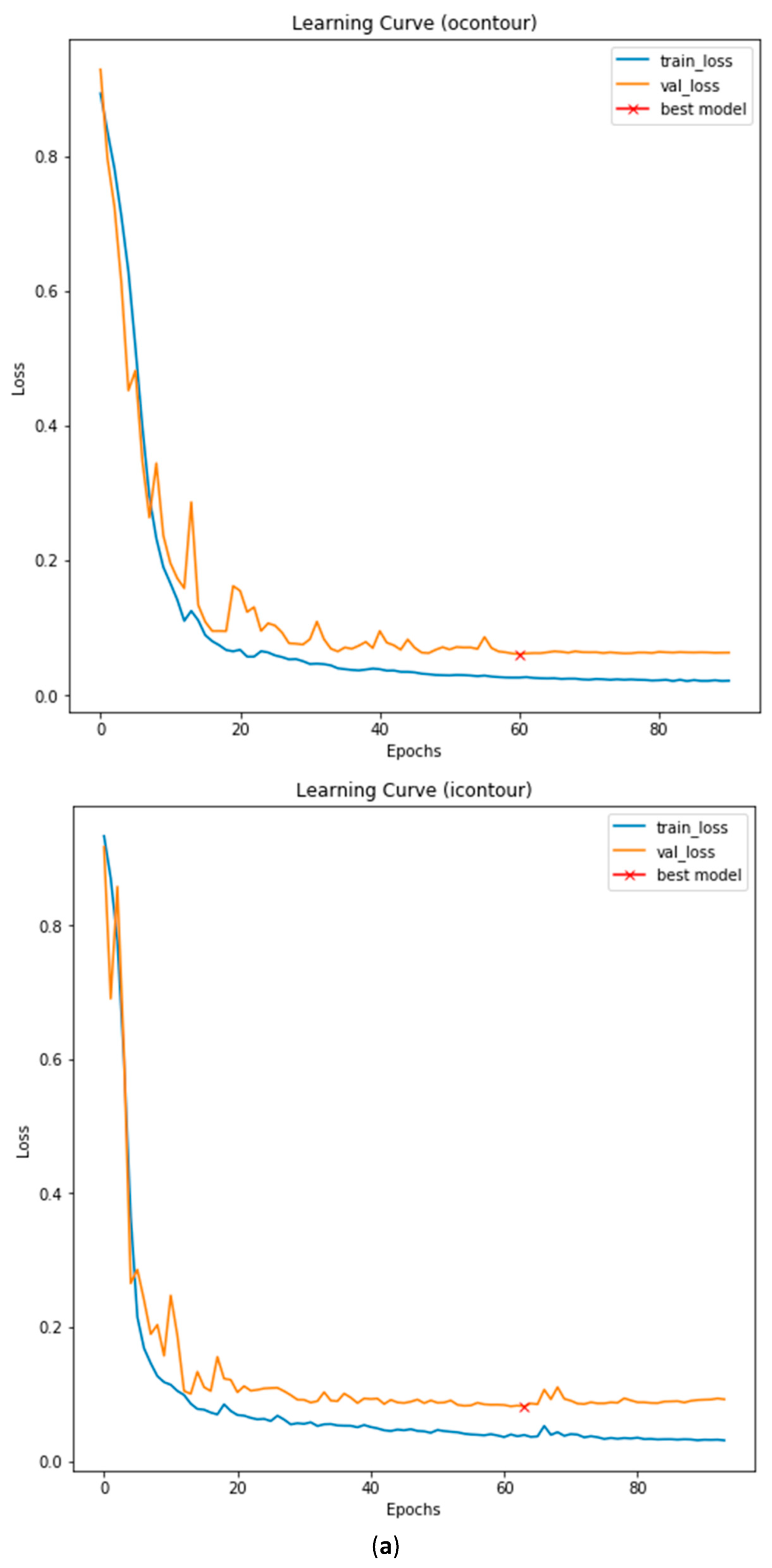
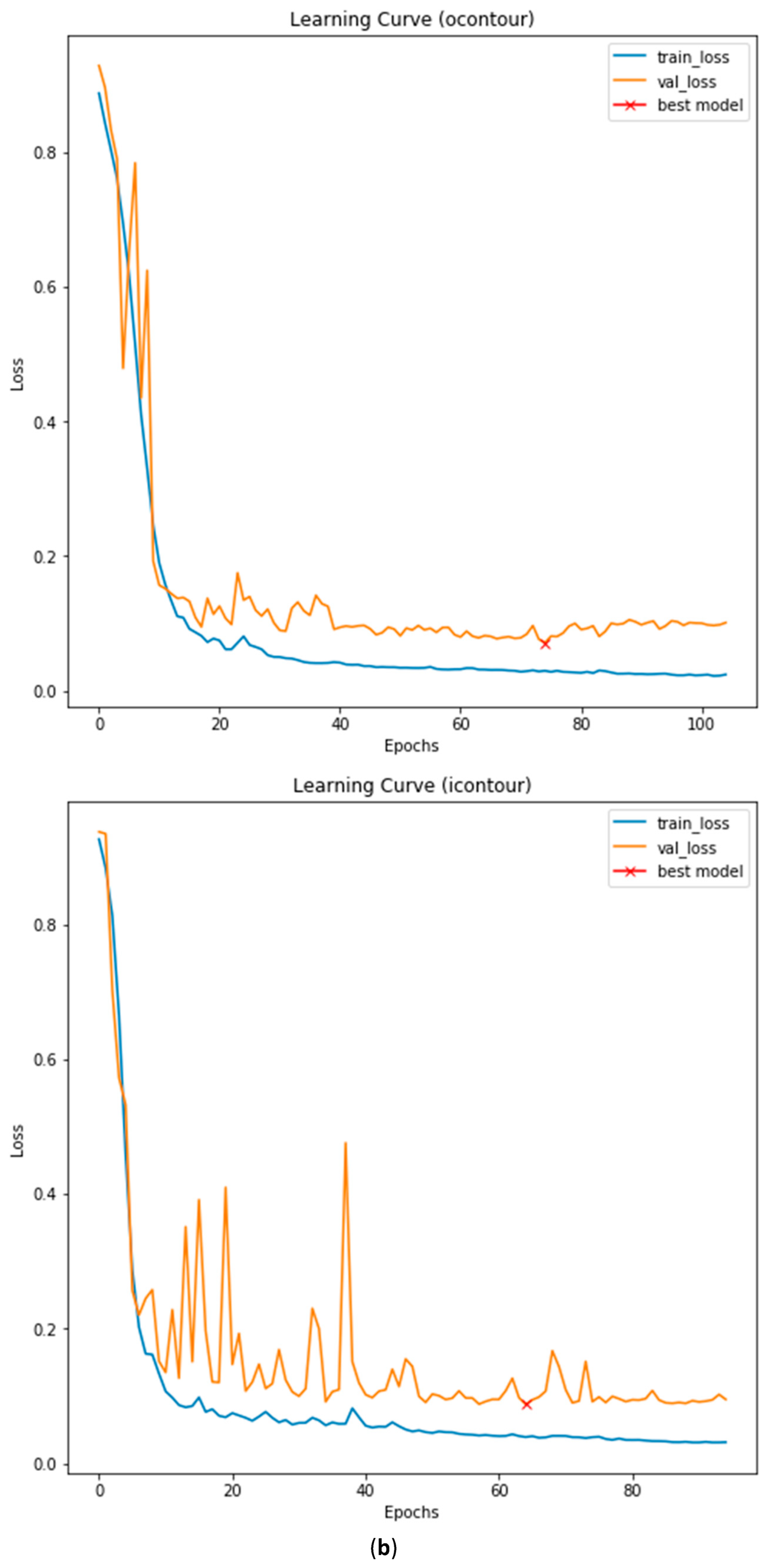
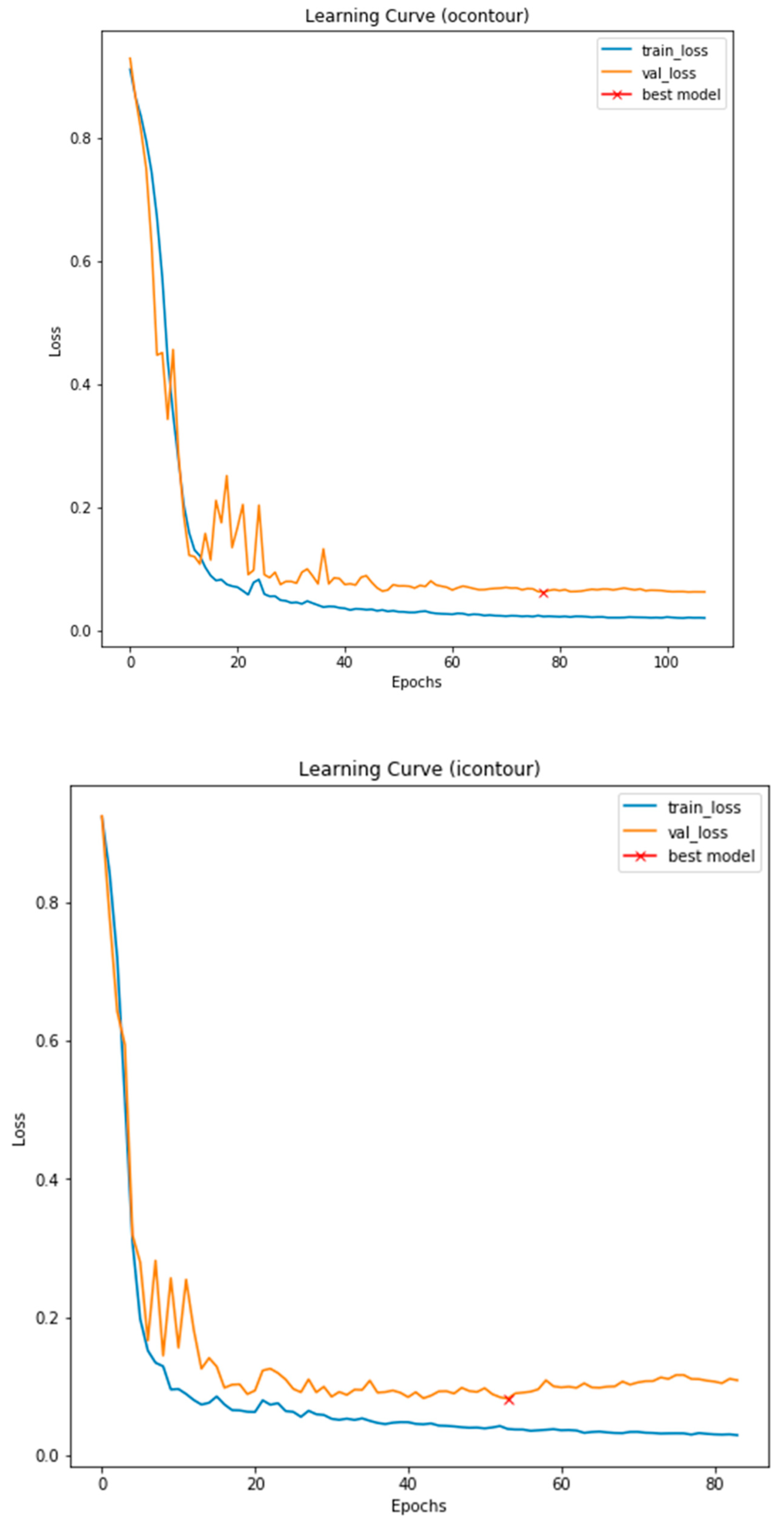
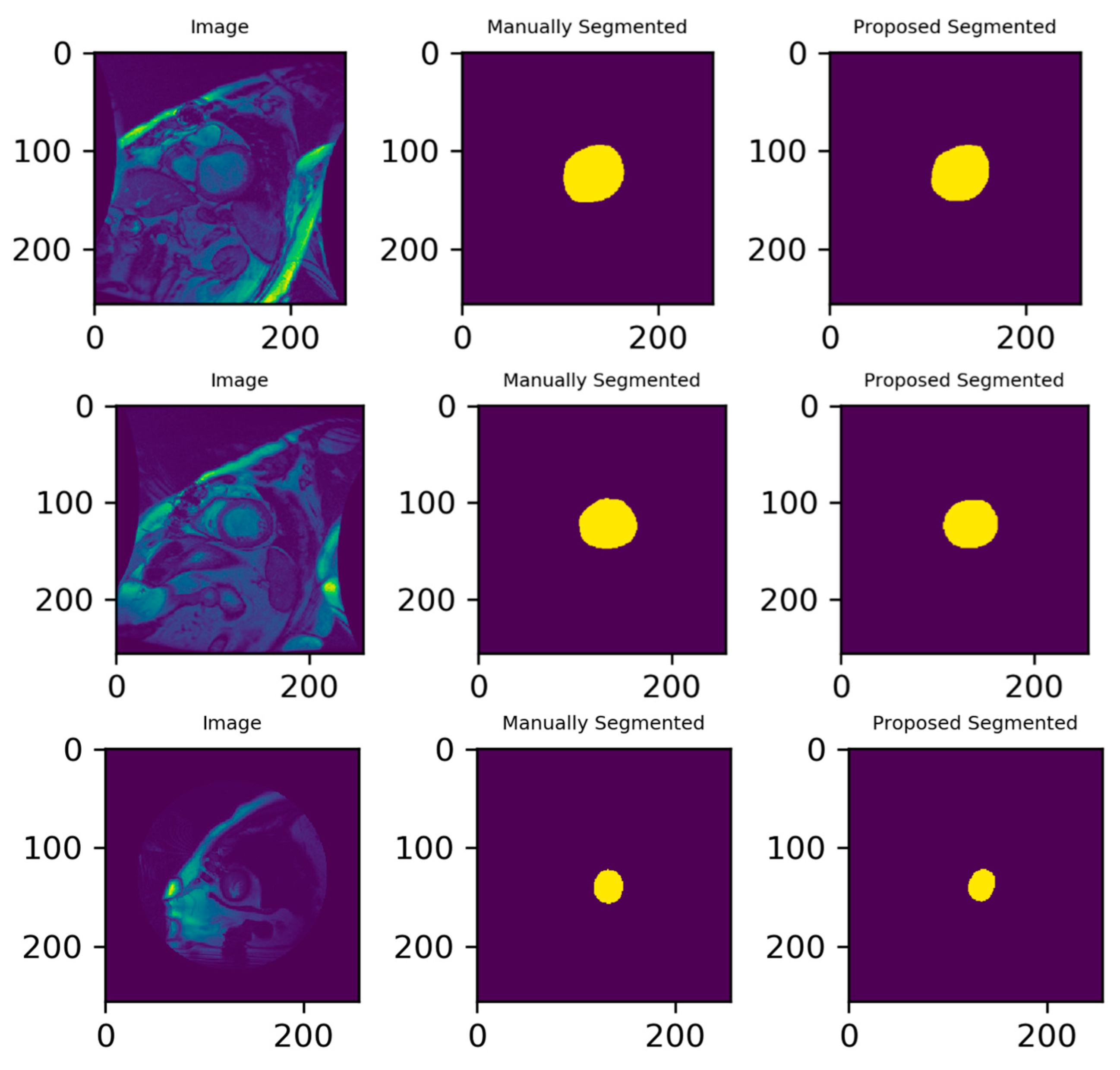
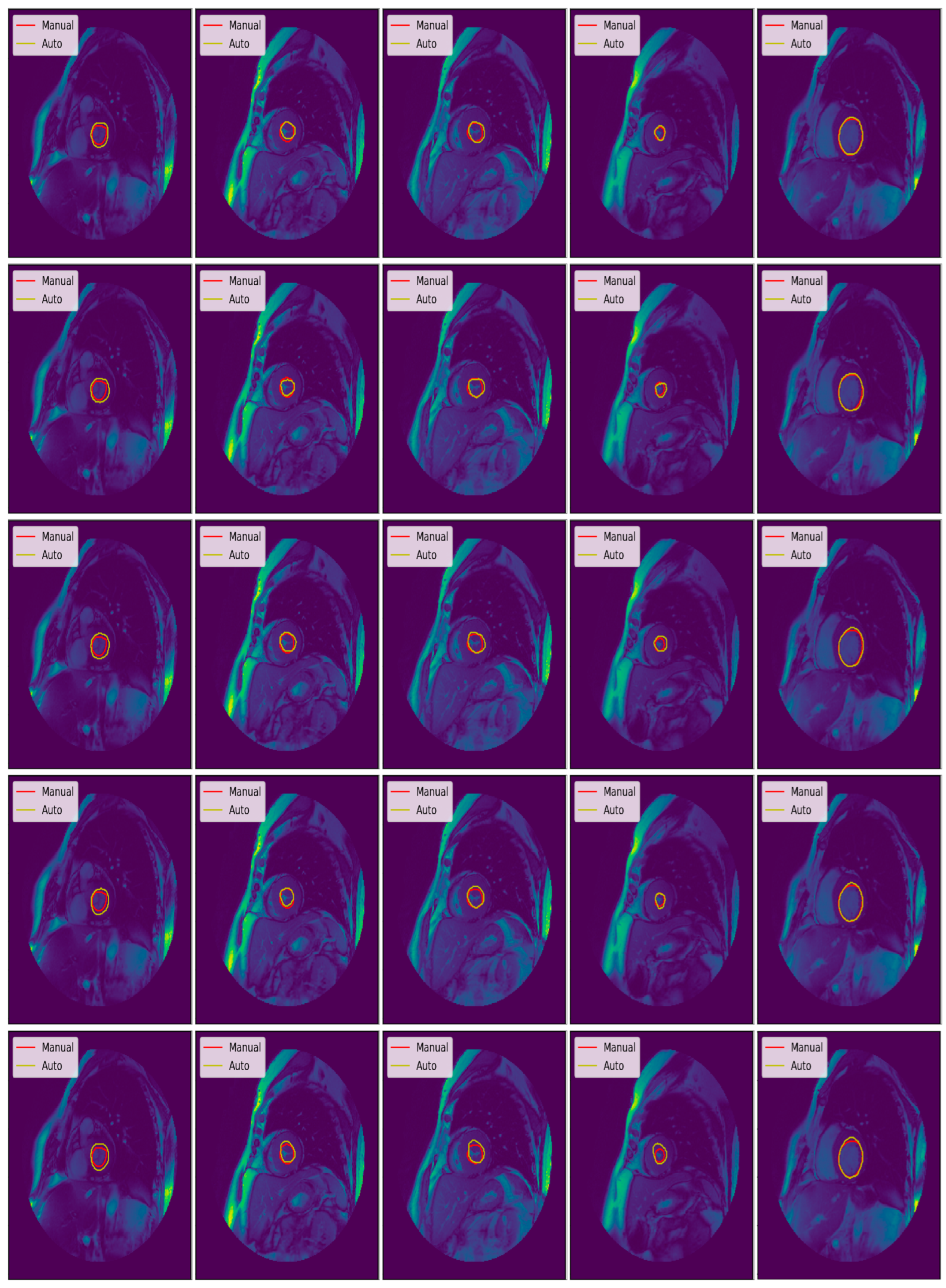
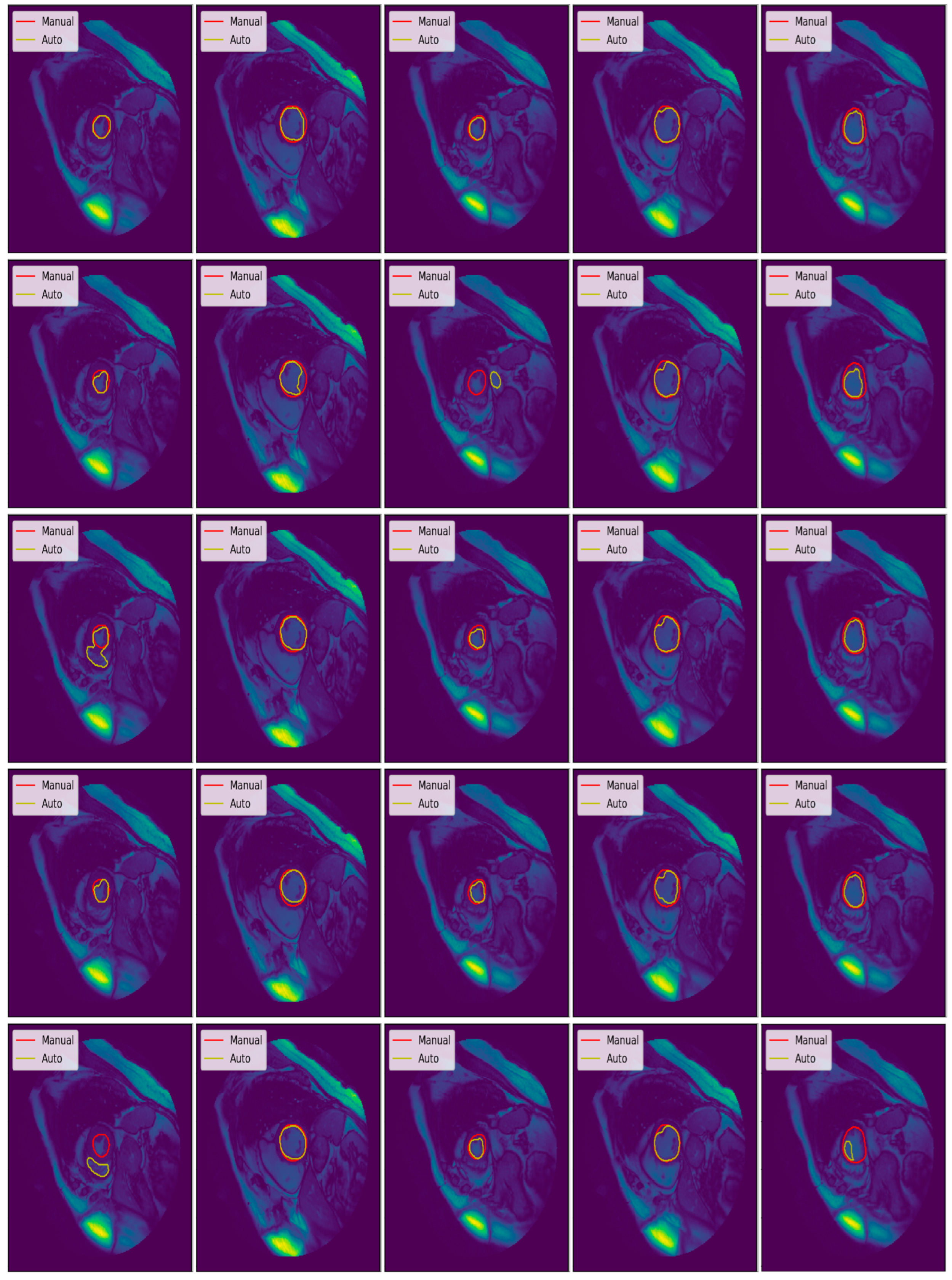
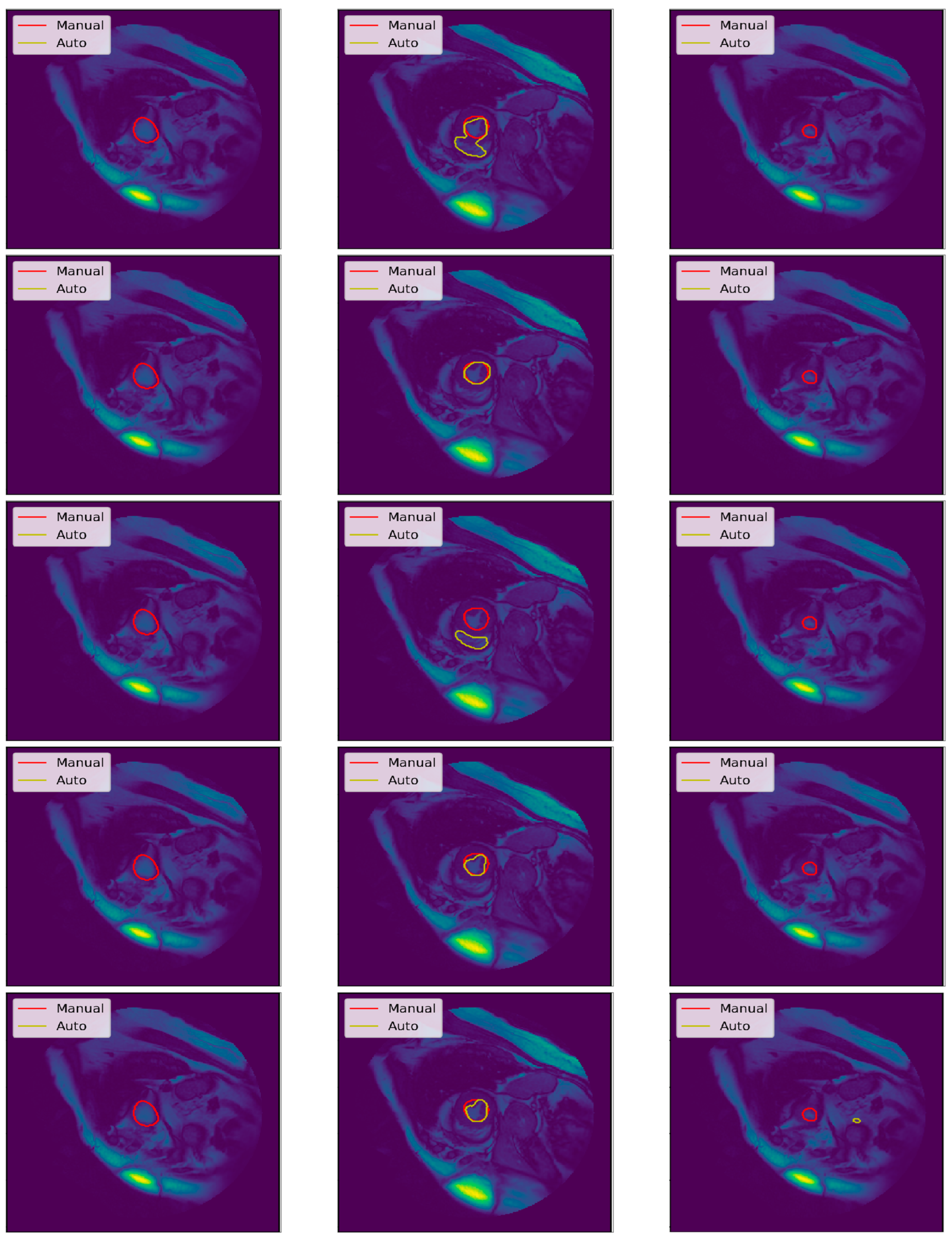
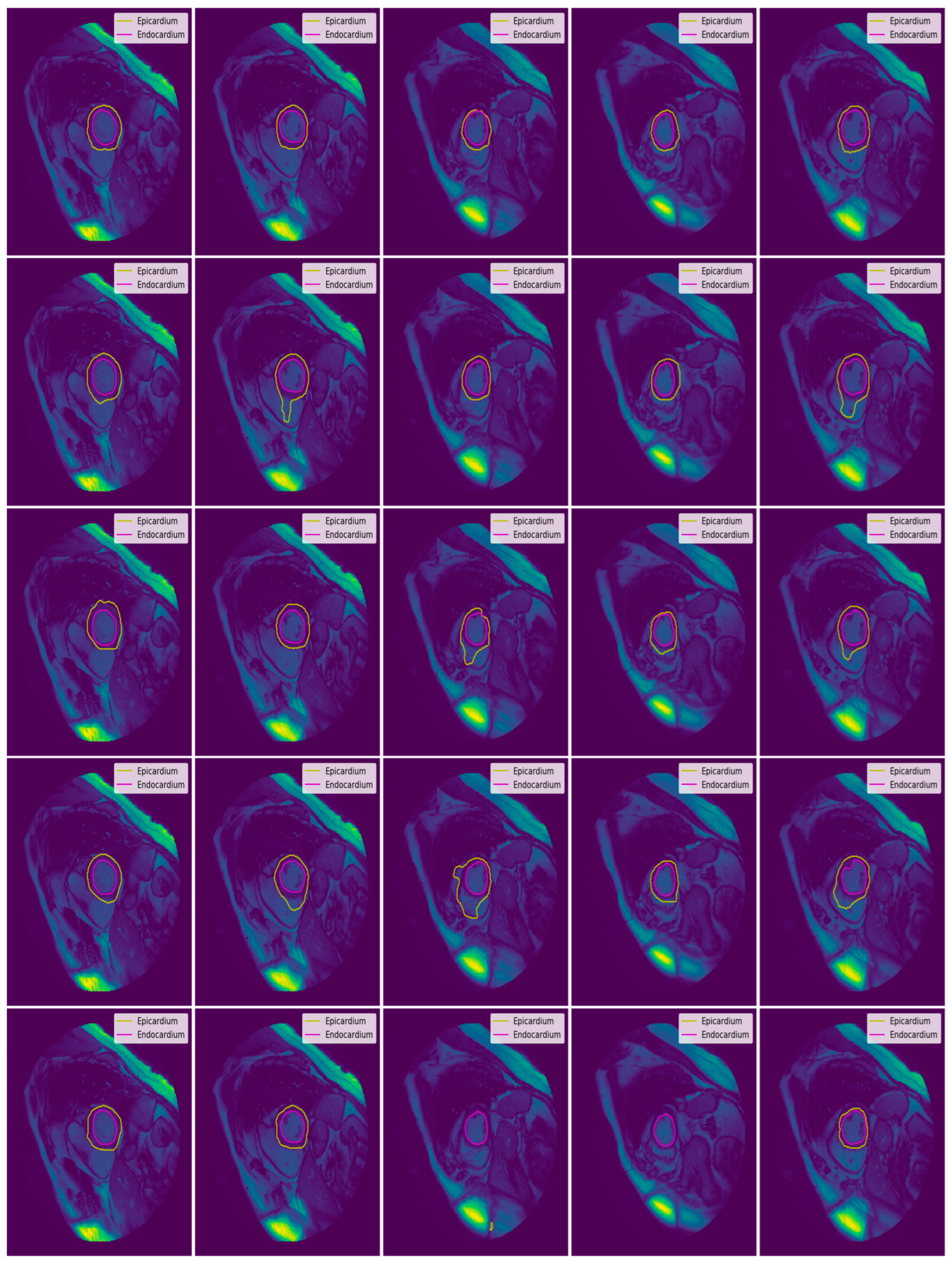
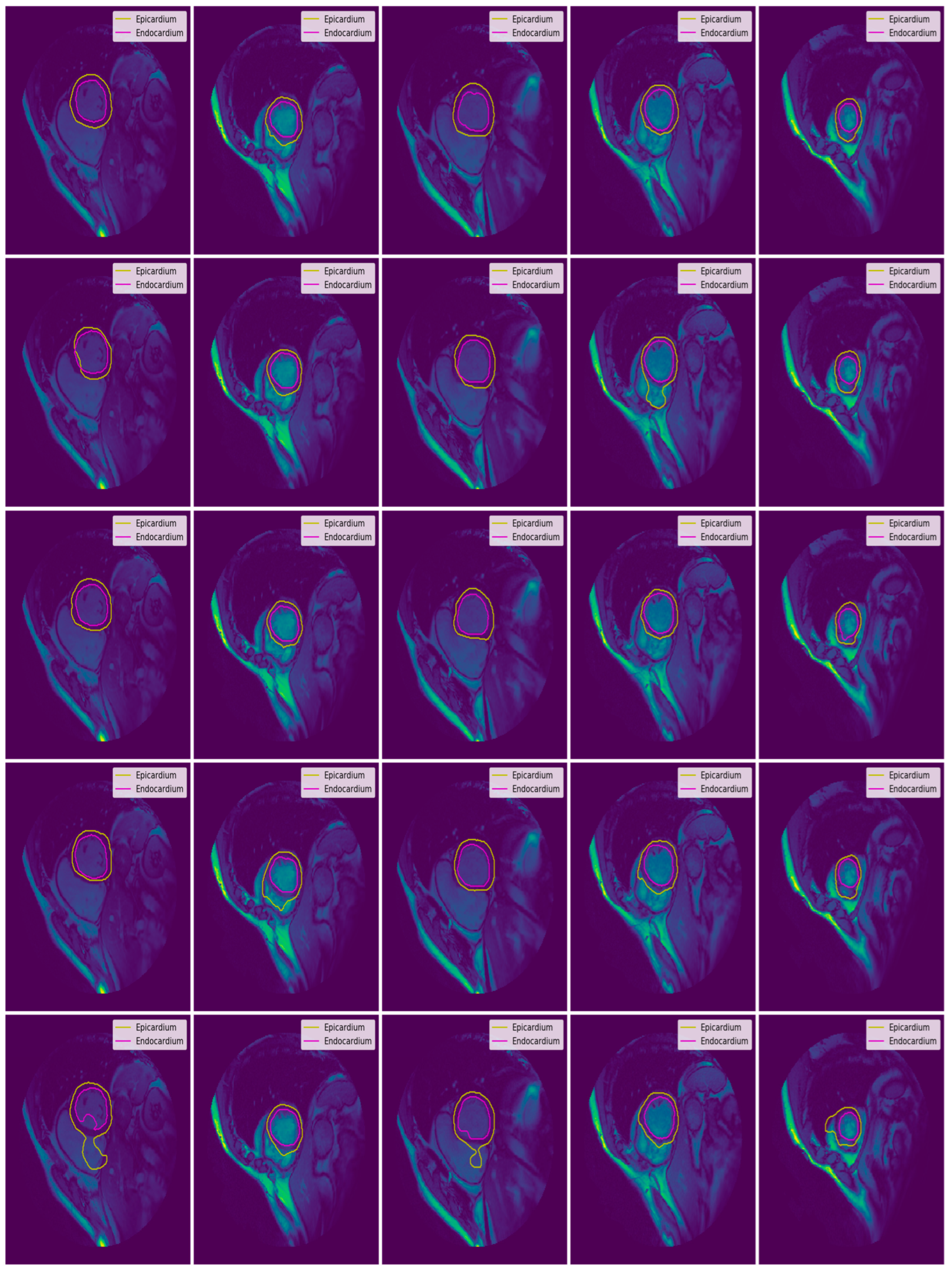
3.3. Illustrated Results
- U-Net with focal Tversky loss;
- U-Net with log-cosh dice loss;
- U-Net with Tversky loss;
- U-Net with dice loss; and
- U-Net with binary cross-entropy loss.
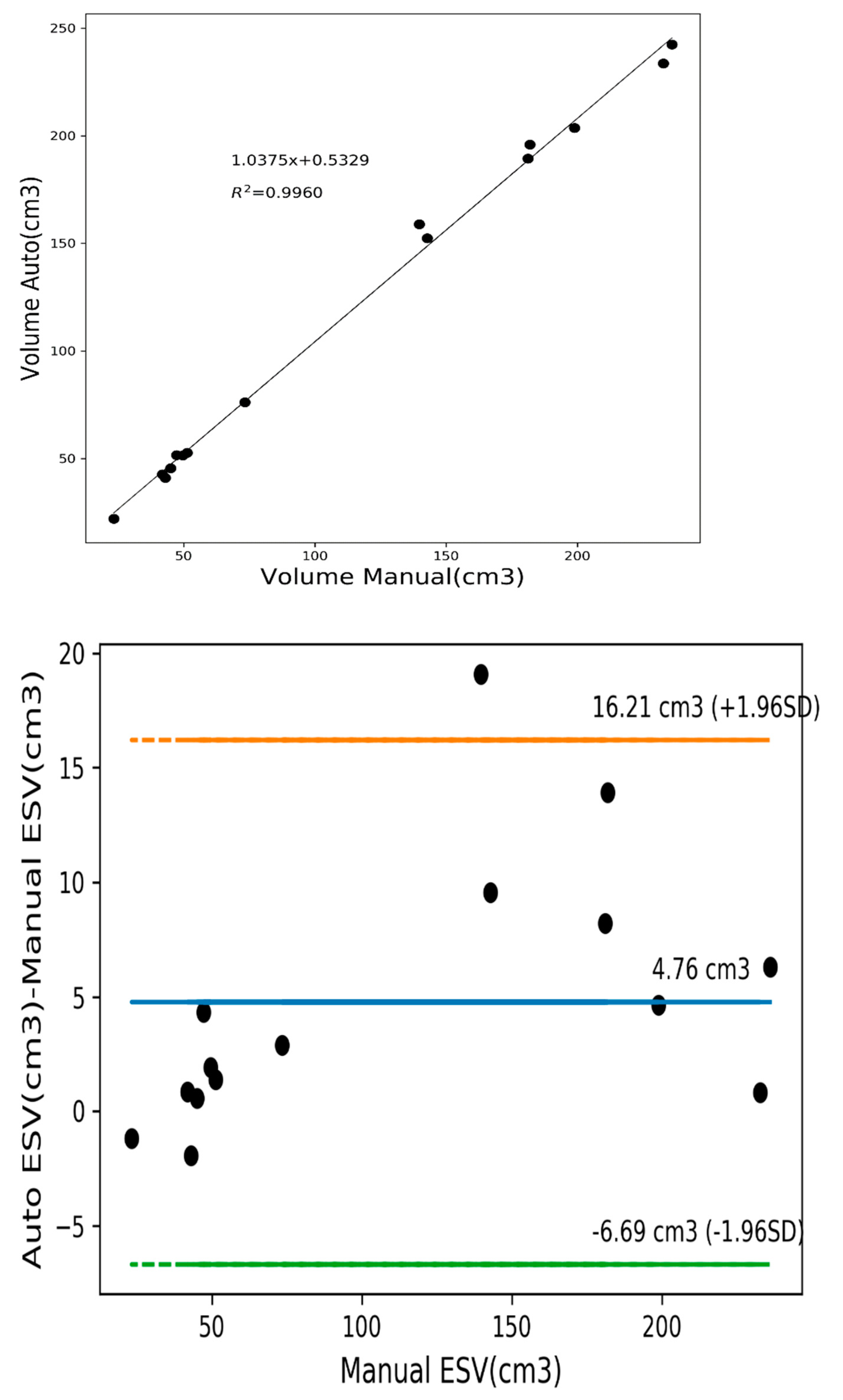
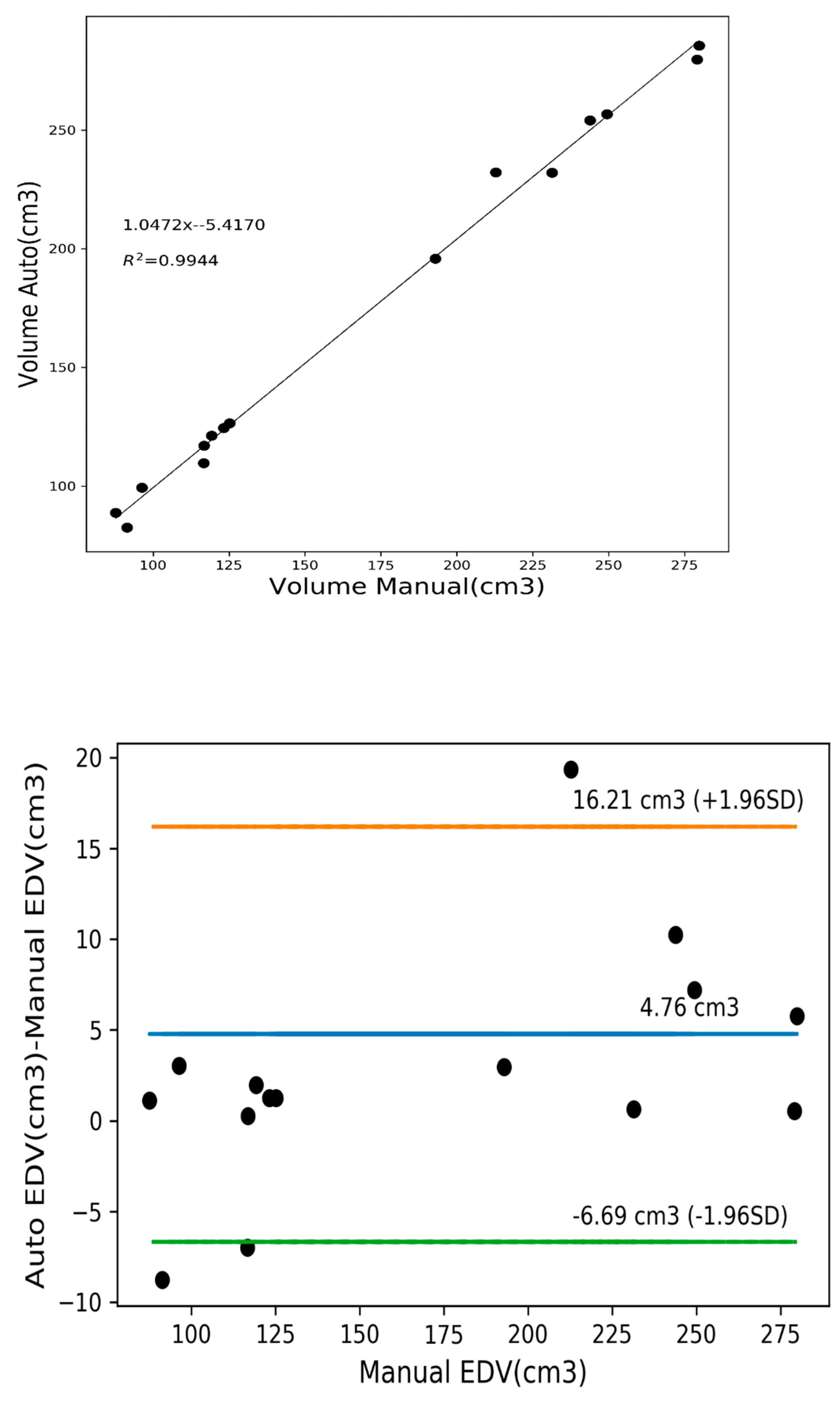
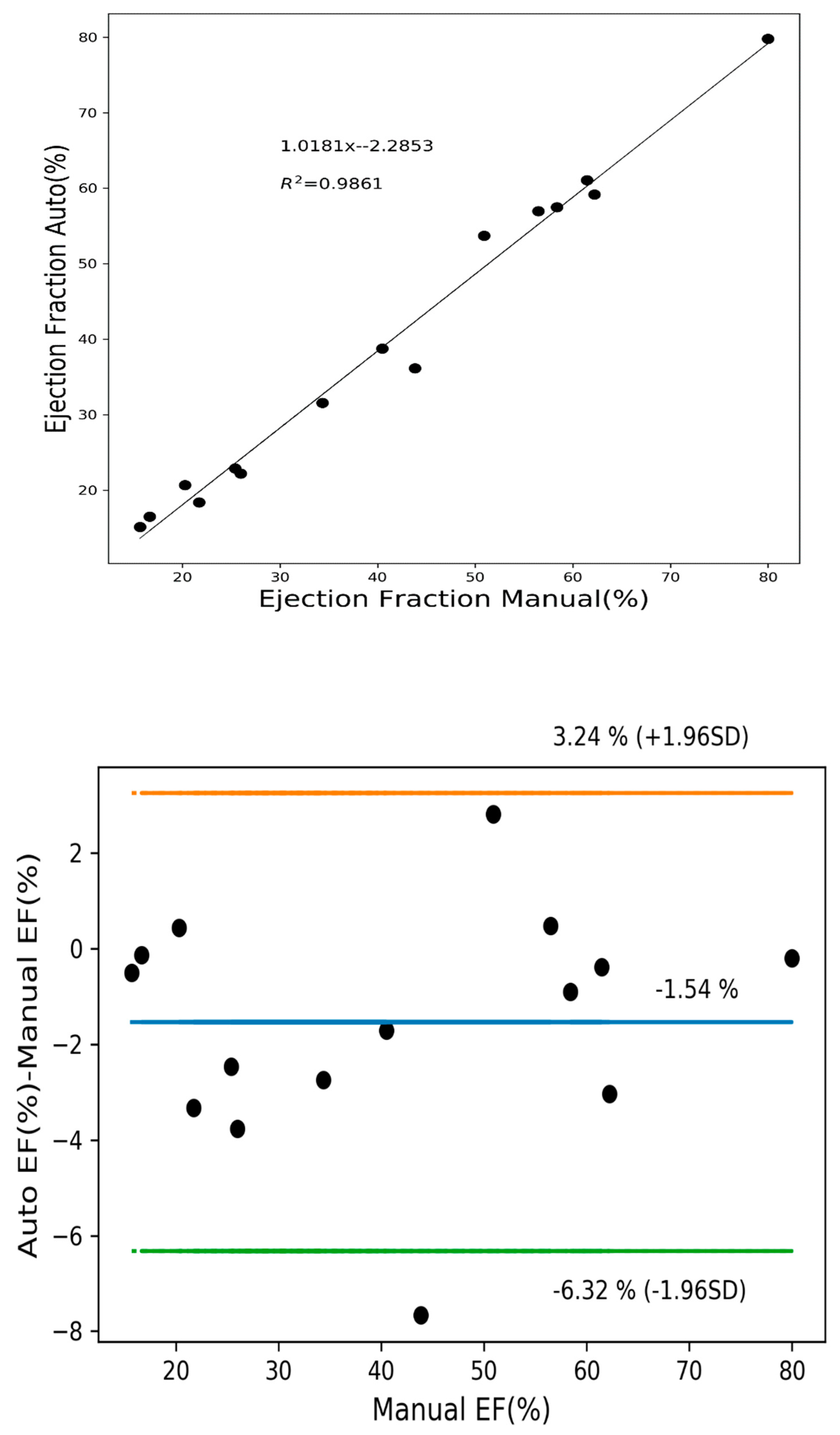
3.4. Quantitative Analysis
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- White, H.D.; Norris, R.M.; Brown, M.A.; Brandt, P.W.; Whitlock, R.M.; Wild, C.J. Left ventricular end-systolic volume as the major determinant of survival after recovery from myocardial infarction. Circulation 1987, 76, 44–51. [Google Scholar] [CrossRef]
- Tan, L.K.; Liew, Y.M.; Lim, E.; McLaughlin, R.A. Convolutional neural network regression for short-axis left ventricle segmentation in cardiac cine MR sequences. Med. Image Anal. 2017, 39, 78–86. [Google Scholar] [CrossRef]
- Sander, J.; de Vos, B.D.; Išgum, I. Automatic segmentation with detection of local segmentation failures in cardiac MRI. Sci. Rep. 2020, 10, 21769. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Leiner, T.; Rueckert, D.; Suinesiaputra, A.; Baeßler, B.; Nezafat, R.; Išgum, I.; Young, A.A. Machine learning in cardiovascular magnetic resonance: Basic concepts and applications. J. Cardiovasc. Magn. Reson. 2019, 21, 61. [Google Scholar] [CrossRef] [PubMed]
- Bernard, O.; Lalande, A.; Zotti, C.; Cervenansky, F.; Yang, X.; Heng, P.A.; Cetin, I.; Lekadir, K.; Camara, O.; Ballester, M.A.; et al. Deep learning techniques for automatic MRI cardiac multi-structures segmentation and diagnosis: Is the problem solved? IEEE Trans. Med. Imaging 2018, 37, 2514–2525. [Google Scholar] [CrossRef] [PubMed]
- Suinesiaputra, A.; Bluemke, D.A.; Cowan, B.R.; Friedrich, M.G.; Kramer, C.M.; Kwong, R.; Plein, S.; Schulz-Menger, J.; Westenberg, J.J.; Young, A.A.; et al. Quantification of LV function and mass by cardiovascular magnetic resonance: Multi-center variability and consensus contours. J. Cardiovasc. Magn. Reson. 2015, 17, 1–8. [Google Scholar] [CrossRef]
- Khened, M.; Kollerathu, V.A.; Krishnamurthi, G. Fully convolutional multi-scale residual DenseNets for cardiac segmentation and automated cardiac diagnosis using ensemble of classifiers. Med. Image Anal. 2019, 51, 21–45. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Fang, Y.; Lai, X. Left ventricle automatic segmentation in cardiac MRI using a combined CNN and U-net approach. Comput. Med. Imaging Graph. 2020, 82, 101719. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Yuwen, C.; Jiang, L.; Xia, Y.; Zhang, Y. Multiscale attention guided U-Net architecture for cardiac segmentation in short-axis MRI images. Comput. Methods Programs Biomed. 2021, 206, 106142. [Google Scholar] [CrossRef] [PubMed]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 60, 84–90. [Google Scholar] [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2015, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Siddique, N.; Paheding, S.; Elkin, C.P.; Devabhaktuni, V. U-net and its variants for medical image segmentation: A review of theory and applications. IEEE Access 2021, 9, 82031–82057. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Moradi, S.; Oghli, M.G.; Alizadehasl, A.; Shiri, I.; Oveisi, N.; Oveisi, M.; Maleki, M.; Dhooge, J. MFP-Unet: A novel deep learning based approach for left ventricle segmentation in echocardiography. Phys. Med. 2019, 67, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Baldeon-Calisto, M.; Lai-Yuen, S.K. AdaResU-Net: Multiobjective adaptive convolutional neural network for medical image segmentation. Neurocomputing 2020, 392, 325–340. [Google Scholar] [CrossRef]
- Khened, M.; Alex, V.; Krishnamurthi, G. Densely connected fully convolutional network for short-axis cardiac cine MR image segmentation and heart diagnosis using random forest. In International Workshop on Statistical Atlases and Computational Models of the Heart; Springer: Cham, Switzerland, 2017; pp. 140–151. [Google Scholar]
- Zotti, C.; Luo, Z.; Humbert, O.; Lalande, A.; Jodoin, P.M. GridNet with automatic shape prior registration for automatic MRI cardiac segmentation. In International Workshop on Statistical Atlases and Computational Models of the Heart; Springer: Cham, Switzerland, 2017; pp. 73–81. [Google Scholar]
- Mehta, R.; Sivaswamy, J. M-net: A convolutional neural network for deep brain structure segmentation. In Proceedings of the 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), Melbourne, Australia, 18–21 April 2017; pp. 437–440. [Google Scholar]
- Baumgartner, C.F.; Koch, L.M.; Pollefeys, M.; Konukoglu, E. An exploration of 2D and 3D deep learning techniques for cardiac MR image segmentation. In International Workshop on Statistical Atlases and Computational Models of the Heart; Springer: Cham, Switzerland, 2017; pp. 111–119. [Google Scholar]
- Patravali, J.; Jain, S.; Chilamkurthy, S. 2D-3D fully convolutional neural networks for cardiac MR segmentation. In International Workshop on Statistical Atlases and Computational Models of the Heart; Springer: Cham, Switzerland, 2017; pp. 130–139. [Google Scholar]
- Isensee, F.; Jaeger, P.F.; Full, P.M.; Wolf, I.; Engelhardt, S.; Maier-Hein, K.H. Automatic cardiac disease assessment on cine-MRI via time-series segmentation and domain specific features. In International Workshop on Statistical Atlases and Computational Models of the Heart; Springer: Cham, Switzerland, 2017; pp. 120–129. [Google Scholar]
- Yang, X.; Bian, C.; Yu, L.; Ni, D.; Heng, P.A. Class-balanced deep neural network for automatic ventricular structure segmentation. In International Workshop on Statistical Atlases and Computational Models of the Heart; Springer: Cham, Switzerland, 2017; pp. 152–160. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In Proceedings of the International Conference on Machine Learning 2015, Lille, France, 6–11 July 2015; pp. 448–456. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Cardiac Atlas Project. Available online: https://www.cardiacatlas.org/studies/sunnybrook-cardiac-data/ (accessed on 15 June 2018).
- Xu, B.; Wang, N.; Chen, T.; Li, M. Empirical evaluation of rectified activations in convolutional network. arXiv 2015, arXiv:1505.00853. [Google Scholar]
- Clevert, D.A.; Unterthiner, T.; Hochreiter, S. Fast and accurate deep network learning by exponential linear units (elus). arXiv 2015, arXiv:1511.07289. [Google Scholar]
- Jadon, S. A survey of loss functions for semantic segmentation. In Proceedings of the 2020 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), Online, 27–29 October 2020; pp. 1–7. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Tran, P.V. A fully convolutional neural network for cardiac segmentation in short-axis MRI. arXiv 2016, arXiv:1604.00494. [Google Scholar]
- Veress, A.; Phatak, N.; Weiss, J. The Handbook of Medical Image Analysis: Segmentation and Registration Models; Springer: New York, NY, USA, 2005; Volume 3. [Google Scholar]
- Queirós, S.; Barbosa, D.; Heyde, B.; Morais, P.; Vilaça, J.L.; Friboulet, D.; Bernard, O.; D’hooge, J. Fast automatic myocardial segmentation in 4D cine CMR datasets. Med. Image Anal. 2014, 18, 1115–1131. [Google Scholar] [CrossRef]
- Ngo, T.A.; Carneiro, G. Left ventricle segmentation from cardiac MRI combining level set methods with deep belief networks. In Proceedings of the 2013 IEEE International Conference on Image Processing, Melbourne, Australia, 15–18 September 2013; pp. 695–699. [Google Scholar]
- Hu, H.; Liu, H.; Gao, Z.; Huang, L. Hybrid segmentation of left ventricle in cardiac MRI using gaussian-mixture model and region restricted dynamic programming. Magn. Reson. Imaging 2013, 31, 575–584. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Hu, H.; Xu, X.; Song, E. Automatic left ventricle segmentation in cardiac MRI using topological stable-state thresholding and region restricted dynamic programming. Acad. Radiol. 2012, 19, 723–731. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Liu, J.; Lee, L.C.; Venkatesh, S.K.; Teo, L.L.; Au, C.; Nowinski, W.L. An image-based comprehensive approach for automatic segmentation of left ventricle from cardiac short axis cine mr images. J. Digit. Imaging 2011, 24, 598–608. [Google Scholar] [CrossRef] [PubMed]
- Irshad, M.; Muhammad, N.; Sharif, M.; Yasmeen, M. Automatic segmentation of the left ventricle in a cardiac MR short axis image using blind morphological operation. Eur. Phys. J. Plus 2018, 133, 148. [Google Scholar] [CrossRef]
- Ngo, T.A.; Lu, Z.; Carneiro, G. Combining deep learning and level set for the automated segmentation of the left ventricle of the heart from cardiac cine magnetic resonance. Med. Image Anal. 2017, 35, 159–171. [Google Scholar] [CrossRef]
- Poudel, R.P.; Lamata, P.; Montana, G. Recurrent fully convolutional neural networks for multi-slice MRI cardiac segmentation. In Reconstruction, Segmentation, and Analysis of Medical Images; Springer: Cham, Switzerland, 2016; pp. 83–94. [Google Scholar]
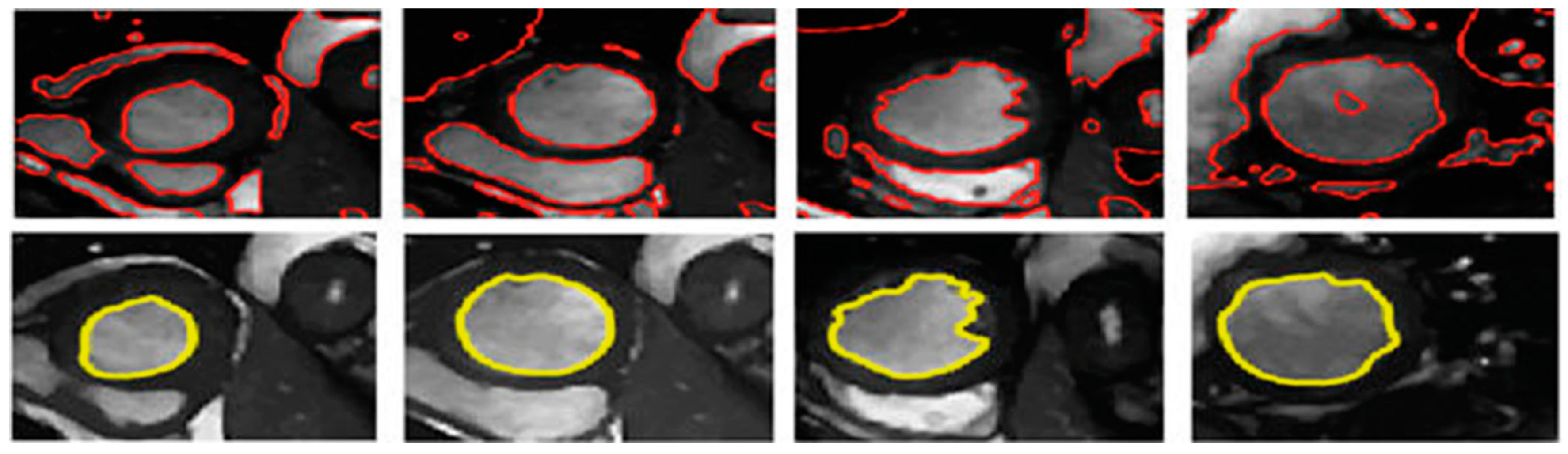





| Set | Pathological Conditions | Cases |
|---|---|---|
| Training | N | 3 |
| HYP | 4 | |
| HF | 4 | |
| HF-I | 4 | |
| Validation | N | 3 |
| HYP | 4 | |
| HF | 4 | |
| HF-I | 4 | |
| Testing | N | 3 |
| HYP | 4 | |
| HF | 4 | |
| HF-I | 4 |
| Evaluation Parameters | N (n = 9) | HYP (n = 12) | HF (n = 12) | HF-I (n = 12) |
|---|---|---|---|---|
| End Diastolic Volume (mL) | 115.69 (36.89) | 114.39 (50.46) | 233.67 (63.21) | 244.92 (86.02) |
| End Systolic Volume (mL) | 43.10 (14.74) | 43.11 (24.50) | 158.28 (56.34) | 174.34 (90.64) |
| Ejection Fraction (%) | 62.93 (3.65) | 62.72 (9.22) | 33.09 (13.07) | 32.01 (12.27) |
| Convolution Layers | Training Weights | Training Time (100 Epochs, 2 GPUs) | Performance (Dice Coefficient) |
|---|---|---|---|
| 18 | 1.9 M | 4 h | 92–93% |
| 23 | 31 M | 6 h | 94–96% |
| 28 | 32 M | 7.5 h | 94–96% |
| Type | Loss Function |
|---|---|
| Distribution-based Loss | Binary Cross-Entropy Weighted Cross-Entropy Balanced Cross-Entropy Focal Loss Distance map derived loss penalty term |
| Region-based Loss | Dice Loss Sensitivity-Specificity Loss Tversky Loss Focal Tversky Loss Log-Cosh Dice Loss |
| Boundary-based Loss | Hausdorff Distance loss Shape aware loss |
| Compounded Loss | Combo Loss Exponential Logarithmic Loss |
| Activation Function | Loss Function | Activation Function | Loss Function |
|---|---|---|---|
| Relu | Binary Cross-Entropy | Elu | Binary Cross-Entropy |
| Dice Loss | Dice Loss | ||
| Log-Cosh Dice Loss | Log-Cosh Dice Loss | ||
| Tversky Loss | Tversky Loss | ||
| Focal Tversky Loss | Focal Tversky Loss |
| Hyperparameters | Value/Name | Hyperparameters | Value/Name |
|---|---|---|---|
| Batch Size | 4 | Dropout | 0.1 |
| Activation Functions | Relu/Elu | Loss Functions | Binary Cross-Entropy Dice Loss Log-Cosh Dice Loss Tversky Loss Focal Tversky Loss |
| Batch Normalization | True |
| Patient Group | Cases | Average Contour Detection (%) | Average Good Contour Detection (%) | Average Perpendicular Distance (mm) | Dice Metrics | ||||
|---|---|---|---|---|---|---|---|---|---|
| Endo | Epi | Endo | Epi | Endo | Epi | Endo | Epi | ||
| N | 9 | 100 | 100 | 96.89 | 95.63 | 1.65 | 2.04 | 0.91 | 0.93 |
| HYP | 12 | 100 | 100 | 95.70 | 89.68 | 1.76 | 2.08 | 0.90 | 0.93 |
| HF-NI | 12 | 100 | 100 | 91.60 | 87.27 | 2.11 | 1.97 | 0.92 | 0.95 |
| HF-I | 12 | 100 | 100 | 97.09 | 97.72 | 1.66 | 1.62 | 0.94 | 0.96 |
| Overall | 45 | 100 | 100 | 95.3 (2.55) | 92.6 (4.91) | 1.79 (0.21) | 1.93 (0.20) | 0.92 (0.02) | 0.94 (0.02) |
| Patient Group | Cases | Average Contour Detection (%) | Average Good Contour Detection (%) | Average Perpendicular Distance (mm) | Dice Metrics | ||||
|---|---|---|---|---|---|---|---|---|---|
| Endo | Epi | Endo | Epi | Endo | Epi | Endo | Epi | ||
| N | 9 | 100 | 100 | 97.86 | 95.43 | 1.72 | 1.97 | 0.91 | 0.94 |
| HYP | 12 | 100 | 100 | 93.93 | 91.23 | 1.76 | 2.00 | 0.90 | 0.94 |
| HF-NI | 12 | 100 | 100 | 86.94 | 83.50 | 1.92 | 1.74 | 0.93 | 0.95 |
| HF-I | 12 | 100 | 100 | 94.75 | 95.45 | 1.68 | 1.90 | 0.94 | 0.95 |
| Overall | 45 | 100 | 100 | 93.4 (4.61) | 91.4 (5.63) | 1.77 (0.11) | 1.90 (0.12) | 0.92 (0.02) | 0.95 (0.01) |
| Patient Group | Cases | Average Contour Detection (%) | Average Good Contour Detection (%) | Average Perpendicular Distance (mm) | Dice Metrics | ||||
|---|---|---|---|---|---|---|---|---|---|
| Endo | Epi | Endo | Epi | Endo | Epi | Endo | Epi | ||
| N | 9 | 100 | 100 | 96.45 | 96.38 | 1.68 | 1.70 | 0.91 | 0.95 |
| HYP | 12 | 100 | 100 | 97.34 | 96.69 | 1.72 | 1.90 | 0.90 | 0.94 |
| HF-NI | 12 | 100 | 100 | 93.05 | 92.85 | 1.87 | 1.78 | 0.93 | 0.95 |
| HF-I | 12 | 100 | 100 | 97.75 | 99.24 | 1.69 | 1.49 | 0.94 | 0.96 |
| Overall | 45 | 100 | 100 | 96.1 (2.14) | 96.3 (2.63) | 1.74 (0.09) | 1.7 (0.17) | 0.92 (0.01) | 0.95 (0.01) |
| Patient Group | Cases | Average Contour Detection (%) | Average Good Contour Detection (%) | Average Perpendicular Distance (mm) | Dice Metrics | ||||
|---|---|---|---|---|---|---|---|---|---|
| Endo | Epi | Endo | Epi | Endo | Epi | Endo | Epi | ||
| N | 9 | 100 | 100 | 96.04 | 84.76 | 1.62 | 1.98 | 0.91 | 0.94 |
| HYP | 12 | 100 | 100 | 96.23 | 85.72 | 1.62 | 2.13 | 0.91 | 0.94 |
| HF-NI | 12 | 100 | 100 | 91.04 | 88.11 | 1.88 | 1.90 | 0.93 | 0.95 |
| HF-I | 12 | 100 | 100 | 97.37 | 97.60 | 1.61 | 1.86 | 0.94 | 0.95 |
| Overall | 45 | 100 | 100 | 95.2 (2.82) | 89.0 (5.87) | 1.68 (0.13) | 1.97 (0.12) | 0.92 (0.02) | 0.95 (0.005) |
| Patient Group | Cases | Average Contour Detection (%) | Average Good Contour Detection (%) | Average Perpendicular Distance (mm) | Dice Metrics | ||||
|---|---|---|---|---|---|---|---|---|---|
| Endo | Epi | Endo | Epi | Endo | Epi | Endo | Epi | ||
| N | 9 | 100 | 100 | 97.75 | 100 | 1.63 | 1.64 | 0.91 | 0.95 |
| HYP | 12 | 100 | 100 | 95.53 | 91.93 | 1.70 | 2.07 | 0.91 | 0.93 |
| HF-NI | 12 | 100 | 100 | 90.71 | 87.64 | 1.91 | 1.91 | 0.93 | 0.95 |
| HF-I | 12 | 100 | 100 | 98.21 | 95.01 | 1.51 | 1.63 | 0.95 | 0.96 |
| Overall | 45 | 100 | 100 | 95.6 (3.43) | 93.65 (5.20) | 1.69 (0.17) | 1.81 (0.21) | 0.92 (0.01) | 0.94 (0.01) |
| Patient Group | Cases | Average Contour Detection (%) | Average Good Contour Detection (%) | Average Perpendicular Distance (mm) | Dice Metrics | ||||
|---|---|---|---|---|---|---|---|---|---|
| Endo | Epi | Endo | Epi | Endo | Epi | Endo | Epi | ||
| N | 9 | 100 | 100 | 98.40 | 93.13 | 1.95 | 2.11 | 0.90 | 0.93 |
| HYP | 12 | 100 | 100 | 95.88 | 95.10 | 1.95 | 2.22 | 0.89 | 0.93 |
| HF-NI | 12 | 100 | 100 | 88.44 | 83.79 | 1.95 | 1.92 | 0.93 | 0.94 |
| HF-I | 12 | 100 | 100 | 96.62 | 92.32 | 1.91 | 2.00 | 0.93 | 0.95 |
| Overall | 45 | 100 | 100 | 94.8 (4.39) | 91.1 (5.00) | 1.94 (0.02) | 2.06 (0.13) | 0.9 (0.20) | 0.93 (0.01) |
| Patient Group | Cases | Average Contour Detection (%) | Average Good Contour Detection (%) | Average Perpendicular Distance (mm) | Dice Metrics | ||||
|---|---|---|---|---|---|---|---|---|---|
| Endo | Epi | Endo | Epi | Endo | Epi | Endo | Epi | ||
| N | 9 | 100 | 100 | 99.26 | 98.88 | 1.72 | 1.88 | 0.91 | 0.94 |
| HYP | 12 | 100 | 100 | 93.32 | 88.49 | 1.64 | 1.99 | 0.91 | 0.94 |
| HF-NI | 12 | 100 | 100 | 93.20 | 80.97 | 2.20 | 1.99 | 0.92 | 0.94 |
| HF-I | 12 | 100 | 100 | 98.31 | 95.51 | 1.67 | 1.85 | 0.94 | 0.95 |
| Overall | 45 | 100 | 100 | 96.02 (3.21) | 90.96 (7.94) | 1.8 (0.26) | 1.92 (0.07) | 0.9 (0.01) | 0.94 (0.005) |
| Patient Group | Cases | Average Contour Detection (%) | Average Good Contour Detection (%) | Average Perpendicular Distance (mm) | Dice Metrics | ||||
|---|---|---|---|---|---|---|---|---|---|
| Endo | Epi | Endo | Epi | Endo | Epi | Endo | Epi | ||
| N | 9 | 100 | 100 | 97.12 | 100 | 2.19 | 1.83 | 0.88 | 0.94 |
| HYP | 12 | 100 | 100 | 91.22 | 92.99 | 2.03 | 2.01 | 0.89 | 0.94 |
| HF-NI | 12 | 100 | 100 | 89.07 | 87.00 | 2.22 | 1.76 | 0.92 | 0.95 |
| HF-I | 12 | 100 | 100 | 94.73 | 92.14 | 1.87 | 1.51 | 0.93 | 0.96 |
| Overall | 45 | 100 | 100 | 93.04 (3.58) | 93.03 (5.34) | 2.07 (0.16) | 1.78 (0.21) | 0.90 (0.02) | 0.95 (0.009) |
| Patient Group | Cases | Average Contour Detection (%) | Average Good Contour Detection (%) | Average Perpendicular Distance (mm) | Dice Metrics | ||||
|---|---|---|---|---|---|---|---|---|---|
| Endo | Epi | Endo | Epi | Endo | Epi | Endo | Epi | ||
| N | 9 | 100 | 100 | 97.28 | 97.22 | 1.94 | 2.15 | 0.90 | 0.93 |
| HYP | 12 | 100 | 100 | 94.69 | 89.78 | 1.77 | 2.17 | 0.90 | 0.93 |
| HF-NI | 12 | 100 | 100 | 93.18 | 84.24 | 2.00 | 2.22 | 0.92 | 0.94 |
| HF-I | 12 | 100 | 100 | 98.73 | 95.02 | 1.69 | 1.90 | 0.94 | 0.95 |
| Overall | 45 | 100 | 100 | 95.97 (2.50) | 91.57 (5.79) | 1.85 (0.14) | 2.11 (0.14) | 0.91 (0.02) | 0.94 (0.009) |
| Patient Group | Cases | Average Contour Detection (%) | Average Good Contour Detection (%) | Average Perpendicular Distance (mm) | Dice Metrics | ||||
|---|---|---|---|---|---|---|---|---|---|
| Endo | Epi | Endo | Epi | Endo | Epi | Endo | Epi | ||
| N | 9 | 100 | 100 | 95.30 | 90.62 | 1.79 | 2.21 | 0.90 | 0.93 |
| HYP | 12 | 100 | 100 | 95.18 | 86.73 | 2.00 | 2.39 | 0.88 | 0.92 |
| HF-NI | 12 | 100 | 100 | 91.93 | 84.41 | 2.20 | 2.26 | 0.92 | 0.94 |
| HF-I | 12 | 100 | 100 | 87.55 | 91.37 | 1.96 | 1.98 | 0.93 | 0.95 |
| Overall | 45 | 100 | 100 | 92.49 (3.64) | 88.28 (3.29) | 1.99 (0.17) | 2.21 (0.17) | 0.90 (0.02) | 0.93 (0.01) |
| Loss Function + Activation Function | Dice Metrics | Average Good Contour Detection (%) | Average Perpendicular Distance (mm) | |||
|---|---|---|---|---|---|---|
| Endo | Epi | Endo | Epi | Endo | Epi | |
| Focal Tversky Loss + Elu | 0.93 (0.01) | 0.96 (0.01) | 96.1 (2.14) | 96.3 (2.63) | 1.74 (0.09) | 1.7 (0.17) |
| Focal Tversky Loss + Relu | 0.92 (0.02) | 0.95 (0.005) | 95.2 (2.82) | 89.0 (5.87) | 1.68 (0.13) | 1.97 (0.12) |
| Tversky Loss + Elu | 0.92 (0.01) | 0.94 (0.01) | 95.6 (3.43) | 93.65 (5.20) | 1.69 (0.17) | 1.81 (0.21) |
| Tversky Loss + Relu | 0.9 (0.20) | 0.93 (0.01) | 94.8 (4.39) | 91.1 (5.00) | 1.94 (0.02) | 2.06 (0.13) |
| Dice Loss + Elu | 0.92 (0.02) | 0.94 (0.02) | 95.3 (2.55) | 92.6 (4.91) | 1.79 (0.21) | 1.93 (0.20) |
| Dice Loss + Relu | 0.92 (0.02) | 0.95 (0.01) | 93.4 (4.61) | 91.4 (5.63) | 1.77 (0.11) | 1.90 (0.12) |
| Log-Cosh Dice Loss + Elu | 0.91 (0.02) | 0.94 (0.009) | 95.97 (2.50) | 91.57 (5.79) | 1.85 (0.14) | 2.11 (0.14) |
| Log-Cosh Dice Loss + Relu | 0.9 (0.02) | 0.93 (0.01) | 92.49 (3.64) | 88.28 (3.29) | 1.99 (0.17) | 2.21 (0.17) |
| Binary Cross-Entropy Loss + Elu | 0.9 (0.01) | 0.94 (0.005) | 96.02 (3.21) | 90.96 (7.94) | 1.8 (0.26) | 1.92 (0.07) |
| Binary Cross-Entropy Loss + Relu | 0.9 (0.02) | 0.95 (0.009) | 93.04 (3.58) | 93.03 (5.34) | 2.07 (0.16) | 1.78 (0.21) |
| Authors | Average Good Contour Detection (%) | Average Perpendicular Distance (mm) | Dice Metrics | |||
|---|---|---|---|---|---|---|
| Endo | Epi | Endo | Epi | Endo | Epi | |
| Proposed (U-Net with Tversky Focal Loss and Elu Activation) | 96.15 | 96.29 | 1.74 | 1.72 | 0.92 | 0.95 |
| Queiros [33] | 92.70 | 95.40 | 1.76 | 1.80 | 0.90 | 0.94 |
| Ngo and Carneiro [34] | 97.91 | - | 2.08 | - | 0.90 | - |
| Hu [35] | 91.06 | 91.21 | 2.24 | 2.19 | 0.89 | 0.94 |
| Liu [36] | 91.17 | 90.78 | 2.36 | 2.19 | 0.88 | 0.94 |
| Huang [37] | 79.20 | 83.90 | 2.16 | 2.22 | 0.89 | 0.93 |
| Irshad [38] | - | - | 2.1 | 3.1 | 0.91 | 0.91 |
| Overall | ||||||
| Proposed (U-Net with Tversky Focal Loss and Elu Activation) | 96.22 | 1.73 | 0.94 | |||
| Ngo and Lu [39] | 95.71 | 2.34 | 0.88 | |||
| Poudel FCN [40] | 94.78 | 2.14 | 0.902 | |||
| Poudel RFCN [40] | 95.34 | 2.05 | 0.9 | |||
| Loss Function + Activation Function | EF | ESV | EDV | |||
|---|---|---|---|---|---|---|
| Manul | Auto | Manual | Auto | Manual | Auto | |
| Focal Tversky Loss + Elu | 38.85 | 36.77 | 111.48 | 117.73 | 182.3 | 186.19 |
| Focal Tversky Loss + Relu | 38.85 | 36.52 | 111.48 | 118.59 | 182.3 | 187.18 |
| Tversky Loss + Elu | 38.85 | 36.87 | 111.48 | 119.88 | 182.3 | 189.88 |
| Tversky Loss + Relu | 38.85 | 36.99 | 111.48 | 119.66 | 182.3 | 189.90 |
| Dice Loss + Elu | 38.85 | 30.73 | 111.48 | 120.33 | 182.3 | 173.73 |
| Dice Loss + Relu | 38.85 | 30.69 | 111.48 | 120.59 | 182.3 | 173.98 |
| Log-Cosh Dice Loss + Elu | 38.85 | 29.68 | 111.48 | 121.05 | 182.3 | 172.13 |
| Log-Cosh Dice Loss + Relu | 38.85 | 29.63 | 111.48 | 121.15 | 182.3 | 172.16 |
| Binary Cross-Entropy Loss + Elu | 38.85 | 28.93 | 111.48 | 122.35 | 182.3 | 172.17 |
| Binary Cross-Entropy Loss + Relu | 38.85 | 28.74 | 111.48 | 123.38 | 182.3 | 173.15 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhan, A.; Mangipudi, P.; Goyal, A. Deep Learning Approach for Automatic Segmentation and Functional Assessment of LV in Cardiac MRI. Electronics 2022, 11, 3594. https://doi.org/10.3390/electronics11213594
Bhan A, Mangipudi P, Goyal A. Deep Learning Approach for Automatic Segmentation and Functional Assessment of LV in Cardiac MRI. Electronics. 2022; 11(21):3594. https://doi.org/10.3390/electronics11213594
Chicago/Turabian StyleBhan, Anupama, Parthasarathi Mangipudi, and Ayush Goyal. 2022. "Deep Learning Approach for Automatic Segmentation and Functional Assessment of LV in Cardiac MRI" Electronics 11, no. 21: 3594. https://doi.org/10.3390/electronics11213594
APA StyleBhan, A., Mangipudi, P., & Goyal, A. (2022). Deep Learning Approach for Automatic Segmentation and Functional Assessment of LV in Cardiac MRI. Electronics, 11(21), 3594. https://doi.org/10.3390/electronics11213594







