A Comprehensive Study on Healthcare Datasets Using AI Techniques
Abstract
1. Introduction
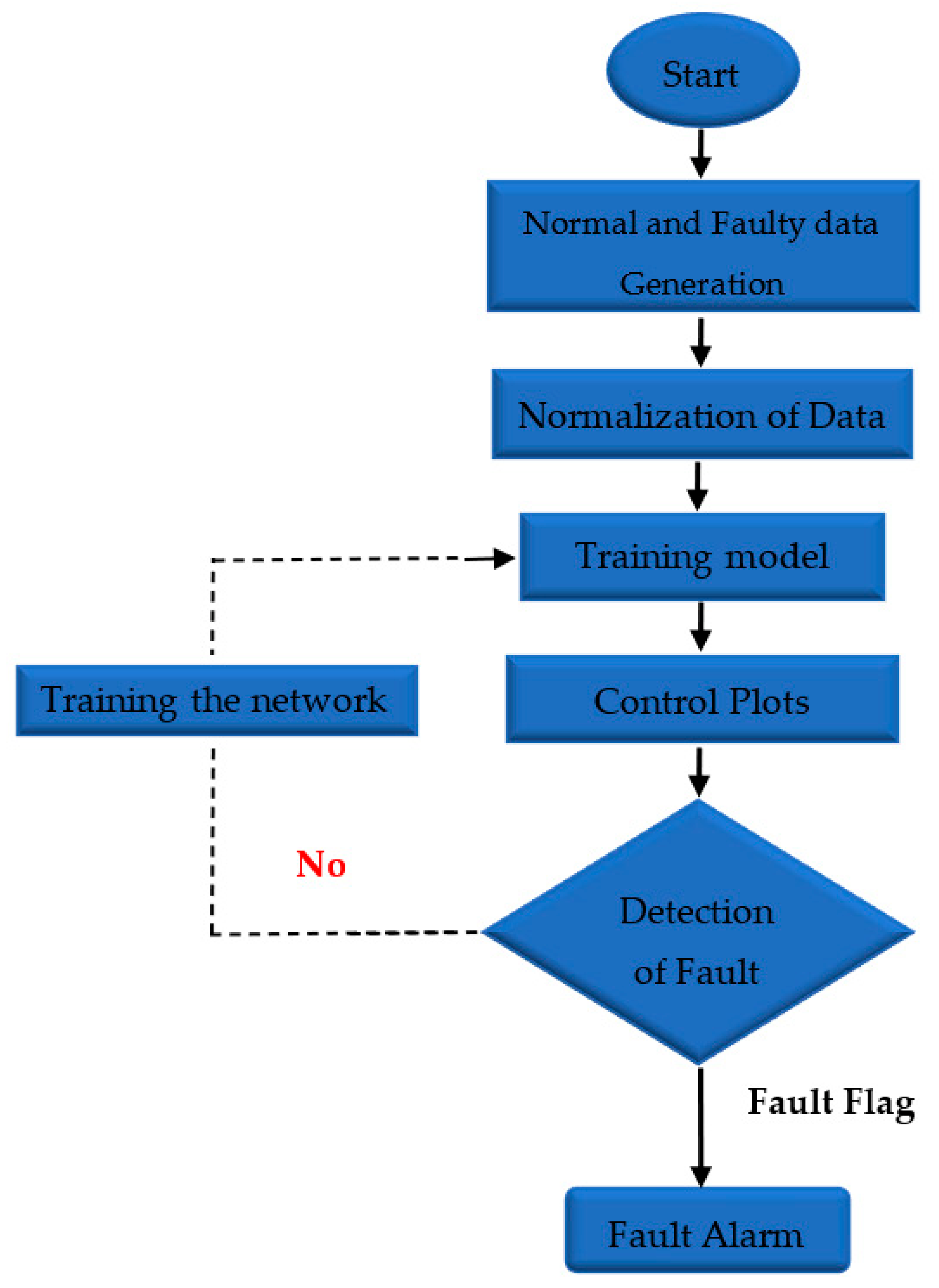
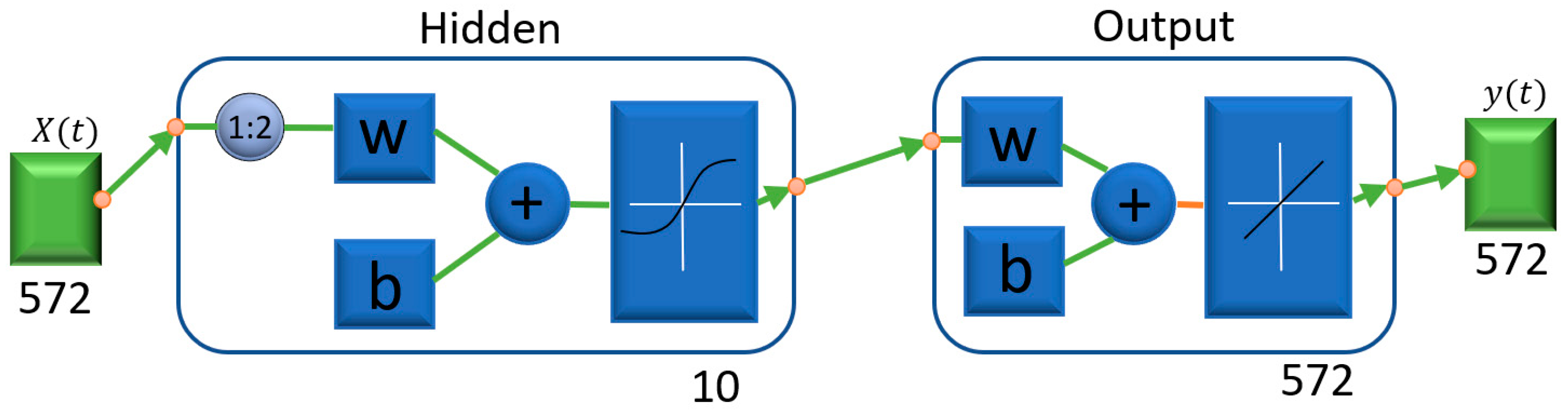
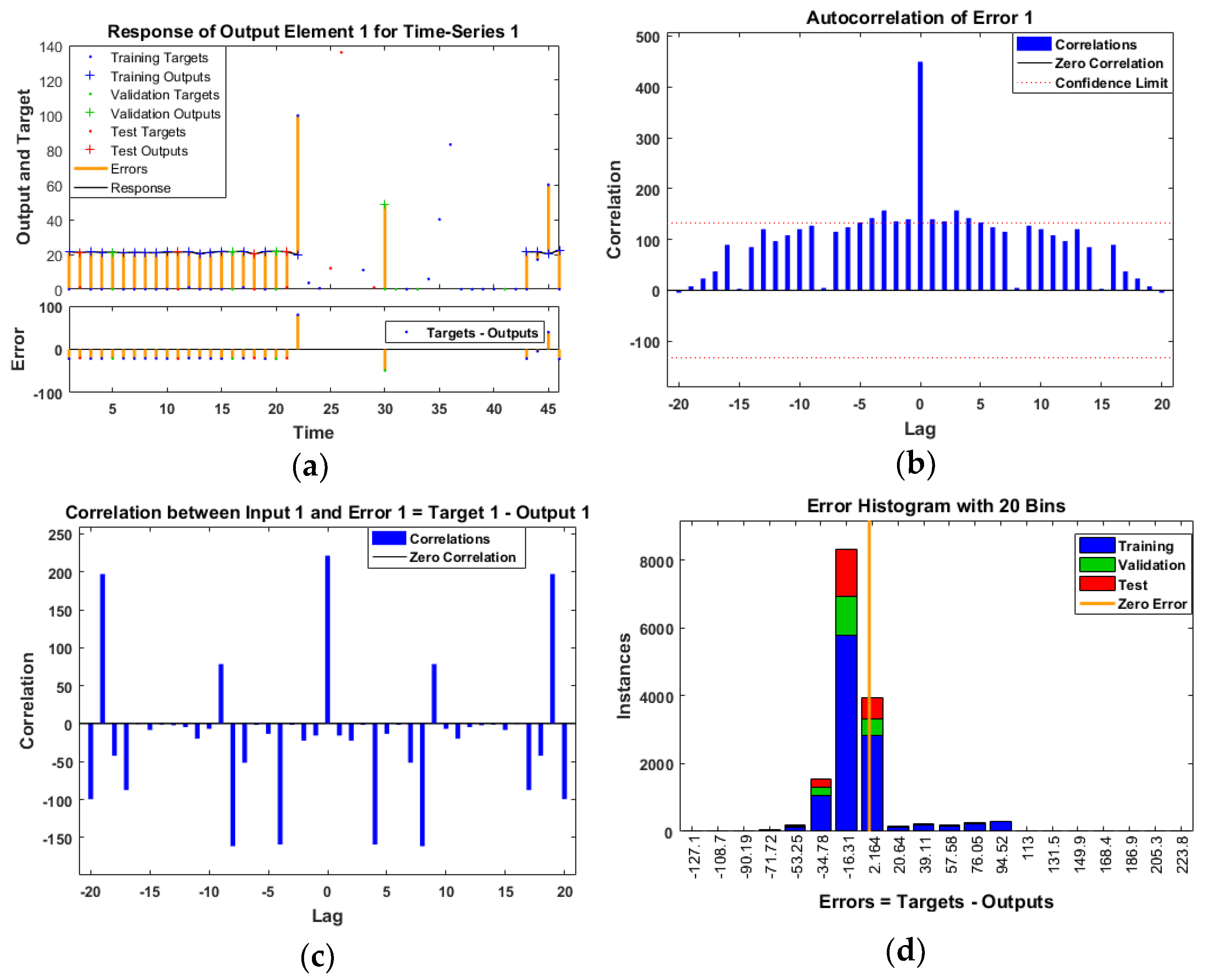
- Data monitoring and fault detection using neural network models are performed with three different algorithms: Levenberg–Marquardt, Bayesian regularization, and Scaled Conjugate Gradient as shown in Table 1. Moreover, in Figure 1 presented process of fault detection, and in the prediction model as shown in Figure 2, there is one input node, one output node, and ten hidden layers. Two delay orders show how the results of the previous two interactions predict incoming data. The number of hidden layers chosen is crucial, since it determines the NARX neural network. It is crucial to note this method in order to access NARX in the hidden layer for the prediction of error data monetization. Additionally, this is a different method of normalizing data.
- The decision-making process uses a machine learning approach to check linking factors between common symptoms and cross-check them with mortality rate. However, the symptoms should be interpreted with a numerical value. It is meaningful to perform mathematical operations on the values of attributes.
2. Related Work
2.1. Transformation
2.2. Healthcare Challenges
Data Matching
3. Methods Employed
3.1. Neural Network Models
3.2. Machine Learning Models
4. Comparative Analysis
5. Experiments and Results
5.1. Trainable Model
5.2. Data Source
5.3. Data Monitoring and Detection
- Set 1: For training: 60% of data points;
- Set 2: For network generalization: 20% of the data was used to test the network, this dataset must be independent of everything;
- Set 3: In order to prevent the network from overtraining, the training must be interrupted. In total, 20% of the data was used to validate the network.
5.4. Machine Learning Approach
6. Discussion
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Berger, M.L.; Sox, H.; Willke, R.J.; Brixner, D.L.; Eichler, H.G.; Goettsch, W.; Madigan, D.; Makady, A.; Schneeweiss, S.; Tarricone, R.; et al. Good practices for real-world data studies of treatment and/or comparative effectiveness: Recommendations from the joint ISPOR-ISPE Special Task Force on real-world evidence in health care decision making. Value Health 2017, 20, 1003–1008. [Google Scholar] [CrossRef] [PubMed]
- Franz, L.; Shrestha, Y.R.; Paudel, B. A deep learning pipeline for patient diagnosis prediction using electronic health records. arXiv 2020, arXiv:2006.16926. [Google Scholar]
- Xu, J.; Glicksberg, B.S.; Su, C.; Walker, P.; Bian, J.; Wang, F. Federated learning for healthcare informatics. J. Healthc. Inform. Res. 2021, 5, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Pavlopoulou, N.; Curry, E. PoSSUM: An Entity-centric Publish/Subscribe System for Diverse Summarization in Internet of Things. ACM Trans. Internet Technol. TOIT 2022, 22, 1–30. [Google Scholar] [CrossRef]
- Liu, X.; Xu, L.Q. Knowledge Graph Building from Real-world Multisource “Dirty” Clinical Electronic Medical Records for Intelligent Consultation Applications. In Proceedings of the 2021 IEEE International Conference on Digital Health (ICDH), Chicago, IL, USA, 5–10 September 2021. [Google Scholar]
- Steorts, R.C.; Ventura, S.L.; Sadinle, M.; Fienberg, S.E. A comparison of blocking methods for record linkage. In Proceedings of the International Conference on Privacy in Statistical Databases, Ibiza, Spain, 17–19 September 2014. [Google Scholar]
- Pérez-Moraga, R.; Forés-Martos, J.; Suay-García, B.; Duval, J.L.; Falcó, A.; Climent, J. A COVID-19 drug repurposing strategy through quantitative homological similarities using a topological data analysis-based framework. Pharmaceutics 2021, 13, 488. [Google Scholar] [CrossRef]
- Hung, T.N.K.; Le, N.Q.K.; Le, N.H.; Van Tuan, L.; Nguyen, T.P.; Thi, C.; Kang, J.H. An AI-based Prediction Model for Drug-drug Interactions in Osteoporosis and Paget’s Diseases from SMILES. Mol. Inform. 2022, 41, 2100264. [Google Scholar] [CrossRef]
- Ouyang, D.; He, B.; Ghorbani, A.; Yuan, N.; Ebinger, J.; Langlotz, C.P.; Heidenreich, P.A.; Harrington, R.A.; Liang, D.H.; Ashley, E.A.; et al. Video-based AI for beat-to-beat assessment of cardiac function. Nature 2020, 580, 252–256. [Google Scholar] [CrossRef]
- Rahman, M.M.; Saha, T.; Islam, K.J.; Suman, R.H.; Biswas, S.; Rahat, E.U.; Hossen, M.R.; Islam, R.; Hossain, M.N.; Mamun, A.A.; et al. Virtual screening, molecular dynamics and structure–activity relationship studies to identify potent approved drugs for COVID-19 treatment. J. Biomol. Struct. Dyn. 2021, 39, 6231–6241. [Google Scholar] [CrossRef]
- Persson, R.; Vasilakis-Scaramozza, C.; Hagberg, K.W.; Sponholtz, T.; Williams, T.; Myles, P.; Jick, S.S. CPRD Aurum database: Assessment of data quality and completeness of three important comorbidities. Pharmacoepidemiol. Drug Saf. 2020, 29, 1456–1464. [Google Scholar] [CrossRef]
- Schmidt, M.; Schmidt, S.A.J.; Adelborg, K.; Sundbøll, J.; Laugesen, K.; Ehrenstein, V.; Sørensen, H.T. The Danish health care system and epidemiological research: From health care contacts to database records. Clin. Epidemiol. 2019, 11, 563. [Google Scholar] [CrossRef]
- Singh, R.P.; Javaid, M.; Haleem, A.; Suman, R. Internet of things (IoT) applications to fight against COVID-19 pandemic. Diabetes Metab. Syndr. Clin. Res. Rev. 2020, 14, 521–524. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Jing, L.; Xu, Y.; Zheng, S.; Gan, Y.; Wen, C. Different Data Mining Approaches Based Medical Text Data. J. Healthc. Eng. 2021, 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Ramadan, B.; Christen, P.; Liang, H.; Gayler, R.W. Dynamic sorted neighborhood indexing for real-time entity resolution. J. Data Inf. Qual. JDIQ 2015, 6, 1–29. [Google Scholar] [CrossRef]
- Rad, M.A.A.; Yazdanpanah, M.J. Designing supervised local neural network classifiers based on EM clustering for fault diagnosis of Tennessee Eastman process. Chemom. Intell. Lab. Syst. 2015, 146, 149–157. [Google Scholar]
- Nozari, H.A.; Shoorehdeli, M.A.; Simani, S.; Banadaki, H.D. Model-based robust fault detection and isolation of an industrial gas turbine prototype using soft computing techniques. Neurocomputing 2012, 91, 29–47. [Google Scholar] [CrossRef]
- Lin, T.; Horne, B.G.; Giles, C.L.; Kung, S.Y. What to remember: How memory order affects the performance of NARX neural networks. In Proceedings of the IEEE International Joint Conference on Neural Networks Proceedings, IEEE World Congress on Computational Intelligence (Cat. No. 98CH36227), Anchorage, AK, USA, 4–9 May 1998. [Google Scholar]
- Isqeel, A.A.; Eyiomika, S.M.J.; Ismaeel, T.B. Consumer Load Prediction Based on NARX for Electricity Theft Detection. In Proceedings of the International Conference on Computer and Communication Engineering (ICCCE), Kuala Lumpur, Malaysia, 26–27 July 2016. [Google Scholar]
- Dzielinski, A. NARX Models Application to Model Based Nonlinear Control. 1999. Available online: https://mathweb.ucsd.edu/~helton/MTNSHISTORY/CONTENTS/2000PERPIGNAN/CDROM/articles/SI20A_2.pdf (accessed on 31 August 2022).
- Lin, T.N.; Giles, C.L.; Horne, B.G.; Kung, S.Y. A delay damage model selection algorithm for NARX neural networks. IEEE Trans. Signal Process. 1997, 45, 2719–2730. [Google Scholar]
- Menezes, J.M.P., Jr.; Barreto, G.A. Long-term time series prediction with the NARX network: An empirical evaluation. Neurocomputing 2008, 71, 3335–3343. [Google Scholar] [CrossRef]
- Rusinov, L.A.; Rudakova, I.V.; Remizova, O.A.; Kurkina, V.V. Fault diagnosis in chemical processes with application of hierarchical neural networks. Chemom. Intell. Lab. Syst. 2009, 97, 98–103. [Google Scholar] [CrossRef]
- Diaconescu, E. The use of NARX neural networks to predict chaotic time series. Wseas Trans. Comput. Res. 2008, 3, 182–191. [Google Scholar]
- Inaoka, H.; Kobayashi, K.; Nebuya, S.; Kumagai, H.; Tsuruta, H.; Fukuoka, Y. Derivation of NARX models by expanding activation functions in neural networks. IEEJ Trans. Electr. Electron. Eng. 2019, 14, 1209–1218. [Google Scholar] [CrossRef]
- Banihabib, M.E.; Ahmadian, A.; Valipour, M. Hybrid MARMA-NARX model for flow forecasting based on the large-scale climate signals, sea-surface temperatures, and rainfall. Hydrol. Res. 2018, 49, 1788–1803. [Google Scholar] [CrossRef]
- Gale, N.K.; Heath, G.; Cameron, E.; Rashid, S.; Redwood, S. Using the framework method for the analysis of qualitative data in multi-disciplinary health research. BMC Med. Res. Methodol. 2013, 13, 1–8. [Google Scholar] [CrossRef]
- Sun, W.; Cai, Z.; Li, Y.; Liu, F.; Fang, S.; Wang, G. Data processing and text mining technologies on electronic medical records: A review. J. Healthc. Eng. 2018, 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Deng, Y.; Li, X.; Naumann, T.; Luo, Y. Natural language processing for EHR-based computational phenotyping. IEEE/ACM Trans. Comput. Biol. Bioinform. 2018, 16, 139–153. [Google Scholar] [CrossRef] [PubMed]
- Jutte, D.P.; Roos, L.L.; Brownell, M.D. Administrative record linkage as a tool for public health research. Annu. Rev. Public Health 2011, 32, 91–108. [Google Scholar] [CrossRef]
- Hellewell, J.; Abbott, S.; Gimma, A.; Bosse, N.I.; Jarvis, C.I.; Russell, T.W.; Munday, J.D.; Kucharski, A.J.; Edmunds, W.J.; Sun, F.; et al. Feasibility of controlling COVID-19 outbreaks by isolation of cases and contacts. Lancet Glob. Health 2020, 8, e488–e496. [Google Scholar] [CrossRef]
- Nakano, J.; Hashizume, K.; Fukushima, T.; Ueno, K.; Matsuura, E.; Ikio, Y.; Ishii, S.; Morishita, S.; Tanaka, K.; Kusuba, Y. Effects of aerobic and resistance exercises on physical symptoms in cancer patients: A meta-analysis. Integr. Cancer Ther. 2018, 17, 1048–1058. [Google Scholar] [CrossRef]
- Koonin, L.M. Novel coronavirus disease (COVID-19) outbreak: Now is the time to refresh pandemic plans. J. Bus. Contin. Emerg. Plan. 2020, 13, 298–312. [Google Scholar]
- Ågerfalk, P.J.; Conboy, K.; Myers, M.D. Information systems in the age of pandemics: COVID-19 and beyond. Eur. J. Inf. Syst. 2020, 29, 203–207. [Google Scholar] [CrossRef]
- Mosavi, N.S.; Santos, M.F. How prescriptive analytics influences decision making in precision medicine. Procedia Comput. Sci. 2020, 177, 528–533. [Google Scholar] [CrossRef]
- Harper, A.; Mustafee, N. Proactive service recovery in emergency departments: A hybrid modelling approach using forecasting and real-time simulation. In Proceedings of the SIGSIM Principles of Advanced Discrete Simulation, Chicago, IL, USA, 29 May 2019. [Google Scholar]
- Mbuh, M.J.; Metzger, P.; Brandt, P.; Fika, K.; Slinkey, M. Application of real-time GIS analytics to support spatial intelligent decision-making in the era of big data for smart cities. EAI Endorsed Trans. Smart Cities 2019, 4, e3. [Google Scholar] [CrossRef]
- Liu, J.; Khattak, A. Informed decision-making by integrating historical on-road driving performance data in high-resolution maps for connected and automated vehicles. J. Intell. Transp. Syst. 2020, 24, 11–23. [Google Scholar] [CrossRef]
- Konchak, C.W.; Krive, J.; Au, L.; Chertok, D.; Dugad, P.; Granchalek, G.; Livschiz, E.; Mandala, R.; McElvania, E.; Park, C.; et al. From testing to decision-making: A data-driven analytics COVID-19 response. Acad. Pathol. 2021, 8, 23742895211010257. [Google Scholar] [CrossRef] [PubMed]
- Bousdekis, A.; Lepenioti, K.; Apostolou, D.; Mentzas, G. A review of data-driven decision-making methods for industry 4.0 maintenance applications. Electronics 2021, 10, 828. [Google Scholar] [CrossRef]
- Lugaresi, G.; Matta, A. Real-time simulation in manufacturing systems: Challenges and research directions. In Proceedings of the Winter Simulation Conference (WSC), Gothenburg, Sweden, 9–12 December 2018. [Google Scholar]
- Liang, J.; Li, Y.; Zhang, Z.; Shen, D.; Xu, J.; Zheng, X.; Wang, T.; Tang, B.; Lei, J.; Zhang, J. Adoption of Electronic Health Records (EHRs) in China during the past 10 years: Consecutive survey data analysis and comparison of sino-american challenges and experiences. J. Med. Internet Res. 2021, 23, e24813. [Google Scholar] [CrossRef]
- Wynberg, E.; van Willigen, H.D.; Dijkstra, M.; Boyd, A.; Kootstra, N.A.; van den Aardweg, J.G.; van Gils, M.J.; Matser, A.; de Wit, M.R.; Leenstra, T.; et al. Evolution of coronavirus disease 2019 (COVID-19) symptoms during the first 12 months after illness onset. Clin. Infect. Dis. 2022, 75, e482–e490. [Google Scholar] [CrossRef]
- Austin, P.C. A critical appraisal of propensity—score matching in the medical literature between 1996 and 2003. Stat. Med. 2008, 27, 2037–2049. [Google Scholar] [CrossRef]
- Brookhart, M.A.; Schneeweiss, S.; Rothman, K.J.; Glynn, R.J.; Avorn, J.; Stürmer, T. Variable selection for propensity score models. Am. J. Epidemiol. 2006, 163, 1149–1156. [Google Scholar] [CrossRef]
- Carfì, A.; Bernabei, R.; Landi, F. Persistent symptoms in patients after acute COVID-19. JAMA 2020, 324, 603–605. [Google Scholar] [CrossRef]
- Lopez-Leon, S.; Wegman-Ostrosky, T.; Perelman, C.; Sepulveda, R.; Rebolledo, P.A.; Cuapio, A.; Villapol, S. More than 50 long-term effects of COVID-19: A systematic review and meta-analysis. Sci. Rep. 2021, 11, 16144. [Google Scholar] [CrossRef]
- Salamanna, F.; Veronesi, F.; Martini, L.; Landini, M.P.; Fini, M. Post-COVID-19 syndrome: The persistent symptoms at the post-viral stage of the disease. A systematic review of the current data. Front. Med. 2021, 8, 653516. [Google Scholar] [CrossRef] [PubMed]
- Law, T.H.; Ng, C.P.; Poi, A.W.H. The sources of the Kuznets relationship between the COVID-19 mortality rate and economic performance. Int. J. Disaster Risk Reduct. 2022, 81, 103233. [Google Scholar] [CrossRef] [PubMed]
- Sze, S.; Pan, D.; Nevill, C.R.; Gray, L.J.; Martin, C.A.; Nazareth, J.; Minhas, J.S.; Divall, P.; Khunti, K.; Abrams, K.R.; et al. Ethnicity and clinical outcomes in COVID-19: A systematic review and meta-analysis. E Clin. Med. 2020, 29, 100630. [Google Scholar] [CrossRef]
- Sayers, A.; Ben-Shlomo, Y.; Blom, A.W.; Steele, F. Probabilistic record linkage. Int. J. Epidemiol. 2016, 45, 954–964. [Google Scholar] [CrossRef]
- Zhao, Y.J.; Xing, X.; Tian, T.; Wang, Q.; Liang, S.; Wang, Z.; Cheung, T.; Su, Z.; Tang, Y.L.; Ng, C.H.; et al. Post COVID-19 mental health symptoms and quality of life among COVID-19 frontline clinicians: A comparative study using propensity score matching approach. Transl. Psychiatry 2022, 12, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Jaro, M.A. Probabilistic linkage of large public health data files. Stat. Med. 1995, 14, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Fellegi, I.P.; Sunter, A.B. A theory for record linkage. J. Am. Stat. Assoc. 1969, 64, 1183–1210. [Google Scholar] [CrossRef]
- Yassin, I.M.; Zabidi, A.; Ali, M.S.; Baharom, R. PSO-Optimized COVID-19 MLP-NARX Mortality Prediction Model. In Proceedings of the IEEE Industrial Electronics and Applications Conference (IEACon), Penang, Malaysia, 22–23 November 2021. [Google Scholar]
- Peng, C.C.; Yeh, C.W.; Wang, J.G.; Wang, S.H.; Huang, C.W. Prediction of LME lead spot price by neural network and NARX model. In Proceedings of the 2nd Eurasia Conference on Biomedical Engineering, Healthcare and Sustainability (ECBIOS), Tainan, Taiwan, 29–31 May 2020. [Google Scholar]
- Bhattacharjee, U.; Chakraborty, M. NARX-Wavelet Based Active Model for Removing Motion Artifacts from ECG. In Proceedings of the International Conference on Computer, Electrical & Communication Engineering (ICCECE), Kolkata, India, 17–18 January 2020. [Google Scholar]
- Wei, H.L. Sparse, interpretable and transparent predictive model identification for healthcare data analysis. In Proceedings of the International Work-Conference on Artificial Neural Networks, Gran Canaria, Spain, 12–14 June 2019. [Google Scholar]
- Chen, S.; Billings, S.A.; Grant, P.M. Non-linear system identification using neural networks. Int. J. Control 1990, 51, 1191–1214. [Google Scholar] [CrossRef]
- Kumpati, S.N.; Kannan, P. Identification and control of dynamical systems using neural networks. IEEE Trans. Neural Netw. 1990, 1, 4–27. [Google Scholar]
- Ljung, L.; Söderström, T. Theory and Practice of Recursive Identification; MIT Press: Cambridge, MA, USA, 1983. [Google Scholar]
- Zemouri, R.; Gouriveau, R.; Zerhouni, N. Defining and applying prediction performance metrics on a recurrent NARX time series model. Neurocomputing 2010, 73, 2506–2521. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, S.; Li, F.; Liu, Z. Fault detection and diagnosis method for cooling dehumidifier based on LS-SVM NARX model. Int. J. Refrig. 2016, 61, 69–81. [Google Scholar] [CrossRef]
- Kong, S.; Li, C.; He, S.; Çiçek, S.; Lai, Q. A memristive map with coexisting chaos and hyperchaos. Chin. Phys. B 2021, 30, 110502. [Google Scholar] [CrossRef]
- Polato, M.; Lauriola, I.; Aiolli, F. A novel boolean kernels family for categorical data. Entropy 2018, 20, 444. [Google Scholar] [CrossRef] [PubMed]
- Bisong, E. Logistic regression. In Building Machine Learning and Deep Learning Models on Google Cloud Platform; Apress: Berkeley, CA, USA, 2019; pp. 243–250. [Google Scholar]
- Du Toit, C.F. The numerical computation of Bessel functions of the first and second kind for integer orders and complex arguments. IEEE Trans. Antennas Propag. 1990, 38, 1341–1349. [Google Scholar] [CrossRef]
- Flynn, J.; Broxton, M.; Debevec, P.; DuVall, M.; Fyffe, G.; Overbeck, R.; Snavely, N.; Tucker, R. Deepview: View synthesis with learned gradient descent. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019. [Google Scholar]
- Al Hamad, M.; Zeki, A.M. Accuracy vs. cost in decision trees: A survey. In Proceedings of the International Conference on Innovation and Intelligence for Informatics, Computing, and Technologies (3ICT), Sakhier, Bahrain, 18–20 November 2018. [Google Scholar]
- Krauss, C.; Do, X.A.; Huck, N. Deep neural networks, gradient-boosted trees, random forests: Statistical arbitrage on the S&P 500. Eur. J. Oper. Res. 2017, 259, 689–702. [Google Scholar]
- Bhuvaneswari, R.; Kalaiselvi, K. Naive Bayesian classification approach in healthcare applications. Int. J. Comput. Sci. Telecommun. 2012, 3, 106–112. [Google Scholar]
- Vembandasamy, K.; Sasipriya, R.; Deepa, E. Heart diseases detection using Naive Bayes algorithm. Int. J. Innov. Sci. Eng. Technol. 2015, 2, 441–444. [Google Scholar]
- Jadhav, S.D.; Channe, H.P. Comparative study of K-NN, naive Bayes and decision tree classification techniques. Int. J. Sci. Res. IJSR 2016, 5, 1842–1845. [Google Scholar]
- Sornalakshmi, M.; Balamurali, S.; Venkatesulu, M.; Navaneetha Krishnan, M.; Ramasamy, L.K.; Kadry, S.; Manogaran, G.; Hsu, C.H.; Muthu, B.A. Hybrid method for mining rules based on enhanced Apriori algorithm with sequential minimal optimization in healthcare industry. Neural Comput. Appl. 2020, 34, 10597–10610. [Google Scholar] [CrossRef]
- Jothi, N.; Husain, W. Data mining in healthcare—A review. Procedia Comput. Sci. 2015, 72, 306–313. [Google Scholar] [CrossRef]
- Manogaran, G.; Lopez, D. Health data analytics using scalable logistic regression with stochastic gradient descent. Int. J. Adv. Intell. Paradig. 2018, 10, 118–132. [Google Scholar] [CrossRef]
- Demir, E. A decision support tool for predicting patients at risk of readmission: A comparison of classification trees, logistic regression, generalized additive models, and multivariate adaptive regression splines. Decis. Sci. 2014, 45, 849–880. [Google Scholar] [CrossRef]
- Khan, N.; Gaurav, D.; Kandl, T. Performance evaluation of Levenberg-Marquardt technique in error reduction for diabetes condition classification. Procedia Comput. Sci. 2013, 18, 2629–2637. [Google Scholar] [CrossRef]
- McCormick, M.; Rubert, N.; Varghese, T. Bayesian regularization applied to ultrasound strain imaging. IEEE Trans. Biomed. Eng. 2011, 58, 1612–1620. [Google Scholar] [CrossRef]
- Paul, B.; Karn, B. Heart Disease Prediction Using Scaled Conjugate Gradient Back Propagation of Artificial Neural Network. 2022. Available online: https://www.researchsquare.com/article/rs-1490110/latest.pdf (accessed on 31 August 2022).
- Taqvi, S.A.; Tufa, L.D.; Zabiri, H.; Maulud, A.S.; Uddin, F. Fault detection in distillation column using NARX neural network. Neural Comput. Appl. 2020, 32, 3503–3519. [Google Scholar] [CrossRef]
- Nishiga, M.; Wang, D.W.; Han, Y.; Lewis, D.B.; Wu, J.C. COVID-19 and cardiovascular disease: From basic mechanisms to clinical perspectives. Nat. Rev. Cardiol. 2020, 17, 543–558. [Google Scholar] [CrossRef]
- Mistry, S.; Wang, L. Efficient Prediction of Heart Disease Using Cross Machine Learning Techniques. In Proceedings of the IEEE Asia-Pacific Conference on Image Processing, Electronics and Computers (IPEC), Dalian, China, 14–16 April 2022. [Google Scholar]
- Shima, D.; Ii, Y.; Yamamoto, Y.; Nagayasu, S.; Ikeda, Y.; Fujimoto, Y. A retrospective, cross-sectional study of real-world values of cardiovascular risk factors using a healthcare database in Japan. BMC Cardiovasc. Disord. 2014, 14, 120. [Google Scholar] [CrossRef]

| Algorithm | Number of Neurons in the Hidden Layer | Test Performance (MSE) |
|---|---|---|
| Levenberg–Marquardt | 5 | |
| Levenberg–Marquardt | 10 | |
| Bayesian Regularization | 5 | |
| Bayesian Regularization | 10 | |
| Scaled Conjugate Gradient | 5 | |
| Scaled Conjugate Gradient | 10 |
| Case | N | Marginal Percentage | |
|---|---|---|---|
| Mortality | Survivor | 351 | 61.6% |
| Expired | 219 | 38.4% | |
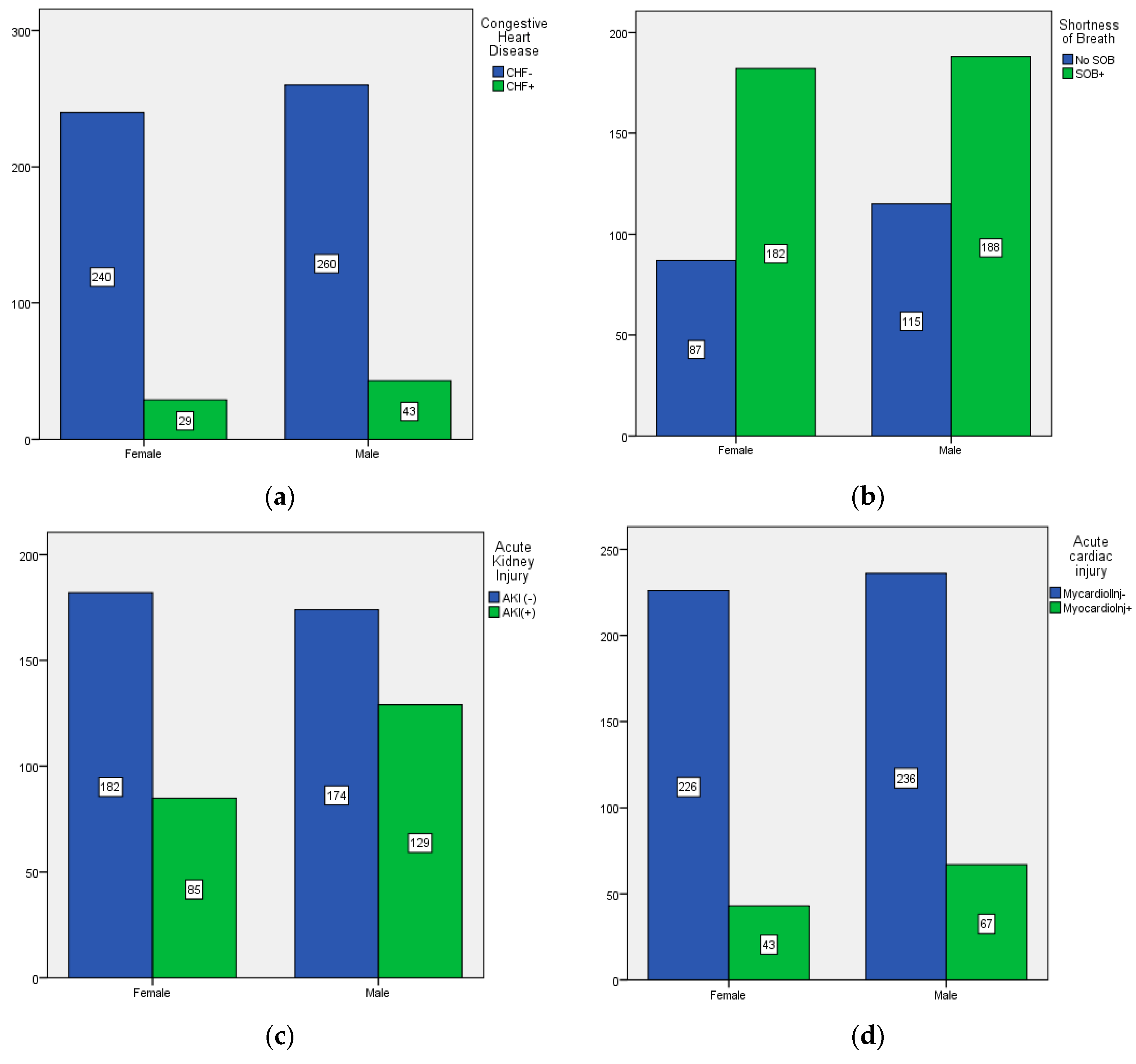
| Congestive Heart Disease | CHF− | 498 | 87.4% |
| CHF+ | 72 | 12.6% | |
| Chronic Obstructive Lung Disease | COPD(−) | 525 | 92.1% |
| COPD(+) | 45 | 7.9% | |
| Asthma | Asthma− | 523 | 91.8% |
| Asthma+ | 47 | 8.2% | |
| Smoking History | SmokeHx− | 489 | 85.8% |
| SmokeHx+ | 81 | 14.2% | |
| Acute cardiac injury | MycardiolInj− | 460 | 80.7% |
| MyocardioInj+ | 110 | 19.3% | |
| Acute Kidney Injury | AKI(−) | 356 | 62.5% |
| AKI(+) | 214 | 37.5% | |
| Valid | 570 | 100.0% | |
| Missing | 2 | - | |
| Total | 572 | - | |
| Subpopulation | 42 a | - | |
| Model | Model Fitting Criteria | Likelihood Ratio Tests | ||
|---|---|---|---|---|
| −2 Log Likelihood | Chi-Square | df | Sig. | |
| Intercept Only | 184.687 | |||
| Final | 107.986 | 76.701 | 6 | 0.000 |
| Effect | Model Fitting Criteria | Likelihood Ratio Tests | ||
|---|---|---|---|---|
| −2 Log Likelihood of Reduced Model | Chi-Square | df | Sig. | |
| Intercept | 107.986 a | 0.000 | 0 | - |
| CHF | 109.536 | 1.551 | 1 | 0.213 |
| COPD | 110.377 | 2.391 | 1 | 0.122 |
| ASTHMA | 107.986 | 0.000 | 1 | 0.988 |
| Smoking | 108.453 | 0.467 | 1 | 0.494 |
| Acute cardiac injury | 131.434 | 23.449 | 1 | 0.000 |
| AKI | 146.448 | 38.463 | 1 | 0.000 |
| Mortality a | B | Std. Error | Wald | df | Sig. | Exp(B) | 95% Confidence Interval for Exp(B) | ||
|---|---|---|---|---|---|---|---|---|---|
| Lower Bound | Upper Bound | ||||||||
| Survivor | Intercept | −1.131 | 0.525 | 4.649 | 1 | 0.031 | - | - | - |
| [CHF = 0.00] | −0.366 | 0.298 | 1.515 | 1 | 0.218 | 0.693 | 0.387 | 1.242 | |
| [CHF = 1.00] | 0 b | - | - | 0 | - | - | - | - | |
| [COPD = 0.000] | 0.549 | 0.355 | 2.391 | 1 | 0.122 | 1.732 | 0.863 | 3.473 | |
| [COPD = 1.00] | 0 b | - | - | 0 | - | - | - | - | |
| [ASTHMA = 0.00] | 0.005 | 0.338 | 0.000 | 1 | 0.988 | 1.005 | 0.518 | 1.949 | |
| [ASTHMA = 1.00] | 0 b | - | - | 0 | - | - | - | - | |
| [Smoking = 0.00] | −0.195 | 0.286 | 0.463 | 1 | 0.496 | 0.823 | 0.470 | 1.442 | |
| [Smoking = 1.00] | 0 b | - | - | 0 | - | - | - | - | |
| [Acutecardiacinjury = 0.00] | 1.124 | 0.236 | 22.720 | 1 | 0.000 | 3.077 | 1.938 | 4.884 | |
| [Acutecardiacinjury = 1.00] | 0 b | - | - | 0 | - | - | - | - | |
| [AKI = 0.00] | 1.152 | 0.188 | 37.693 | 1 | 0.000 | 3.164 | 2.190 | 4.569 | |
| [AKI = 1.00] | 0 b | - | - | 0 | - | - | - | - | |
| Case | Cluster | |
|---|---|---|
| 1 | 2 | |
| Shortness of Breath | 0.00 | 1.00 |
| Chronic Obstructive Lung Disease | 1.00 | 0.00 |
| Asthma | 0.00 | 1.00 |
| Smoking History | 1.00 | 0.00 |
| Acute cardiac injury | 0.00 | 1.00 |
| Ventilation | 0.00 | 1.00 |
| ICU Admission | 0.00 | 1.00 |
| Clinical status on last day 5/12 | 0.00 | 5.00 |
| Case | Cluster | |
|---|---|---|
| 1 | 2 | |
| Shortness of Breath | 0.63 | 0.80 |
| Chronic Obstructive Lung Disease | 0.00 | 0.00 |
| Asthma | 0.08 | 0.11 |
| Smoking History | 0.14 | 0.11 |
| Acute cardiac injury | 0.00 | 0.00 |
| Ventilation | 0.22 | 0.48 |
| ICU Admission | 0.17 | 0.50 |
| Clinical status on last day | 0.42 | 5.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mistry, S.; Wang, L.; Islam, Y.; Osei, F.A.J. A Comprehensive Study on Healthcare Datasets Using AI Techniques. Electronics 2022, 11, 3146. https://doi.org/10.3390/electronics11193146
Mistry S, Wang L, Islam Y, Osei FAJ. A Comprehensive Study on Healthcare Datasets Using AI Techniques. Electronics. 2022; 11(19):3146. https://doi.org/10.3390/electronics11193146
Chicago/Turabian StyleMistry, Sunit, Lili Wang, Yousuf Islam, and Frimpong Atta Junior Osei. 2022. "A Comprehensive Study on Healthcare Datasets Using AI Techniques" Electronics 11, no. 19: 3146. https://doi.org/10.3390/electronics11193146
APA StyleMistry, S., Wang, L., Islam, Y., & Osei, F. A. J. (2022). A Comprehensive Study on Healthcare Datasets Using AI Techniques. Electronics, 11(19), 3146. https://doi.org/10.3390/electronics11193146









