Fast Self-Attention Deep Detection Network Based on Weakly Differentiated Plant Nematodess
Abstract
:1. Introduction
2. Materials and Methods
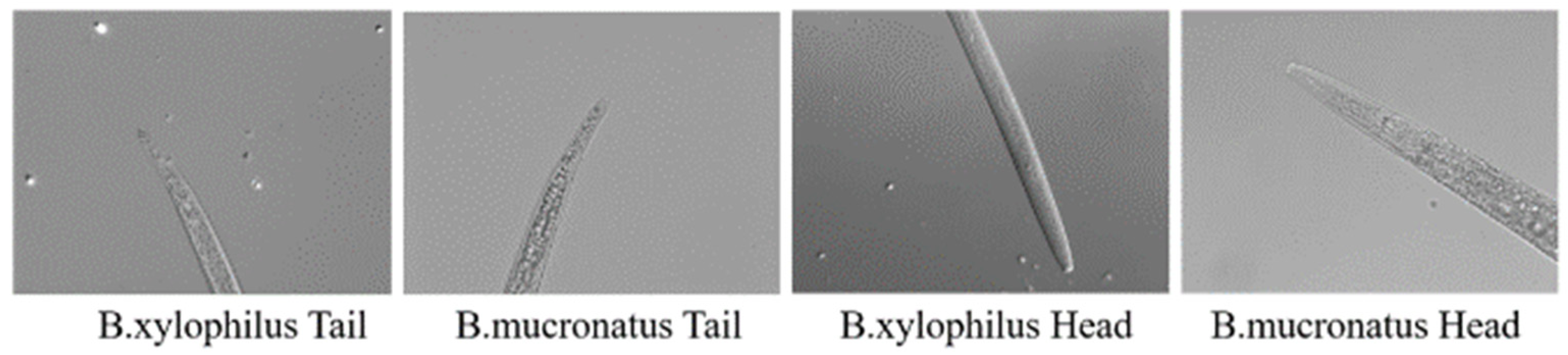
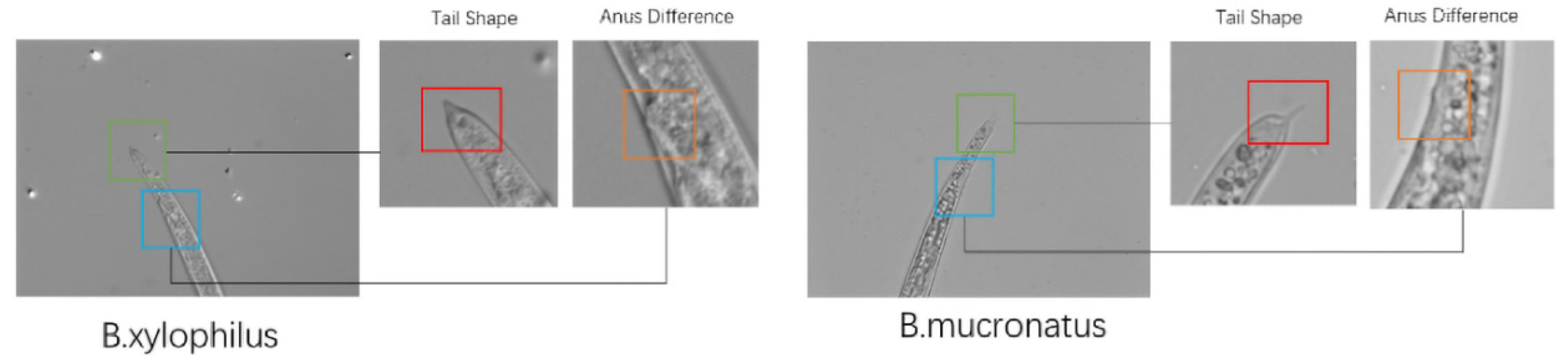
2.1. Dataset
2.1.1. Dataset
2.1.2. Traditional Detection Methods
2.2. Methods
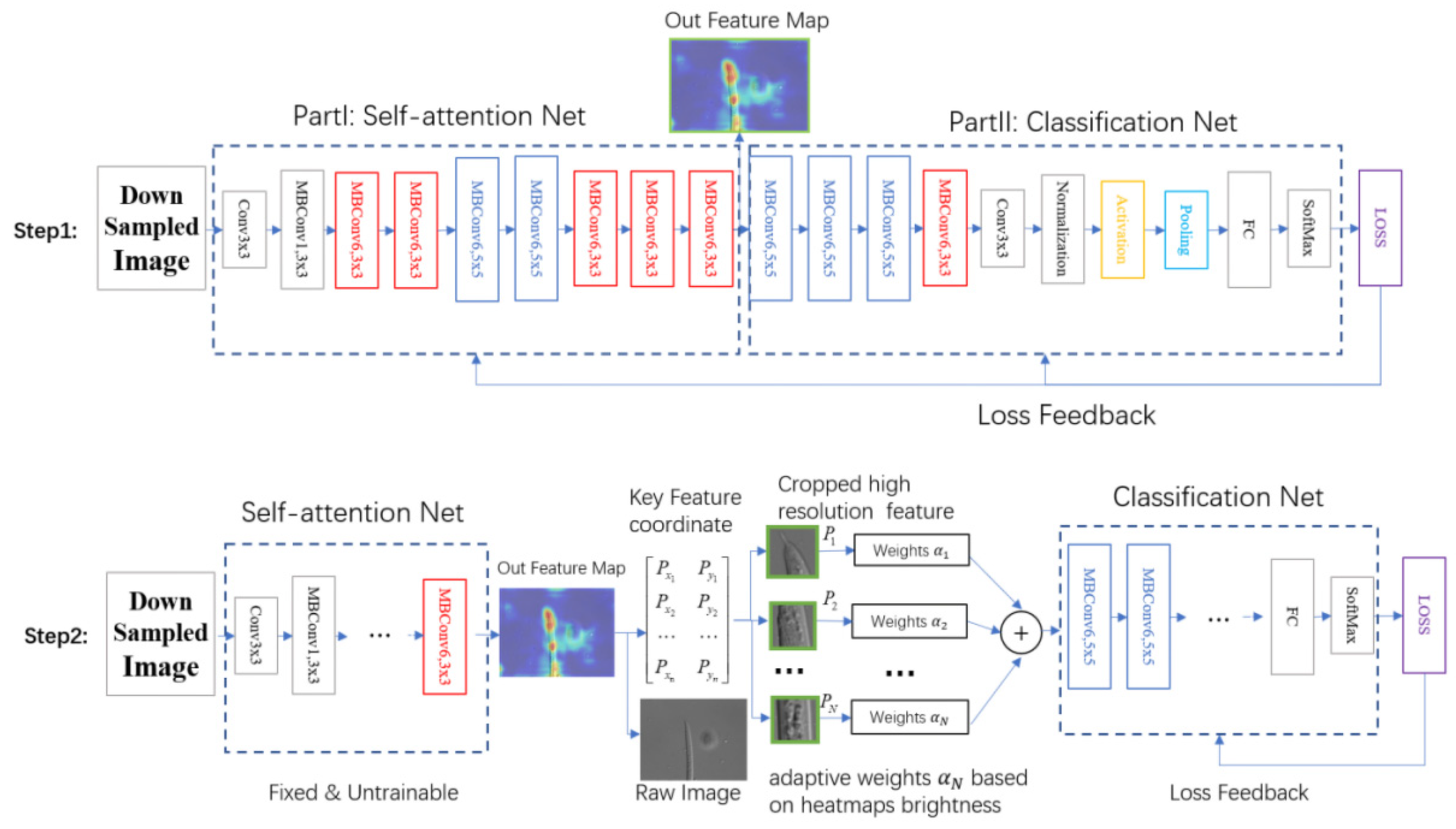
2.2.1. Part A: Self-Attention Feature Network
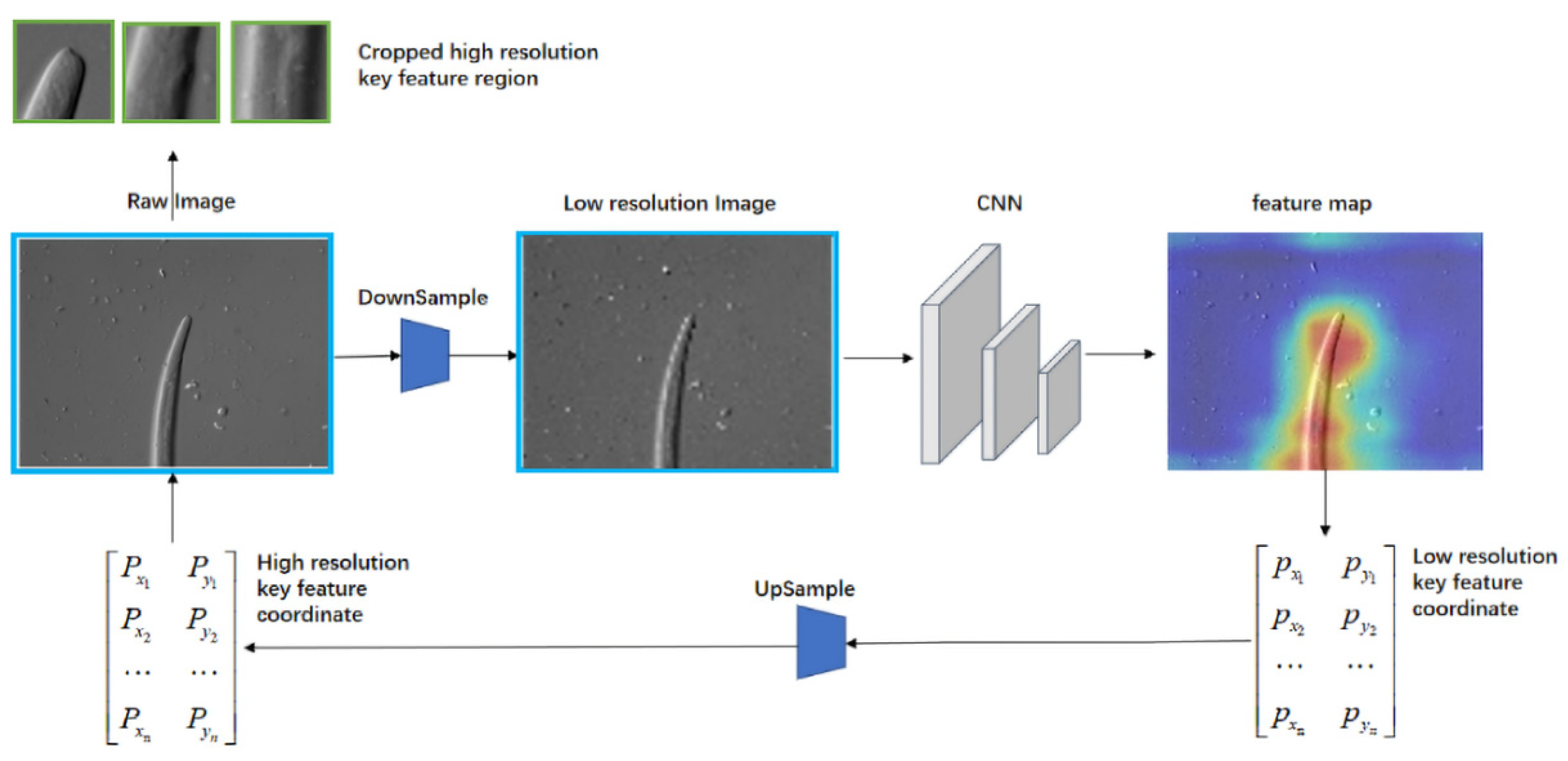
2.2.2. Small-Scale Fast Focus Method
2.2.3. Multi-Feature Joint Classification Network Based on Adaptive Weights of Heatmap Brightness
3. Results
3.1. Experiment Environment Match
3.2. Dataset
3.3. Performance Comparison
3.3.1. Results of Self-Attention Network
3.3.2. Efficiency and Accuracy Improvement
4. Conclusions
- The detection accuracy of our proposed method reaches 99%.
- Compared with other methods, our method is the fastest.
- We constructed a more complete high-resolution training dataset involving 24 species of Bursaphelenchus xylophilus and other common hybrid species with a total amount of data exceeded 10,000.
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nicol, J.M.; Turner, S.J.; Coyne, D.L.; den Nijs, L.; Hockland, S.; Maafi, Z.T. Current nematode threats to world agriculture. Genom. Mol. Genet. Plant-Nematode Interact. 2011, 21–43. [Google Scholar] [CrossRef]
- Bernard, G.C.; Egnin, M.; Bonsi, C. The impact of plant-parasitic nematodes on agriculture and methods of control. Nematol. -Concepts Diagn. Control. 2017, 10, 121–151. [Google Scholar]
- Hallmann, J.; Subbotin, S.A. Methods for extraction, processing and detection of plant and soil nematodes. Plant Parasit. Nematodes Subtrop. Trop. Agric. 2018, 87–119. [Google Scholar]
- Liu, M.; Wang, X.; Liu, X. Classification of multi-focal nematode image stacks using a projection based multilinear approach. In Proceedings of the 2017 IEEE International Conference on Image Processing (ICIP), Beijing, China, 17–20 September 2017; IEEE: New York, NY, USA, 2018; pp. 595–599. [Google Scholar]
- Chou, Y.; Lee, D.J.; Zhang, D. Edge Detection Using Convolutional Neural Networks for Nematode Development and Adaptation Analysis. In Proceedings of the International Conference on Computer Vision Systems; Springer: Cham, Switzerland, 2017; pp. 228–238. Available online: https://books.google.com.hk/books?hl=zh-CN&lr=&id=rHg5DwAAQBAJ&oi=fnd&pg=PA228&ots=IXnEZSxKS7&sig=wvjgp3c9kWq58ZEiIGUZoDTT6Qs&redir_esc=y#v=onepage&q&f=false (accessed on 19 October 2022).
- Liu, M.; Wang, X.; Zhang, H. Taxonomy of multi-focal nematode image stacks by a CNN based image fusion approach. Comput. Methods Programs Biomed. 2018, 156, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Braun-Kiewnick, A.; Kiewnick, S. Real-time PCR, a great tool for fast identification, sensitive detection and quantification of important plant-parasitic nematodes. Eur. J. Plant Pathol. 2018, 152, 271–283. [Google Scholar] [CrossRef]
- Kurmi, Y.; Gangwar, S.; Agrawal, D.; Kumar, S.; Srivastava, H.S. Leaf image analysis-based crop diseases classification. Signal Image Video Process. 2021, 15, 589–597. [Google Scholar] [CrossRef]
- Li, X.; Yu, L.; Chang, D.; Ma, Z.; Cao, J. Dual cross-entropy loss for small-sample fine-grained vehicle classification. IEEE Trans. Veh. Technol. 2019, 8, 4204–4212. [Google Scholar] [CrossRef]
- Abadi, M.; Agarwal, A.; Barham, P. Tensorflow: Large-scale machine learning on heterogeneous distributed systems. arXiv 2016, arXiv:1603.04467. [Google Scholar]
- Huang, J.; Lu, X.; Chen, L.; Sun, H.; Wang, S.; Fang, G. Accurate Identification of Pine Wood Nematode Disease with a Deep Convolution Neural Network. Remote Sens. 2022, 14, 913. [Google Scholar] [CrossRef]
- Doshi, R.A.; King, R.L.; Lawrence, G.W. Wavelet-SOM in feature extraction of hyperspectral data for classification of nematode species. In Proceedings of the International Geoscience and Remote Sensing Symposium, Barcelona, Spain, 23–28 July 2007; IEEE: New York, NY, USA, 2008; pp. 2818–2821. [Google Scholar] [CrossRef]
- Romain, T.; Van Linh, L.E.; Heloïse, V.; Alain, B.; Marie, B.; Eric, G.; Laurent, F.; Nicolas, P. Image based species identification of Globodera quarantine nematodes using computer vision and deep learning. Comput. Electron. Agric. 2021, 186, 106058. [Google Scholar]
- Uhlemann, J.; Cawley, O.; Kakouli-Duarte, T. Nematode Identification using Artificial Neural Networks. In Proceedings of the 1st International Conference on Deep Learning Theory and Applications-DeLTA, Paris, France, 8–10 July 2020; pp. 13–22. [Google Scholar] [CrossRef]
- Carneiro, R.; de Oliveira Lima, F.S.; Correia, V.R. Methods and tools currently used for the identification of plant parasitic nematodes. Nematol. -Concepts Diagn. Control 2017, 19. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.B.; Sun, J. R-CNN: Towards real-time object detection with region proposal networks. Adv. Neural Inf. Process. Syst. 2015, 28, 91–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Redmon, J.; Divvala, S.; Girshick, R. You only look once: Unified, real-time object detection. In Proceedings of the Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; IEEE: New York, NY, USA, 2016; pp. 779–788. [Google Scholar]
- Schandry, N.; Becker, C. Allelopathic plants: Models for studying plant–interkingdom interactions. Trends Plant Sci. 2020, 25, 176–185. [Google Scholar] [CrossRef]
- Holladay, B.H.; Willett, D.S.; Stelinski, L.L. High throughput nematode counting with automated image processing. BioControl 2016, 61, 177–183. [Google Scholar] [CrossRef]
- Chen, Y.; Li, W.; Sakaridis, C. Domain adaptive faster r-cnn for object detection in the wild. In Proceedings of the Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; IEEE: New York, NY, USA, 2018; pp. 3339–3348. [Google Scholar]
- Qin, J.; Wang, B.; Wu, Y. Identifying Pine Wood Nematode Disease Using UAV Images and Deep Learning Algorithms. Remote Sens. 2021, 13, 162. [Google Scholar] [CrossRef]
- Lee, S.; Park, S.; Baek, G. Detection of damaged pine tree by the pine wilt disease using UAV Image. Korean J. Remote Sens. 2019, 35, 359–373. [Google Scholar]
- Chen, L.; Strauch, M.; Daub, M. A CNN framework based on line annotations for detecting nematodes in microscopic images. In Proceedings of the 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; IEEE: New York, NY, USA, 2020; pp. 508–512. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. Computer Science. airXiv 2014, arXiv:1409.1556. [Google Scholar]
- Wu, J.; Zhang, Q.; Liu, M.; Xiao, Z.; Zhang, F.; Geng, L.; Liu, Y.; Wang, W. Diabetic macular edema grading based on improved Faster R-CNN and MD-ResNet. Signal Image Video Processing 2021, 15, 743–751. [Google Scholar] [CrossRef]












| Methods | Annotation Time | Model Parameters | Training Time | Detect Time | Accuracy |
|---|---|---|---|---|---|
| Expert Network | 2 Day | 44 M | 7.5 h | 1.8 s | 0.93 |
| Ours | 0 | 11 M | 3 h | 1.1 s | 0.995 |
| Areas Number | 1 | 2 | 3 | 4 | ||||
|---|---|---|---|---|---|---|---|---|
| Ratio | - | - | 1:1 | 0.54:0.46 | 1:1:1 | 0.42:0.36:0.22 | 1:1:1:1 | 0.37:0.32:0.20:0.11 |
| Accuracy | 0.9895 | 0.9895 | 0.9950 | 0.9953 | 0.9922 | 0.9934 | 0.9905 | 0.992 |
| Method | Training Method | Model Parameters | Annotation Time | Training Time | Detect Time | Acctracy |
|---|---|---|---|---|---|---|
| TOLO | ImageNet | 20.8 M | 2 Day | 5 h | 1.3 s | 0.84 |
| No-ImageNet | 0.62 | |||||
| Faster RCNN | ImageNet | 30 M | 2 Day | 6 h | 1.5 s | 0.88 |
| No-ImageNet | 0.64 | |||||
| Expert Network | ImageNet | 44 M | 2 Day | 7.5 h | 1.8 s | 0.93 |
| No-ImageNet | 0.72 | |||||
| Ours | ImageNet | 11 M | 0 | 3 h | 1.12 s | 0.995 |
| No-ImageNet | 0.98 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhuang, J.; Liu, Y.; Xu, N.; Zhu, Y.; Xiao, J.; Gu, J.; Mao, T. Fast Self-Attention Deep Detection Network Based on Weakly Differentiated Plant Nematodess. Electronics 2022, 11, 3497. https://doi.org/10.3390/electronics11213497
Zhuang J, Liu Y, Xu N, Zhu Y, Xiao J, Gu J, Mao T. Fast Self-Attention Deep Detection Network Based on Weakly Differentiated Plant Nematodess. Electronics. 2022; 11(21):3497. https://doi.org/10.3390/electronics11213497
Chicago/Turabian StyleZhuang, Jiayan, Yangming Liu, Ningyuan Xu, Yi Zhu, Jiangjian Xiao, Jianfeng Gu, and Tianyi Mao. 2022. "Fast Self-Attention Deep Detection Network Based on Weakly Differentiated Plant Nematodess" Electronics 11, no. 21: 3497. https://doi.org/10.3390/electronics11213497
APA StyleZhuang, J., Liu, Y., Xu, N., Zhu, Y., Xiao, J., Gu, J., & Mao, T. (2022). Fast Self-Attention Deep Detection Network Based on Weakly Differentiated Plant Nematodess. Electronics, 11(21), 3497. https://doi.org/10.3390/electronics11213497






