Diagnosis of Breast Cancer with Strongly Supervised Deep Learning Neural Network
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
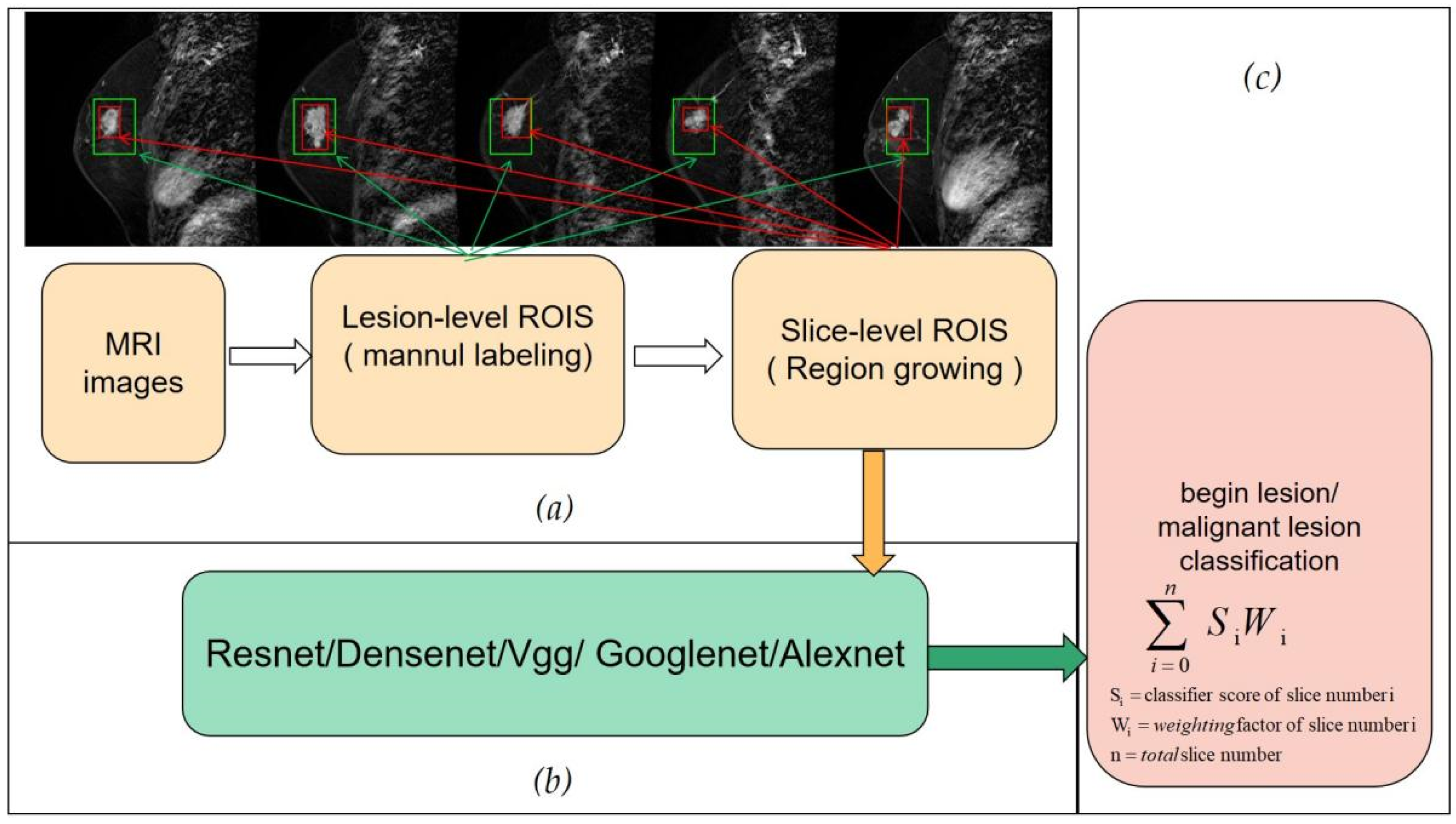
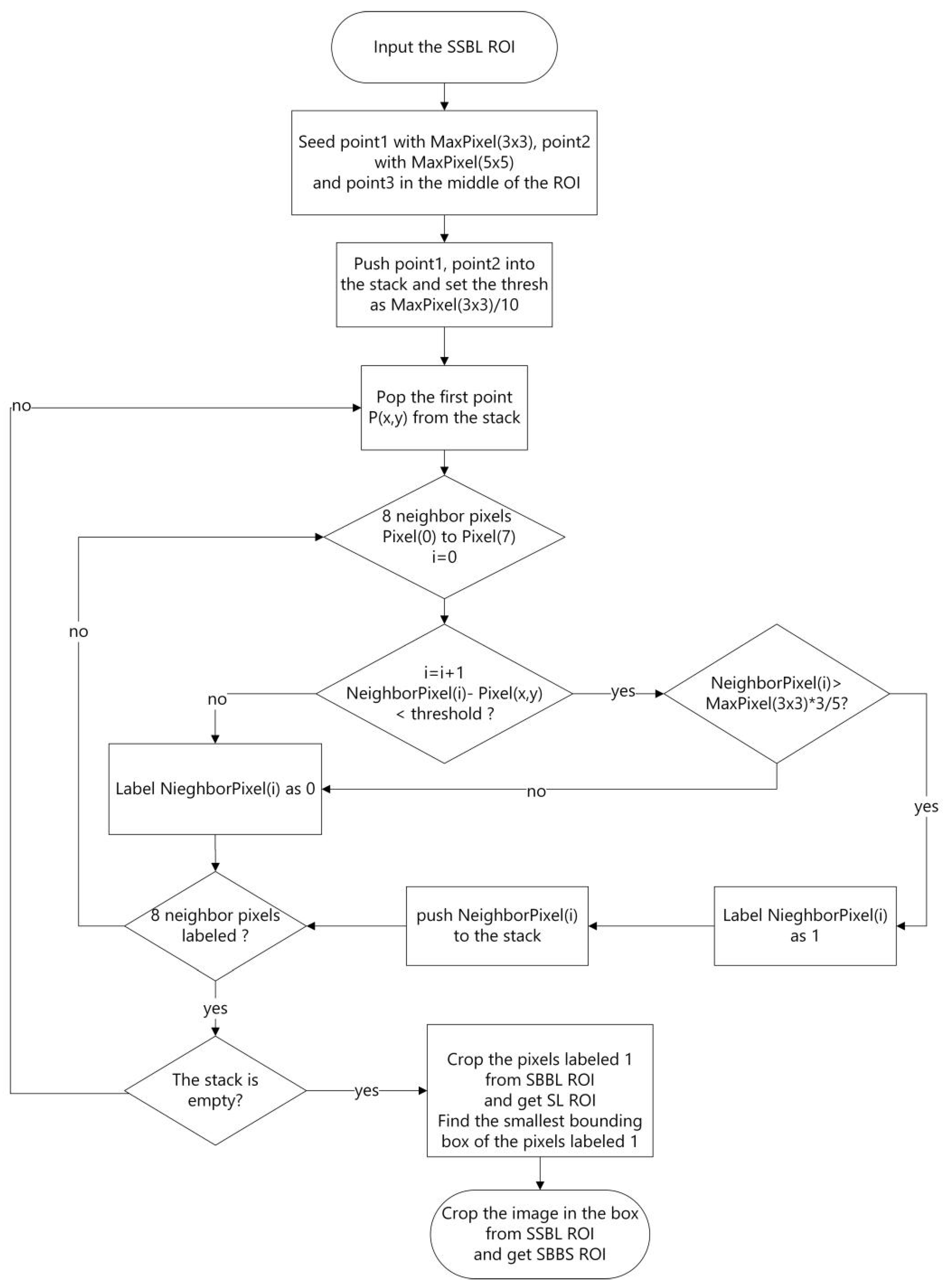
2.2.1. Slice-Level ROI
2.2.2. Slice-Level Training and Classification
2.2.3. Lesion-Level Diagnosis
3. Results
3.1. Performance Metrics
- True positive (TP): predicted positive in positive samples (malignant lesions).
- True negative (TN): predicted negative in negative samples (benign lesions).
- False positive (FP): predicted positive in negative samples.
- False negative (FN): predicted negative in positive samples.
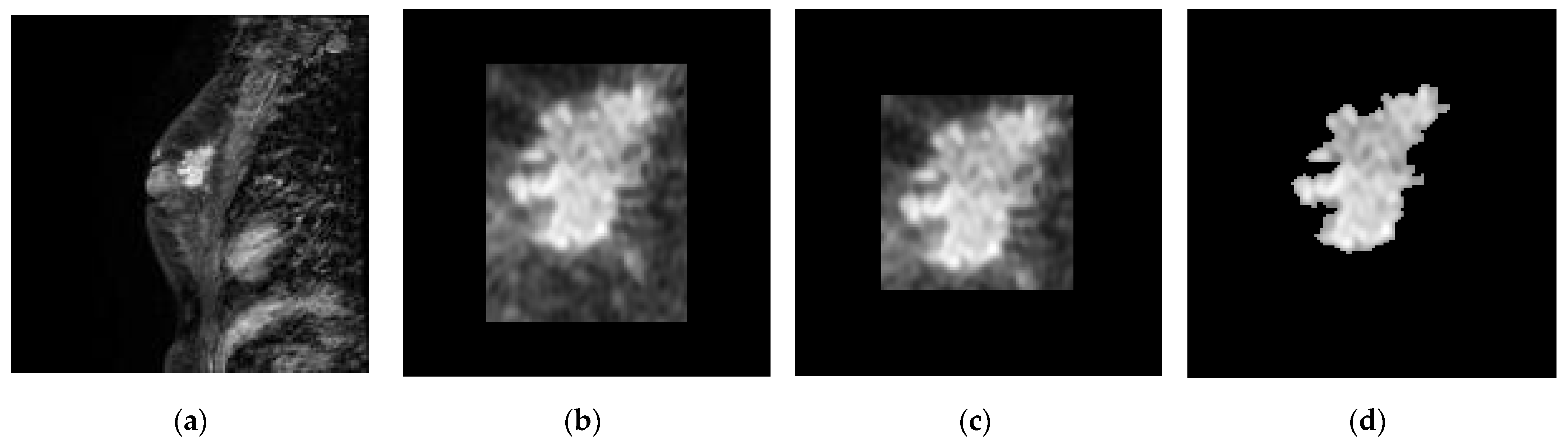
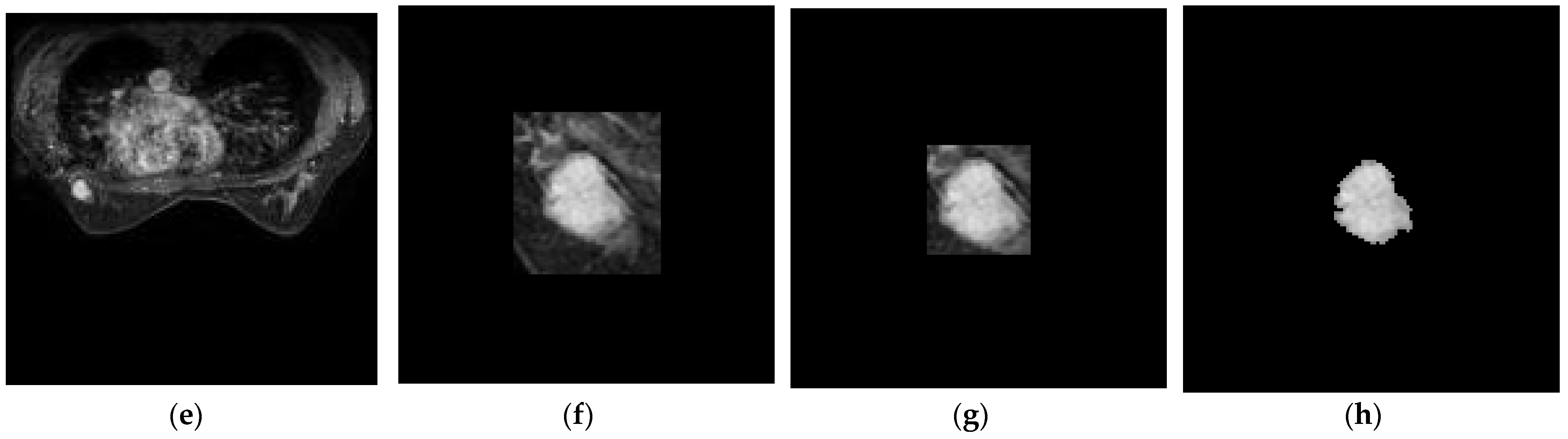
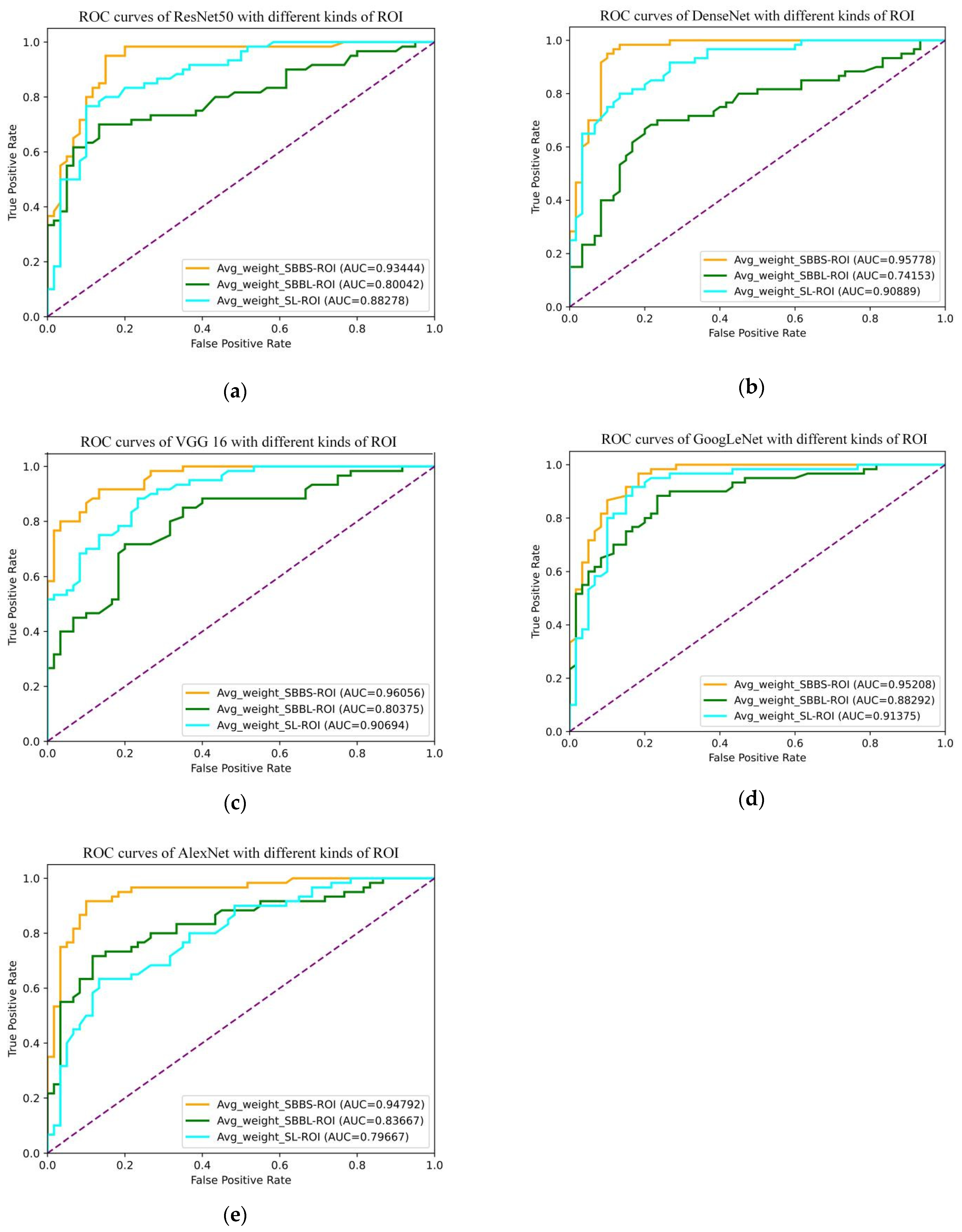
3.2. Slice-Level ROI
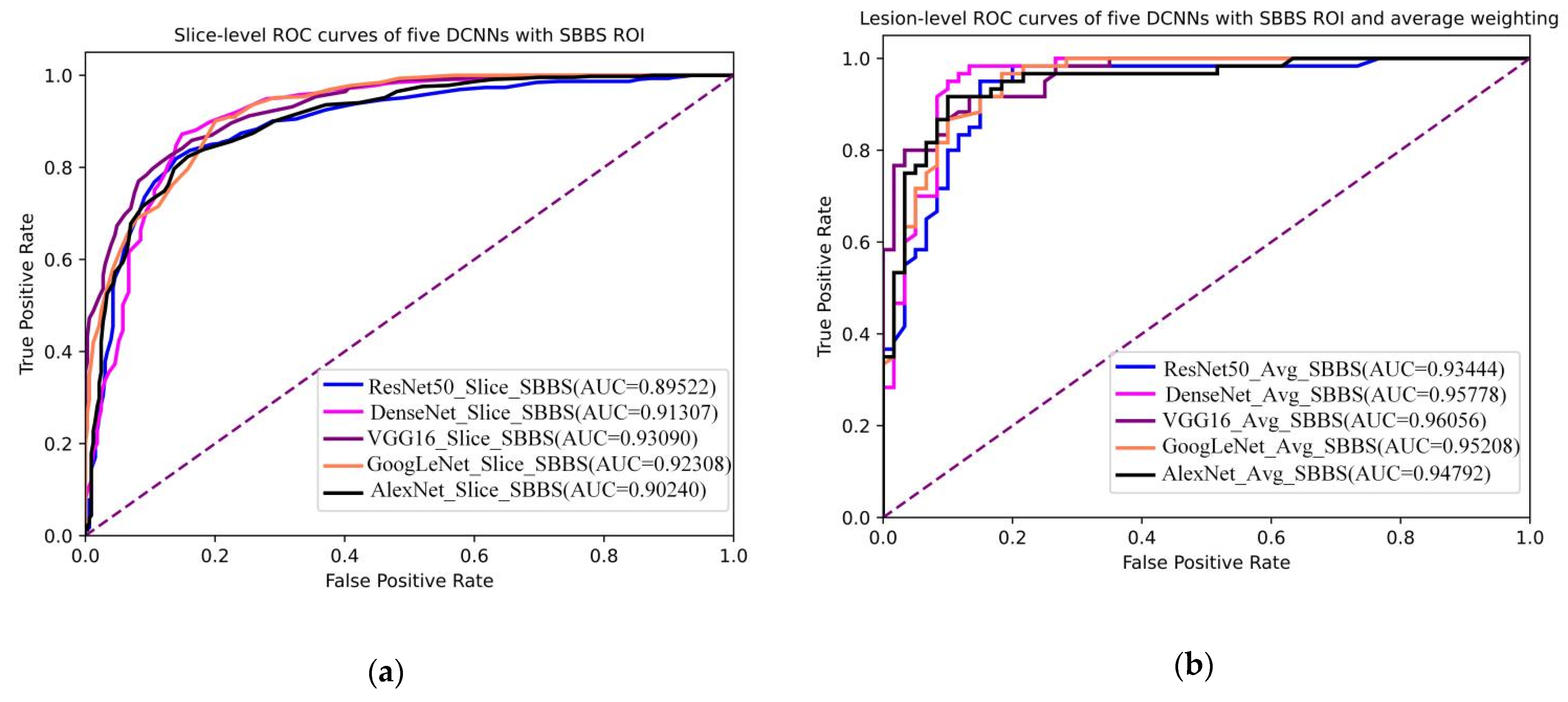
3.3. Lesion-Level Diagnosis
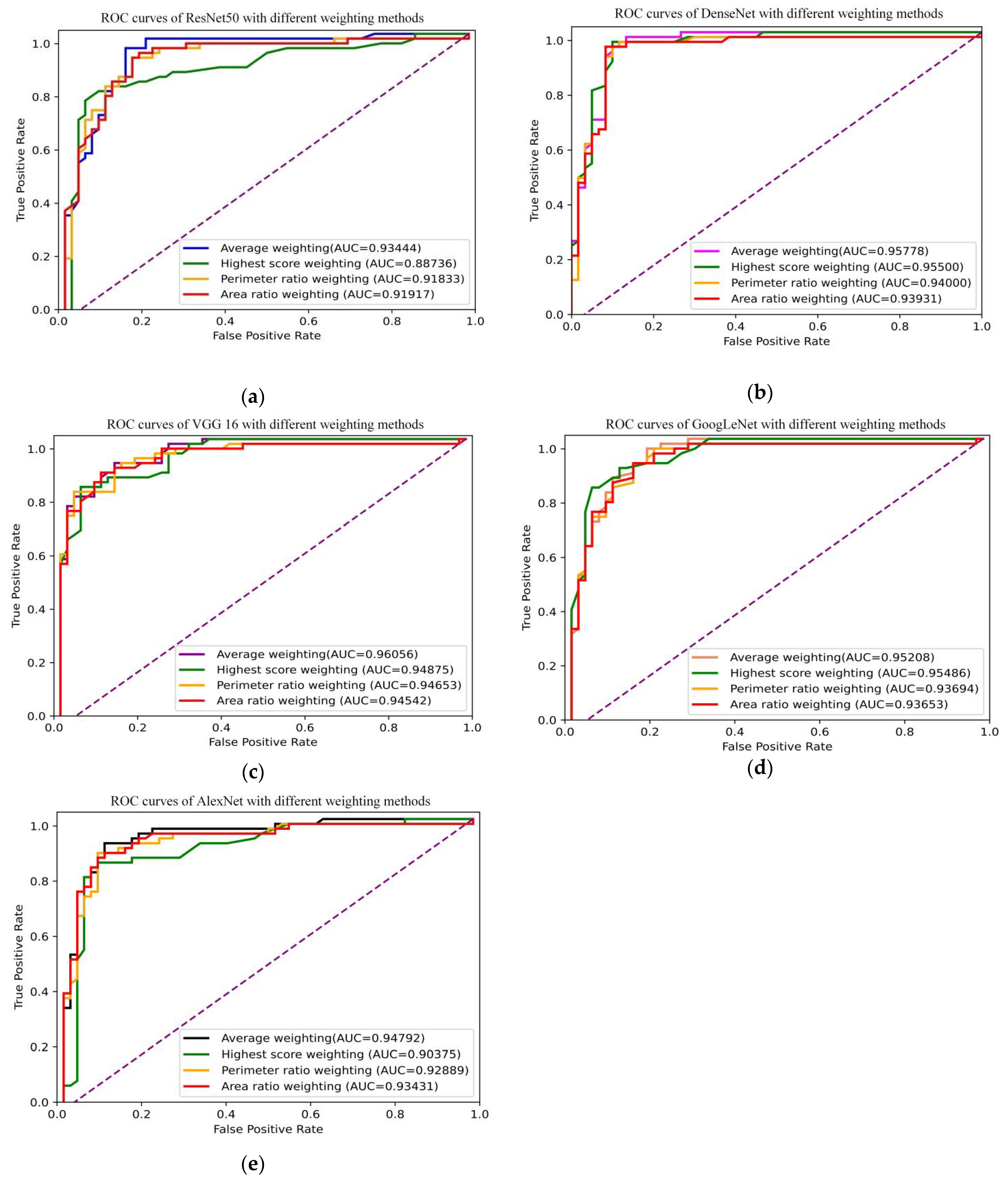
3.4. DCNNs Combined with Slice-Level ROI and Lesion-Level Diagnosis
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Latest Global Cancer Data: Cancer Burden Rises to 19.3 Million New Cases and 10.0 Million Cancer Deaths in 2020. Available online: https://www.iarc.fr/fr/news-events/ (accessed on 15 December 2020).
- DeSantis, C.E.; Ma, J.; Goding Sauer, A.; Newman, L.A.; Jemal, A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA A Cancer J. Clin. 2017, 67, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Jaglan, P.; Dass, R.; Duhan, M. Breast Cancer Detection Techniques: Issues and Challenges. J. Inst. Eng. India Ser. B 2019, 100, 379–386. [Google Scholar] [CrossRef]
- Elmore, J.G.; Armstrong, K.; Lehman, C.D.; Fletcher, S.W. Screening for breast cancer. JAMA 2005, 293, 1245–1256. [Google Scholar] [CrossRef] [PubMed]
- Zeeshan, M.; Salam, B.; Khalid, Q.S.B.; Sayani, R. Diagnostic Accuracy of Digital Mammography in the Detection of Breast Cancer. Cureus 2018, 10, e2448. [Google Scholar] [CrossRef] [PubMed]
- Pisano, E.D.; Gatsonis, C.; Hendrick, E.; Yaffe, M.; Baum, J.K.; Acharyya, S.; Conant, E.F.; Fajardo, L.L.; Bassett, L.; D’Orsi, C.; et al. Diagnostic Performance of Digital versus Film Mammography for Breast-Cancer Screening. N. Engl. J. Med. 2005, 353, 1773–1783. [Google Scholar] [CrossRef] [PubMed]
- Türkbey, B.; Thomasson, D.; Pang, Y.; Bernardo, M.; Choyke, P.L. The role of dynamic contrast-enhanced MRI in cancer diagnosis and treatment. Diagn. Interv. Radiol. 2010, 16, 186–192. [Google Scholar] [PubMed]
- Liu, P.-F.; Krestin, G.P.; Huch, R.A.; Göhde, S.C.; Caduff, R.F.; Debatin, J.F. MRI of the uterus, uterine cervix, and vagina: Diagnostic performance of dynamic contrast-enhanced fast multiplanar gradient-echo imaging in comparison with fast spin-echo T2-weighted pulse imaging. Eur. Radiol. 1998, 8, 1433–1440. [Google Scholar] [CrossRef]
- Jager, G.J.; Ruijter, E.T.; Van De Kaa, C.A.; De La Rosette, J.J.; Oosterhof, G.O.; Thornbury, J.R.; Ruijs, S.H.; Barentsz, J.O. Dynamic TurboFLASH subtraction technique for contrast-enhanced MR imaging of the prostate: Correlation with histopathologic results. Radiology 1997, 203, 645–652. [Google Scholar] [CrossRef]
- Ocak, I.; Bernardo, M.; Metzger, G.; Barrett, T.; Pinto, P.; Albert, P.S.; Choyke, P.L. Dynamic contrast-enhanced MRI of prostate cancer at 3 T: A study of pharmacokinetic parameters. AJR Am. J. Roentgenol. 2007, 189, 192–201. [Google Scholar] [CrossRef]
- Tartar, M.; Comstock, C.E.; Kipper, M.S. CHAPTER 2—Evaluation of the Symptomatic Patient: Diagnostic Breast Imaging. Breast Cancer Imaging 2008, 2008, 38–75. [Google Scholar]
- Wei, M.; Du, Y.; Wu, X.; Su, Q.; Zhu, J.; Zheng, L.; Lv, G.; Zhuang, J. A benign and Malignant Breast Tumor Classification Method via Efficiently Combining Texture and Morphological Features on Ultrasound Images. Comput. Math. Methods Med. 2020, 2020, 5894010. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Zhu, D.; Chen, D.; Li, L.; Shao, Y. A computer-aided diagnosis system for dynamic contrast-enhanced MR images based on level set segmentation and ReliefF feature selection. Comput. Math. Methods Med. 2015, 2015, 450531. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, J.; Gao, J.; Liu, S.; Liu, X.; Zhao, Z.; Guo, D.; Dan, G. A comprehensive hierarchical classification based on multi-features of breast DCE-MRI for cancer diagnosis. Med. Biol. Eng. Comput. 2020, 58, 2413–2425. [Google Scholar] [CrossRef]
- Cai, H.; Peng, Y.; Ou, C.; Chen, M.; Li, L. Diagnosis of Breast Masses from Dynamic Contrast-Enhanced and Diffusion-Weighted MR: A Machine Learning Approach. PLoS ONE 2014, 9, e87387. [Google Scholar] [CrossRef] [PubMed]
- Dalmiş, M.U.; Gubern-Mérida, A.; Vreemann, S.; Bult, P.; Karssemeijer, N.; Mann, R.; Teuwen, J. Artificial Intelligence–Based Classification of Breast Lesions Imaged with a Multiparametric Breast MRI Protocol with Ultrafast DCE-MRI, T2, and DWI. Investig. Radiol. 2019, 54, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Jiao, H.; Jiang, X.; Pang, Z.; Lin, X.; Huang, Y.; Li, L. Deep Convolutional Neural Networks-Based Automatic Breast Segmentation and Mass Detection in DCE-MRI. Comput. Math. Methods Med. 2020, 2020, 2413706. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.Z.; Swintelski, C.; Sun, S.; Siddique, M.; Desperito, E.; Jambawalikar, S.; Ha, R. Weakly Supervised Deep Learning Approach to Breast MRI Assessment. Acad. Radiol. 2021, 29, 166–172. [Google Scholar] [CrossRef]
- Zhou, J.; Luo, L.; Dou, Q.; Chen, H.; Chen, C.; Li, G.; Jiang, Z.; Heng, P.A. Weakly supervised 3D deep learning for breast cancer classification and localization of the lesions in MR images. J. Magn. Reson. Imaging 2019, 50, 1144–1151. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, Y.; Chang, K.; Lee, K.E.; Wang, O.; Li, J.; Lin, Y.; Pan, Z.; Chang, P.; Chow, D.; et al. Diagnosis of Benign and Malignant Breast Lesions on DCE-MRI by Using Radiomics and Deep Learning with Consideration of Peritumor Tissue. J. Magn. Reson. Imaging 2019, 51, 798–809. [Google Scholar] [CrossRef]
- He, K.; Zhang, Y.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Andrew Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. NIPS 2012, 60, 84–90. [Google Scholar]
- Hojjatoleslami, S.A.; Kittler, J. Region growing: A new approach. IEEE Trans. Image Process. A Publ. IEEE Signal Proces. Soc. 1998, 7, 1079–1084. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Whitney, H.M.; Giger, M.L. A deep learning methodology for improved breast cancer diagnosis using multiparametric MRI. Sci. Rep. 2020, 10, 10536. [Google Scholar] [CrossRef] [PubMed]
| References | Dataset | Classification Method Used | ROI | Features | Diagnosis Metrics | Limitations |
|---|---|---|---|---|---|---|
| [12] | Ultrasound images: 184 benign 264 malignant | SVM | Whole Ultrasound | Radiomics features | Lesion-level (one slice) | Professional instruments required Features chosen with subjective bias Discarded the other slices |
| [13] | DCE-MRI: 42 benign 78 malignant | SVM | Whole DCE-MRI | Radiomics features | Lesion-level (one slice) | Small dataset Professional instruments required Features chosen with subjective bias Discarded the other slices |
| [14] | DCE-MRI: 40 benign 73 malignant | SVM | Whole DCE-MRI | Radiomics features | Lesion-level (one slice) | Small dataset Professional instruments required Discarded the other slices |
| [15] | DCE-MRI: 85 benign 149 malignant | SVM | Whole DCE-MRI | DWI Features + Radiomics features | Lesion-level (one slice) | Small dataset Professional instruments required Discarded the other slices |
| [16] | DCE-MRI: 149 benign 368 malignant | Random Forest | Whole DCE-MRI | Radiomics features + Feature maps extracted by DCNN | Lesion-level (one slice) | Rough ROI affect the performance (the highest value was 0.85) Discarded the other slices |
| [17] | DCE-MRI: 75 lesions | Unet++ and Faster RCNN | Segmented breast | Feature maps extracted by DCNN | Lesion-level (one slice) | Small dataset Without diagnosis |
| [18] | DCE-MRI: 438 patients, 3167 slices | ResNet101 | Whole DCE-MRI | Feature maps extracted by DCNN | Slice-level | Rough ROI DCNN training and testing with slice-level data, can not stand for the performance in lesion-level diagnosis |
| [19] | DCE-MRI: 506 benign 1031 malignant | 3D DenseNet | Segmented breast | Feature maps extracted by DCNN | 3D lesion-level (one slice) | Rough ROI Discarded the other slices |
| [20] | DCE-MRI: 88 benign 139 malignant | ResNet50/Random Forest | Lesion level smallest bounding box | Feature maps extracted by DCNN | 3D lesion-level (slice with max score) | Lesion-level ROI Lesion-level diagnosis with the max score slice (Discarded the other slices) |
| Dataset | Training | Validation | Testing | Subtotal |
|---|---|---|---|---|
| Benign lesions | 162 | 45 | 60 | 267 |
| Malign lesions | 115 | 45 | 60 | 220 |
| Benign slices | 1278 | 259 | 329 | 1866 |
| Malign slices | 1053 | 341 | 453 | 1847 |
| Lesion Type | Lesion Name | Number |
|---|---|---|
| Malignant | ||
| Invasive cancer | 142 | |
| Ductal carcinoma in situ | 53 | |
| Semi-invasive carcinoma in situ | 8 | |
| Premium Executive Internal Cancer | 5 | |
| Invasive lobular carcinoma | 5 | |
| low-grade intraductal carcinoma | 1 | |
| Intermediate-grade intraductal carcinoma | 1 | |
| Intraductal papillary carcinoma | 1 | |
| Invasive intraductal carcinoma | 1 | |
| Squamous cell carcinoma | 1 | |
| Eczematous breast cancer | 1 | |
| Calcified intraductal carcinoma | 1 | |
| Benign | ||
| Fibroadenoma | 160 | |
| Fibrocystic breast disease | 39 | |
| Intraductal papilloma | 33 | |
| Adenopathy | 7 | |
| Papillary hyperplasia | 4 | |
| Hyperplastic nodule | 3 | |
| Lymphoma infiltration | 3 | |
| Lymphoma | 3 | |
| Chronic inflammation | 2 | |
| Fibroadenosis with lymphoid tissue | 2 | |
| Chronic granulomatous inflammation | 1 | |
| Complex sclerosing lesions | 1 | |
| Glandular hyperplasia with myoepithelial hyperplasia, | 1 | |
| Interstitial fibrous collagen hyperplasia | 1 | |
| Myoepithelial tumor | 1 | |
| Calcified nodules | 1 | |
| Dermal fibroma | 1 | |
| Compliant with BI-RADS Class II | 1 | |
| Compliant with BI-RADS Class III | 1 | |
| Fibroadenoma with tubular adenoma | 1 | |
| Epithelial hyperplasia | 1 |
| DCNN | ROI Type | Threshold | Sensitivity | Specificity | Accuracy | AUC |
|---|---|---|---|---|---|---|
| DenseNet | SBBS | 0.520 | 0.950 | 0.900 | 0.925 | 0.958 |
| SBBL | 0.500 | 0.700 | 0.767 | 0.733 | 0.742 | |
| SL | 0.500 | 0.800 | 0.867 | 0.833 | 0.909 | |
| VGG16 | SBBS | 0.480 | 0.917 | 0.867 | 0.892 | 0.961 |
| SBBL | 0.510 | 0.717 | 0.783 | 0.750 | 0.804 | |
| SL | 0.470 | 0.867 | 0.767 | 0.817 | 0.907 | |
| ResNet50 | SBBS | 0.460 | 0.950 | 0.850 | 0.900 | 0.934 |
| SBBL | 0.520 | 0.700 | 0.867 | 0.783 | 0.800 | |
| SL | 0.490 | 0.767 | 0.900 | 0.833 | 0.883 | |
| GoogLeNet | SBBS | 0.490 | 0.967 | 0.817 | 0.892 | 0.952 |
| SBBL | 0.450 | 0.883 | 0.767 | 0.825 | 0.883 | |
| SL | 0.500 | 0.867 | 0.883 | 0.850 | 0.914 | |
| AlexNet | SBBS | 0.470 | 0.917 | 0.900 | 0.909 | 0.948 |
| SBBL | 0.530 | 0.700 | 0.883 | 0.792 | 0.837 | |
| SL | 0.550 | 0.633 | 0.867 | 0.750 | 0.797 | |
| Average | SBBS | 0.484 | 0.940 | 0.867 | 0.903 | 0.951 |
| SBBL | 0.502 | 0.740 | 0.813 | 0.777 | 0.813 | |
| SL | 0.502 | 0.787 | 0.857 | 0.817 | 0.882 |
| DCNN | Weighting Methods | Threshold | Sensitivity | Specificity | Accuracy | AUC |
|---|---|---|---|---|---|---|
| ResNet50 | Average | 0.460 | 0.950 | 0.850 | 0.900 | 0.934 |
| Highest score | 0.440 | 0.800 | 0.900 | 0.850 | 0.887 | |
| Area ratio | 0.440 | 0.917 | 0.833 | 0.875 | 0.919 | |
| Perimeter ratio | 0.440 | 0.917 | 0.833 | 0.875 | 0.918 | |
| DenseNet | Average | 0.520 | 0.950 | 0.900 | 0.925 | 0.958 |
| Highest score | 0.530 | 0.967 | 0.900 | 0.933 | 0.955 | |
| Area ratio | 0.520 | 0.950 | 0.917 | 0.933 | 0.939 | |
| Perimeter ratio | 0.520 | 0.950 | 0.900 | 0.925 | 0.940 | |
| VGG16 | Average | 0.480 | 0.917 | 0.867 | 0.892 | 0.961 |
| Highest score | 0.570 | 0.830 | 0.950 | 0.892 | 0.949 | |
| Area ratio | 0.480 | 0.900 | 0.867 | 0.883 | 0.945 | |
| Perimeter ratio | 0.480 | 0.900 | 0.867 | 0.883 | 0.947 | |
| GoogLeNet | Average | 0.490 | 0.967 | 0.817 | 0.892 | 0.952 |
| Highest score | 0.600 | 0.900 | 0.883 | 0.892 | 0.959 | |
| Area ratio | 0.490 | 0.933 | 0.800 | 0.867 | 0.937 | |
| Perimeter ratio | 0.490 | 0.967 | 0.780 | 0.875 | 0.937 | |
| AlexNet | Average | 0.470 | 0.917 | 0.900 | 0.909 | 0.948 |
| Highest score | 0.480 | 0.850 | 0.917 | 0.883 | 0.904 | |
| Area ratio | 0.470 | 0.883 | 0.867 | 0.875 | 0.934 | |
| Perimeter ratio | 0.470 | 0.883 | 0.883 | 0.883 | 0.929 | |
| Average Score | Average | 0.484 | 0.940 | 0.867 | 0.903 | 0.951 |
| Highest score | 0.524 | 0.869 | 0.910 | 0.890 | 0.930 | |
| Area ratio | 0.480 | 0.917 | 0.857 | 0.887 | 0.935 | |
| Perimeter ratio | 0.480 | 0.923 | 0.853 | 0.888 | 0.934 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gui, H.; Su, T.; Pang, Z.; Jiao, H.; Xiong, L.; Jiang, X.; Li, L.; Wang, Z. Diagnosis of Breast Cancer with Strongly Supervised Deep Learning Neural Network. Electronics 2022, 11, 3003. https://doi.org/10.3390/electronics11193003
Gui H, Su T, Pang Z, Jiao H, Xiong L, Jiang X, Li L, Wang Z. Diagnosis of Breast Cancer with Strongly Supervised Deep Learning Neural Network. Electronics. 2022; 11(19):3003. https://doi.org/10.3390/electronics11193003
Chicago/Turabian StyleGui, Haitian, Tao Su, Zhiyong Pang, Han Jiao, Lang Xiong, Xinhua Jiang, Li Li, and Zixin Wang. 2022. "Diagnosis of Breast Cancer with Strongly Supervised Deep Learning Neural Network" Electronics 11, no. 19: 3003. https://doi.org/10.3390/electronics11193003
APA StyleGui, H., Su, T., Pang, Z., Jiao, H., Xiong, L., Jiang, X., Li, L., & Wang, Z. (2022). Diagnosis of Breast Cancer with Strongly Supervised Deep Learning Neural Network. Electronics, 11(19), 3003. https://doi.org/10.3390/electronics11193003






