Abstract
Deep learning approaches have been explored for surgical tool classification in laparoscopic videos. Convolutional neural networks (CNN) are prominent among the proposed approaches. However, concerns about the robustness and generalisability of CNN approaches have been raised. This paper evaluates CNN generalisability across different procedures and in data from different surgical settings. Moreover, generalisation performance to new types of procedures is assessed and insights are provided into the effect of increasing the size and representativeness of training data on the generalisation capabilities of CNN. Five experiments were conducted using three datasets. The DenseNet-121 model showed high generalisation capability within the dataset, with a mean average precision of 93%. However, the model performance diminished on data from different surgical sites and across procedure types (27% and 38%, respectively). The generalisation performance of the CNN model was improved by increasing the quantity of training videos on data of the same procedure type (the best improvement was 27%). These results highlight the importance of evaluating the performance of CNN models on data from unseen sources in order to determine their real classification capabilities. While the analysed CNN model yielded reasonably robust performance on data from different subjects, it showed a moderate reduction in performance for different surgical settings.
1. Introduction
Computer-assisted interventions (CAIs) have been improved by ongoing technological developments to meet the demands of advancing surgical modalities. Thus, the operative environment amid progressing advancements has become complicated. Therefore, processing and integrating data flows from diverse technologies is necessary to enrich surgical practice rather than expose human operators to overly complex information streams [1,2,3,4]. In turn, this may improve the workflow in the operating room (OR) by supporting the surgeon in making decisions, anticipating possible complications, and enhancing cooperation between multidisciplinary OR teams [1,5,6]. Furthermore, surgical workflow recognition can benefit OR resource management optimisation [2,6,7], automatic report generation [1,8,9], surgeon training [6], and operative skill assessment [1,6,10].
Accurate surgical tool detection is a key factor in recognising surgical activities and conceptualizing surgical workflow [6,11]. Research has been conducted to identify surgical tools using different techniques and methodologies. In early approaches, tool use signals were acquired using radio-frequency identification (RFID) systems [12]. This technique requires installation of specific sensors and instruments that may cause interference in the intervention workflow. Thus, image-based laparoscopic video signal approaches have been investigated as modern alternatives. Visual features in different colour spaces can be employed to separate tool pixels and identify tool types [13,14]. Other studies have used features of ORB (Oriented FAST and Rotated BRIEF), SIFT (Scale Invariant Feature Transform), and SURF (Speeded Up Robust Features) to classify surgical tools [15,16].
The expansion of deep learning approaches in object classification tasks has directed medical researchers to the exploration of convolutional neural networks (CNNs). However, the paucity of labelled datasets has hindered wider exploration of the potential of CNNs for analysing laparoscopic images. The Cholec80 dataset, made available to researchers in 2017, contains labelled laparoscopic videos of 80 surgeries [17]. The first utilisation of Cholec80 was carried out by training a CNN model, EndoNet, to learn visual features for recognising surgical tools and phases [17]. Subsequent studies alleviated the imbalanced dataset problem by applying loss weights and resampling strategies [18,19,20]. In addition to spatial information captured by the CNN, other studies have leveraged temporal dependencies along the video sequence using long short-term memory (LSTM) [20,21,22], convolutional LSTM [23], gated recurrent unit (GRU) [24], or graph convolutional networks (GCNs) [25]. The proposed methods in the previous studies show good performance for detecting and classifying surgical tools. However, evaluation is generally carried out using images belonging to a single dataset, in particular, datasets recorded at one hospital and which contain only one type of procedure. Thus, robustness in terms of the ability to identify tools in different datasets has not been investigated yet.
Development of CAIs requires labelled laparoscopic videos. However, such labelled videos are time-intensive to acquire. Furthermore, validating methodologies requires data from multiple sites, further exacerbating the cost of data. Therefore, medical datasets are relatively small and of ambiguous value with respect to different procedures, surgeons, or hospitals. Transfer learning has been proposed as a solution to remedy the small data size issue by leveraging learned knowledge of object detection tasks using the large-scale dataset ImageNet [26] in the medical domain [17]. Nevertheless, there is a need to determine the generalisability of CNN models despite the low diversity of datasets.
The generalisation capability of deep learning models has been addressed in studies performed on laparoscopic videos. Ross reported a decrease in the performance of tool segmentation by more than 50% when the testing images were obtained from a different hospital than the training images [27]. In the 2019 Robust Medical Instrument Segmentation (ROBUST-MIS) challenge, the segmentation performance of proposed approaches dropped when they were evaluated with images of different procedure types [28]. Bar et al. studied the generalisation of a deep model consisting of CNN-LSTM for surgical phase recognition. A reduction of about 10% in accuracy was reported on videos from an unseen hospital [29]. In prior preliminary studies, the generalisation performance of deep learning approaches was evaluated across different datasets for classifying surgical tools [30,31].
In this work, we evaluated the generalisation capability of a CNN model to (1) new subjects, (2) different OR locations, and (3) different procedure types. Moreover, we studied model generalisation with respect to the size and variability of training data. State-of-the-art CNN models (mainly DenseNet-121) and laparoscopic images from cholecystectomy and gynaecology surgeries were used to carry out the study. Laparoscopic videos of gynaecological procedures were recorded and labelled for surgical tool presence. The cholecystectomy data was obtained from two publicly available datasets.
The novel contributions of this paper are: (1) Evaluation of CNN generalisation for surgical tool classification in data of new subjects; (2) Robustness assessment of CNN for surgical tool classification in multiple source data and multiple types of laparoscopic procedures; (3) Determination of the generalisation capability of CNNs with respect to the size of training data; (4) Evaluation of the effect of CNN training with multi-site data on model generalisability; and (5) Systematic evaluation across three datasets of cholecystectomy and gynaecology laparoscopic videos.
2. Materials and Methods
2.1. Dataset Description
Three datasets were used to conduct this study: Cholec80 [17], EndovisChole [32], and Gyna08. Cholec80 and EndovisChole are publicly available datasets containing laparoscopic videos of cholecystectomy procedures. The Gyna08 dataset contains laparoscopic videos of genecology procedures, and was recorded and labelled specifically for this study.
These datasets contained several surgical tools used to conduct laparoscopic procedures. A few of the surgical tools had different shapes in the different surgical centres, while others had multiple distinct subtypes to execute different surgical actions during the procedure. While the various surgical tool subtypes can be visually distinguished, they typically share similar forms. However, certain surgical tools differed significantly in appearance across the available datasets. Therefore, those surgical tools that did not have a similar shape across the datasets were not considered in the study of generalisability across OR settings. By visually inspecting the tools in the three datasets, five tool classes (grasper, hook, scissors, irrigator, and bag) were identified to be the similar tools across the datasets. It is worth noting that each of these tool classes in each dataset can have different subtypes of tools which have similar functionality and different shapes, e.g., the grasper class in Gyna08 (Figure 1). All the surgical tools in the Cholec80 dataset were considered to study generalisation to new subjects within the dataset. Figure 2 shows the occurrences of surgical tools considered in this work in all three datasets. The percentage of tool occurrence was calculated for every procedure by dividing the number of images of each tool over the total number of images in the procedure. Then, the mean and standard deviation of the calculated occurrence percentages were computed over all procedures in each dataset (see Table 1).
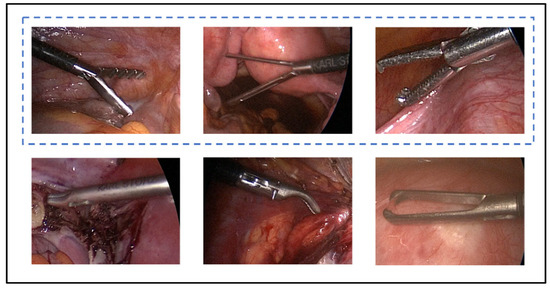
Figure 1.
Different types of graspers in the Gyna08 dataset. The tools in the blue dashed box form the main grasper class.
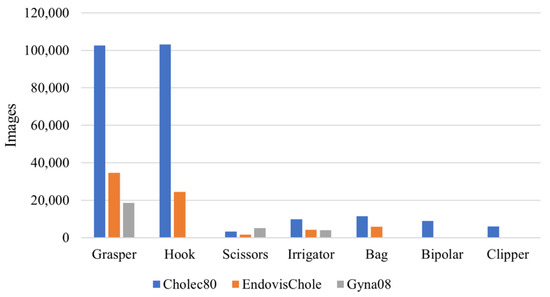
Figure 2.
Distribution of surgical tools in Cholec80, EndovisChole, and Gyna08 datasets.

Table 1.
Mean and standard deviation of the percentage of tool usage in procedures in the Cholec80, EndovisChole, and Gyna08 datasets.
2.1.1. Cholec80 Dataset
This dataset was made publicly available in 2017. It consists of 80 videos of cholecystectomy procedures performed by thirteen surgeons. The data were collected at the University Hospital of Strasburg, France with an acquisition rate of 25 Hz. Three videos have a resolution of 1920 × 1080, while the resolution for the other videos is 854 × 480. The median duration of the videos is 34.9 min (min: 12.3, max: 99.9). The dataset was manually labelled for surgical tools at 1 Hz and surgical phases at 25 Hz. The labels for surgical tools were binary, based on whether at least half of the tool tip appeared in the scene. Seven surgical tools were used to perform Cholec80 procedures, namely, grasper, hook, scissors, irrigator, specimen bag, bipolar, and clipper. The number of images and the frequency of their use in the procedures for every surgical tool are shown in Figure 2 and Table 1, respectively.
2.1.2. EndovisChole Dataset
This dataset was introduced in the sub-challenge “Surgical Workflow and Skill Analysis” within the Endoscopic Vision (EndoVis) challenge 2019. The dataset consists of 33 videos of cholecystectomy procedures collected at three hospitals in Germany. The organising team of the challenge split the data into training and testing sets. The training data include 24 videos of cholecystectomy procedures, which were released in 2021. Twelve videos in the training set were recorded at Heidelberg University Hospital (Heidelberg, Germany) and twelve were obtained from Salem Hospital (Heidelberg, Germany). The resolution and recording rate for the training videos in the Endovis challenge 2019 are presented in Table 2. The testing set of the Endovis challenge 2019 consists nine videos recorded at Heidelberg University Hospital (three videos), Salem Hospital (three videos), and GRN-hospital Sinsheim, Sinsheim, Germany (three videos). To the best of our knowledge, the testing set has not been released, and thus only the training data, termed the EndovisChole dataset, were used in our study. The median length of procedures in the EndovisChole dataset is 32.9 min (min: 21.2, max: 85.1).

Table 2.
Resolution and recording rate for the released training set of Endovis challenge 2019.
Labels for surgical tools, actions, and phases are provided for every frame. There were 21 surgical tools used in the EndovisChole procedures. The labels for these tools are included in the dataset, and the frames of each video are continuously labelled with the tool classes. If a tool shaft appears in a frame and is not manually determinable by searching preceding frames, the label “undefined instrument shaft” is assigned for that tool. However, if the tool tip can be recognised from the preceding frames, a tool presence label is assigned to that frame. The tools are grouped into seven categories based on tool function; for example, the grasper category has twelve subtypes of grasping tools. The tool category is provided for every frame.
In this study, frames at 1 Hz were used. Only tools that had similar appearance to corresponding tools in the Cholec80 and Gyna08 datasets were included. Thus, the curved atraumatic grasper, toothed grasper, fenestrated toothed grasper, atraumatic grasper, atraumatic grasper short, and flat grasper were all used under the grasper class. Additionally, the electric hook, scissors, irrigator, and specimen bag were considered when conducting the experiments in this work.
2.1.3. Gyna08 Dataset
Eight gynaecological procedures were recorded at Schwarzwald-Baar Clinic in Villingen-Schwenningen, Germany. The study received ethical approval from the ethics commission of Furtwangen University (application Nr. 19-0306LEKHFU) and consent forms were obtained from all subjects by the anaesthesiologist. A data acquisition framework [33] was implemented in an integrated operating theatre (OR1, KARL STORZ) to record laparoscopic procedure data. Laparoscopic videos were recorded at 25 Hz with a resolution of 1920 × 1080. The procedures commenced after inserting the laparoscopic camera inside the patient’s body and lasted until the camera was removed from the body. However, the surgical team may have taken the laparoscopic camera out during the procedure to clean its lenses. To ensure anonymisation, all frames which were captured when the camera was not in the trocar were replaced with white images. The median duration of the procedures was 81.5 min (min: 23.0, max: 150.9).
The gynaecological videos were labelled for surgical tool presence at 1 Hz. Tool labelling was performed by three engineers. The tools were labelled as present in the image if any part of the tool tip appeared in the scene. However, if the tool tip was not in the scene or was covered by an anatomical structure, the tool was labelled as not present in the image. However, certain surgical tools, such as the irrigator, did not have a characteristic tip, in which case the shaft was sufficient to identify the tool type. Such surgical tools were labelled based on the presence of the shaft.
In total, 26 surgical tools were present in the videos. However, these tools can be grouped into main classes with respect to functionality. For instance, several types of tools were used to perform grasping, for such diverse purposes as grasping tissues or holding a surgical needle. Figure 1 shows six types of graspers used in the procedures in the Gyna08 dataset. However, only three of those types, which are shown in Figure 1 in the blue dashed box, were similar to the graspers used in the other datasets, and thus formed the grasper class.
2.2. Experiments
Five experiments were carried out to study the generalisation capability of the DenseNet-121 model [34]. The generalisation performance of the model was evaluated in the first three experiments. The last two experiments were conducted to explore factors that may improve model generalisation. A summary description of the conducted experiments can be found in Table 3 and Figure 3. Table 3 shows the data used for training and testing the CNN models, the model names, and the number of surgical tools considered.

Table 3.
Summary of the conducted experiments.
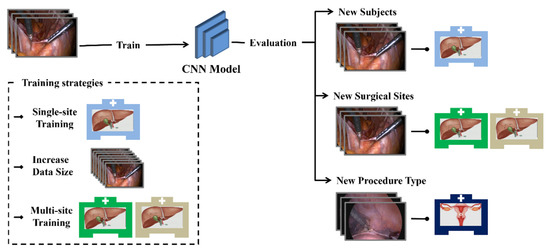
Figure 3.
Diagram of the conducted experiments.
2.2.1. Experiment 1
Model generalisation across subjects was studied. This is a typical training/testing CNN validation approach. Initially, forty videos of Cholec80 dataset formed the training set, while the remaining videos were used for testing model performance. This model was named CNN-40S. Secondly, a CNN model (CNN-80S) was trained on data from all videos of the Cholec80 dataset. CNN-80S was trained on images randomly selected across the Cholec80 dataset without consideration for isolating certain patients. Five-fold cross-validation was performed for both the CNN-80S and CNN-40S models. To ensure a fair comparison, an equal number of images (~92,512 ± 4480 images) was used for training CNN-40S and CNN-80S in each fold. Thus, both models were trained on the same size and type of data obtained from the same surgical location for classifying the seven surgical tools of Cholec80 dataset. However, CNN-40S was limited to validation in patients not seen prior, and CNN-80S had no such limitation.
2.2.2. Experiment 2
CNN generalisation to new surgical sites was evaluated. A CNN model (CNN-40-Target) was trained on forty videos of Cholec80 dataset to classify tools as grasper, hook, scissors, irrigator, and bag. Then, the performance of the model was evaluated on the EndovisChole dataset containing cholecystectomy images recorded at other surgical locations. To ensure the reliability of the results, five-fold cross-validation was conducted by randomly selecting 40 videos from the Cholec80 dataset for training.
2.2.3. Experiment 3
In this experiment, CNN generalisation to different types of procedures was evaluated. The model CNN-40-Target trained on cholecystectomy images from Cholec80 dataset was tested on images of gynaecological procedures. Similar to experiment 2, only the grasper, hook, scissors, irrigator, and bag tools were considered.
2.2.4. Experiment 4
Experiment 4 investigated the influence of the amount of training data on model generalisability. Several training runs were conducted on the DenseNet-121 model using the Cholec80 dataset with varying amounts of training data. Training data ranged from 1 to 70 videos. However, the classification performance was evaluated on three fixed sets of data to ensure a fair comparison. The testing sets were the last ten videos from the Cholec80, EndovisChole, and Gyna08 datasets. Thus, only the surgical tools appearing in all these sets were considered.
2.2.5. Experiment 5
This experiment compared the generalisation performance of a CNN model that was trained with data from multiple sites (MST-CNN) to a CNN model that was trained with data from a single site (SST-CNN). The EndovisChole dataset (~55 k images) was used for MST-CNN model training; 25 videos from the Cholec80 dataset (~55 k images) were used to train the SST-CNN model. Thus, both models had the same data size and type of procedure (cholecystectomy) for training. The generalisation capability of the trained models was then evaluated using cross-dataset validation. In addition to the DenseNet-121 model, Experiment 5 used EfficientNet-B0 [35], ResNet-50 [36], and VGG-16 [37] for broad indication of the validity of results.
2.3. CNN Model
An ablation study was conducted on several state-of-the-art CNN models for classifying surgical tools in laparoscopic images. The CNN models were VGG-16, ResNet-50, EfficientNet-B0, and DenseNet-121. The models had different architectures with varied depths and different numbers of convolutional layers. To determine the best candidate architecture for further analysis, a preliminary analysis was undertaken. Each model was trained on forty videos from the Cholec80 dataset and evaluated on the remaining videos. The models were trained for ten epochs using the training setup defined in Section 2.4. The best tool classification performance was achieved by the ResNet-50 and DenseNet-121 models, with a mean average precision of ~92%. VGG-16 and Efficient-B0 showed slightly inferior performance of 89% and 90%, respectively. Hence, the DenseNet-121 model was used to perform experiments 1 to 5, whereas the other three architectures were only used in Experiment 5.
The DenseNet-121 model is composed of four convolutional blocks, called dense blocks, with a global average pooling layer and a fully-connected layer on top. Every layer in a dense block is connected to all succeeding layers of the block. These dense connections enhance data transfer within the block and enable features of previous layers to be fused, improving utilisation of information learned at different layers [34]. The dense blocks are connected through convolutional and pooling layers.
2.4. Training Setup
The CNN model initialised with ImageNet weights [26] was employed. The last fully-connected layer in the model was replaced with another layer suitable for surgical tool classification. The new fully-connected layer was randomly initialised and had a number of classification nodes equal to the number of tool classes. Therefore, the number of nodes was set to seven in experiment 1 and to five in the other experiments. The sigmoid activation function was used in the classification layer, as the task is a multi-label binary classification one.
The loss was computed using the binary cross-entropy function (see Equation (1)). The loss of each surgical tool was weighted according to the number of tool images in the training data. This training approach was employed to reduce the influence of imbalanced distribution of the training data (see Figure 2 and Table 1) [19].
where is the computed loss for a particular tool, is the batch size, is the sigmoid function, is the binary label, and is the prediction confidence. Model training was conducted using the Adam optimiser [38] with an initial learning rate of 10−4. The classification layer had a higher learning rate of 2 × 10−3. The images of the training set were shuffled every epoch. The experiments were conducted using the Keras framework. An NVIDIA GeForce RTX 2080Ti graphics processing unit (GPU) was used.
2.5. Model Evaluation
Average precision (AP) was used to evaluate the classification performance of the CNN models. AP refers to the area under the precision–recall curve. Moreover, Gradient-weighted Class Activation Mapping (Grad-CAM) [39] was employed to depict the image areas relevant to model prediction. This method was based on backpropagating gradient information of a particular class into the final convolutional layer in the model. The visualisations of localisation maps help to understand the models’ focus and explain their prediction decisions.
3. Results
Generalisation performance of the DenseNet-121 model to new subjects within the same dataset (Experiment 1) is shown in Figure 4. This figure presents a comparison between the tool classification performance of CNN-80S and CNN-40S within (at novel frames) and across subjects, respectively. Figure 4 shows the mean and standard deviation of five-fold cross-validation using CNN-80S and CNN-40S. The results show a drop in model performance across subjects. Figure 5 presents class-discriminative activation maps generated by the CNN-80S and CNN-40S models for an image from the testing set. In this example, CNN-80S was superior to CNN-40S in identifying graspers despite the partial appearance of the tool. Figure 6 shows another example in which the CNN-80S did not focus on the right region (the tool region), as shown by the activation maps.
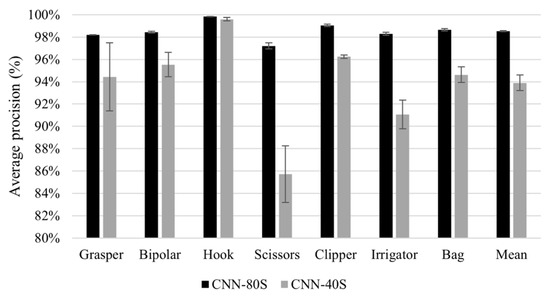
Figure 4.
Results of generalisation to new subjects. Average precision of tool presence detection for all tools yielded by CNN-80S model (back bars) and CNN-40S model (grey bars). Note the truncated scale of the y-axis.
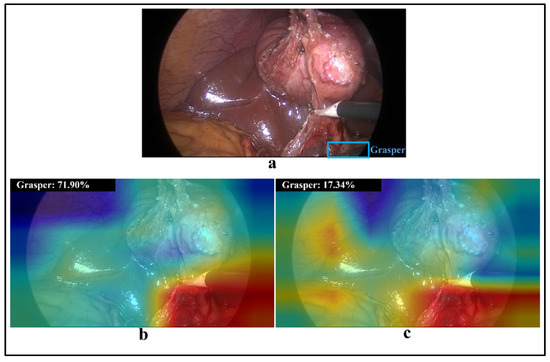
Figure 5.
(a) A laparoscopic image containing a hook and a partial grasper (in blue box); (b,c) visualisations of class activation maps and prediction probability for class grasper obtained by CNN-80S and CNN-40S models, respectively.
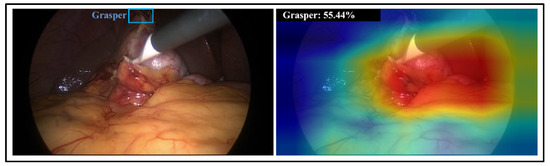
Figure 6.
(left) a laparoscopic image containing a hook and grasper (in blue box) appearing in the dark area; (right) visualisation of class activation maps and prediction probability for grasper class obtained by CNN-80S model.
The results of model generalisation to new surgical sites and types of procedures (Experiments 2 and 3) are presented in Figure 7. The results include model performance on unseen cholecystectomy data from the same surgical site (green bars), from other surgical sites (blue bars), and on gynaecological data (red bars). The model exhibits a reasonable generalisation ability to EndovisChole data, except perhaps for scissors. However, poor generalisation performance to gynaecological data (Gyna08 dataset) was observed. In fact, the surgical tools in the Cholec80 and Gyna08 dataset had distinct appearances, and thus recognition was inhibited. Figure 8 shows one of the trocars which was used to insert surgical tools during certain procedures in the Gyna08 dataset. The trocar looks quite similar to the irrigator used in procedures in the Cholec80 dataset, causing incorrect classification of such images as containing an irrigator.
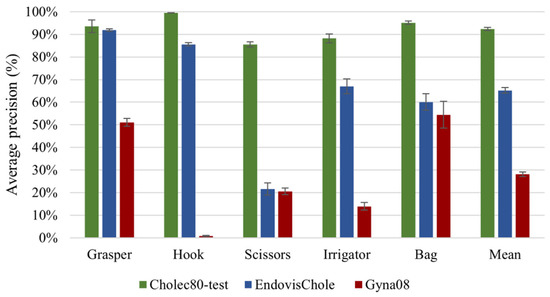
Figure 7.
Results of generalisation to other surgical locations and types of procedures.
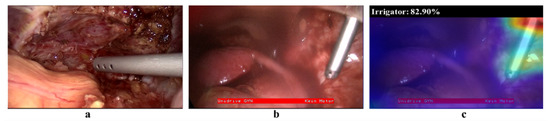
Figure 8.
(a) Image of an irrigator from the Cholec80 dataset, (b) a trocar from the Gyna08 dataset, and (c) visualisation of activation maps and prediction probability of irrigator class in (b).
The relationship between the size of the training set and model performance (Experiment 4) is presented in Figure 9, Figure 10 and Figure 11. Figure 9 shows the mean average precision over all tools on the last ten videos of the Cholec80, EndovisChole, and Gyna08 datasets. The performance for each surgical tool (grasper, hook, scissors, irrigator, and bag) on the EndovisChole and Gyna08 datasets is shown in Figure 10 and Figure 11, respectively. Generalisability to the same type of procedure exhibits a clear tendency towards improvement when increasing the amount of training data.
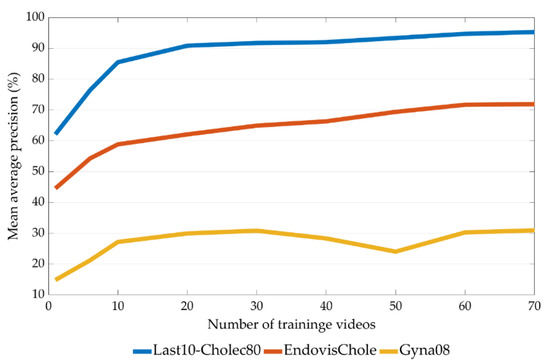
Figure 9.
Mean average precision on the tools in the last ten videos of the Cholec80, EndovisChole and Gyna08 datasets as a function of training set size.
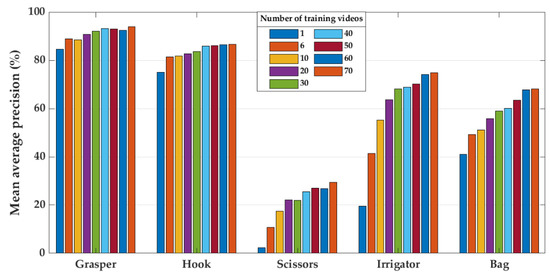
Figure 10.
Model performance on the EndovisChole dataset for grasper, hook, scissors, irrigator, and bag when using increasing number of training videos.
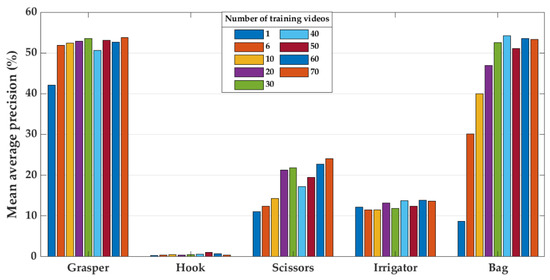
Figure 11.
Model performance on the Gyna08 dataset for grasper, hook, scissors, irrigator, and bag when using increased number of training videos.
Figure 12 shows the generalisation performance using single-site data and multi-site data (Experiment 5). The generalisation performance is shown as the mean and standard deviation of average precision over the four CNN architectures (DenseNet-121, EfficientNet-B0, ResNet-50, and VGG-16).
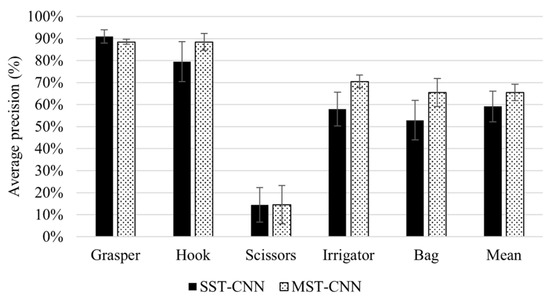
Figure 12.
Generalisation performance of four CNN architectures using single-site and multi-site training. The figure shows the mean and standard deviation of average precision with the DenseNet-121, EfficientNet-B0, ResNet-50, and VGG-16 models.
4. Discussion
This work evaluated the generalisation capability of CNN models for tool classification from laparoscopic videos. The study involved two aspects of generalisation: across subjects in the same dataset, and across datasets from different sources. The cross-dataset evaluation examined model performance on data from new surgical locations and on laparoscopic data from different procedure types. Moreover, we explored the influence of size and variability of training data on the generalisation performance of CNN models. Five experiments were conducted for surgical tool classification using three sources of data, the Cholec80, EndovisChole, and Gyna08 datasets (see Table 3).
The results of Experiment 1 imply that the CNN model is generalisable to new subjects. The CNN-80S and CNN-40S models achieved high classification performance, with an mAP of 98.5% and 93.8%, respectively (Figure 4). There was a drop of approximately 5% in the classification performance over all tools when the classification was undertaken in new subjects from the same procedure and location. However, certain surgical tools had lower generalisation performance than other tools. Model performance for grasper, scissors, and irrigator on unseen subjects decreased by 3.76%, 10.65%, and 8.98%, respectively. A retrospective analysis was conducted to investigate the causes of these reductions in classification performance. Thus, misclassified images belonging to the testing sets of both models (CNN-80S and CNN-40S) were examined. In many false classification samples with the CNN-40S model, the corresponding surgical tool is obscured by image disturbances such as blood or smoke, or the tool only partially appears in the scene. Nevertheless, the CNN-80S model yielded correct classifications for most of these challenging samples. Hence, it can be concluded that using training data with a higher subject variation in terms of procedure and surgical location improves model robustness. Figure 5a shows an image with only a small part of a grasper in the frame. The CNN-40S model was not able to detect the grasper in this image, while the CNN-80S model recognised it with a prediction probability of 72%. The region of the corresponding tool was solely engaged in the prediction of the CNN-80S model, as demonstrated by the activation maps in Figure 5b. On the contrary, the activation maps of CNN-40S involve image regions not related to the corresponding tool (Figure 5c).
In certain samples, the activation maps of CNN-80S mismatch the tool region even though the model provides a correct classification. In these samples, the classification decision was based on tissue regions rather than the region of the corresponding tool. An example of this is presented in Figure 6. This phenomenon occurs when the training set includes images preceding or succeeding the examined image in the laparoscopic video that show a similar scene. Although the laparoscopic camera moves during the intervention, it becomes relatively static when the surgeon executes a surgical action. Consequently, neighbouring images in certain parts of the video can have an almost identical scene.
Experiment 2 explored model generalisation to new surgical locations. The CNN-40-Target model achieved a mAP value of 65% on the EndovisChole dataset (see blue bars in Figure 7). Compared with its performance on the Cholec80 data, its classification performance for the target tools dropped by 27% (Figure 7). This model showed a high generalisation capability for the most consistently observed tools (grasper and hook), with an AP of 92% and 86%, respectively. There was a greater degradation of performance for the other tools in the EndovisChole dataset. Nevertheless, the model generally retained reasonable classification performance, with AP > 60% for the irrigator and bag. However, scissors had a low classification result in the EndovisChole dataset (22%), showing poor model generalisation for tools that were under-represented in the training set (Figure 2). Thus, while acceptable generalisation is possible across surgical locations, this is only the case for tools that are well-presented in the training data. According to the reported results for tool presence detection in the Endovis challenge 2019 [32], CNNs were purported to have high generalisation capability to new surgical sites when procedures of the same type were performed using identical surgical tools at different sites. The CNN-based methods submitted to the challenge did not show a drop in performance on data from new surgical sites (the GRN-hospital Sinsheim), as the surgical tools used in Sinsheim Hospital are identical to those used in Salem Hospital (represented in the training data).
Experiment 3 indicates a lower classification performance on data across procedure types. The mAP on the different tools dropped by 38% when the CNN-40-Target model was tested on the gynaecological images in the Gyna08 dataset as compared with the mAP on the EndovisChole dataset (cholecystectomy data). The highest drops in model performance were for the hook and irrigator. In particular, variation in tool incidence across the datasets makes comparing performance on the tools with disparate incidence unfair in these sets. For example, the Gyna08 dataset has few samples containing a hook (146 images). The hook samples were presented only in one procedure, forming about 4% of the Gyna08 dataset. On the contrary, a hook appears in 56% and 44% of the Cholec80 and EndovisChole datasets, respectively. Therefore, the number of hook samples was very small in the gynaecology surgery data compared to the cholecystectomy data. To investigate the effect of data incidence on evaluation performance, a small set was generated to achieve a specific incidence of hooks from the Gyna08 dataset. The incidence rate of hooks was 56% in both the newly generated set and the Cholec80 dataset. The average precision for the hook class on this small subset was 57%, which was much higher than the performance on the Gyna08 dataset (1%). Thus, tool incidence in evaluation sets affects performance criteria. Nevertheless, the model performance on the new set of gynaecological data was 30% lower than the performance on cholecystectomy data. Overall, it can be inferred that while the implemented CNN model has generalisation capability across procedure types, it is important to consider tool incidence variation across sets.
The low classification performance on the irrigator in the Gyna08 dataset was due to the presence of a trocar. In particular, the trocar had a shape that was quite similar to the irrigator in certain procedures. Hence, the model classified trocar images as containing an irrigator, and therefore the false positive rate of the irrigator class increased. Figure 8 shows two images form the Gyna08 dataset. Figure 8a,b shows images with an irrigator and a trocar, respectively. The activation map of Figure 8b for the irrigator class is shown along with the yielded prediction probability (82.9%) in Figure 8c.
These results imply that the type of procedure affects the generalisability of CNN models. Data from a new procedure type can contain different surgical tools and/or a different surgical region with distinct anatomical structures. Additionally, the surgical actions or order of their execution can differ. All such factors limit model generalisation across procedure types.
The results of Experiment 4 imply that training the CNN model on increasing quantities of data enhances generalisability to data from other sources (Figure 9). The classification performance was improved by about 27% on the EndovisChole dataset after expanding the Cholec80 training set from a single video to 70 videos (Figure 9). However, lesser improvement was reported on the Gyna08 dataset, about 16% (Figure 9). Nonetheless, it can be concluded that increasing the amount of training data can enhance generalisability, especially to the same type of procedure. However, performance was affected inconsistently by the various surgical tools analysed. For instance, performance for the grasper and hook was only marginally affected by increasing data size. On the other hand, the AP for the bag and scissors in the EndovisChole dataset increased by 27%. Moreover, the irrigator in the EndovisChole dataset was classified with an increase in AP from 20% to 75% (Figure 10). This trend was not observed in classification results of the irrigator in the Gyna08 dataset (Figure 11) due to the high false positive rate caused by the presence of a trocar, which appears similar to irrigator (Figure 8). The best improvement in classification results on Gyna08 dataset was reported for the bag, with an improvement in AP of 45% (Figure 11).
Because certain surgical tools are used more frequently than others (Figure 2), it requires much more surgical data to enable CNN models to accumulate a broad range of knowledge regarding their geometry in an image. In particular, the accumulation rate of images with scissors is less than for other tools. Hence, the AP when classifying scissors showed a continuous improvement with the use of more videos for training (Figure 10). However, the AP for scissors was less than 30% in both testing sets (EndovisChole and Gyna08 datasets, Figure 10 and Figure 11). Thus, it can be expected that more data enables better identification of the tool type. On the other hand, the bag and irrigator had a higher occurrence rate than the scissors (Table 1). Both tools had similar sample incidences (Figure 2), and therefore, both tools had a roughly similar improvement trend in classification performance on the EndovisChole dataset when the training set was expanded. Figure 10 implies that thirty training videos (containing ~4 k images of positive instances for the bag and the irrigator) are required to achieve a generalisation performance greater than 60% to data of the same procedure type. The grasper and hook images in the first six videos of the Cholec80 dataset (~9 k tool images) were sufficient to achieve AP of 89% and 81%, respectively, on the EndovisChole dataset, and training with more videos showed diminishing returns in model performance (~5%). These results imply that CNN model tool classification performance is improved when the quantity of training samples containing that tool increases. However, this improvement becomes saturated at a certain quantity.
The impact of training a CNN model on data from multiple surgical sites on generalisation performance was investigated in Experiment 5. To produce a broad analysis, Experiment 5 was conducted on various CNN models with different depths and numbers of parameters. Figure 12 shows that the multi-site training achieved higher mean AP on all tools compared to single-site training. Training with data from different surgical sites enhances data representativeness, and therefore improves model generalisability. The improvement in generalisation performance was reported for the hook, irrigator, and bag, with 8.93%, 12.56%, and 12.56% improvements, respectively. However, multiple-site training did not improve the performance for the scissors. Both training strategies (single- and multi-site training) failed to train the models for detecting scissors due to the very small number of samples belonging to this tool in the training sets.
Within the surgical domain, many factors can cause differences between training and testing data that CNN-based models find difficult to parse. In this study, variations in data arose due to differences in anatomy between subjects, different types of procedures, varied subtypes of surgical tools, variations in the instrumentation used in different clinics (and to an extent even within a single clinic, as in the Gyna08 procedures), and diverse surgical manoeuvres executed by different surgeons. The experimental results demonstrate the sensitivity of model generalisability to such variations. However, the results show that increasing diversity within training data generally provides more representative and discriminative information, thereby strengthening model learning and improving generalisability and applicability to new clinical data. The quantity of representative data is vital for deep learning. Nevertheless, the quality and reliability of data labelling is critical to enable correct learning as well. To date, labelling laparoscopic videos has been performed manually, and is therefore is quite time-consuming and prone to errors. The datasets used in this work were labelled by different annotators. This factor adds an additional source of variation between the datasets. This variation was an inevitable limitation in this study.
5. Conclusions
This study emphasises the need for robust evaluation of the capabilities of deep learning models before deploying them as assistive systems in the surgical environment. While the CNN models analysed in this research yielded reasonably robust performance on data from new subjects, they showed a moderate reduction in performance for different surgical settings. This reduction in performance was exacerbated across procedure types. In addition, this research elucidates improvements in generalisation performance with increasing quantities of training data.
Author Contributions
Conceptualization, T.A.A., N.A.J. and K.M.; methodology, T.A.A. and N.A.J.; software, T.A.A. and N.A.J.; validation, T.A.A. and N.A.J.; formal analysis, T.A.A., N.A.J., P.D.D., T.N. and K.M.; investigation, T.A.A. and N.A.J.; resources, K.M.; data curation, T.A.A. and N.A.J.; writing—original draft preparation, T.A.A.; writing—review and editing, N.A.J., P.D.D., T.N. and K.M.; visualization, T.A.A.; supervision, K.M. and T.N.; project administration, K.M.; funding acquisition, K.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the German Federal Ministry of Research and Education (BMBF) under grant CoHMed/IntelliMed, grant number 13FH5I05IA.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of Furtwangen University (application Nr. 19-0306LEKHFU, 1 July 2019).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study by the anaesthesiologist.
Data Availability Statement
The data presented in this study were composed of two publicly available datasets (Cholec80 and EndoVis challenge 2019) and one unpublished dataset (Gyna08 dataset). The Cholec80 dataset is available (http://camma.u-strasbg.fr/datasets/ (accessed on 22 March 2017)) from the publisher upon request. The EndoVis challenge 2019 dataset is available at (https://www.synapse.org/#!Synapse:syn18824884/files/ (accessed on 12 September 2019)). The Gyna08 dataset is available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Shi, X.; Jin, Y.; Dou, Q.; Heng, P.-A. LRTD: Long-range temporal dependency based active learning for surgical workflow recognition. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 1573–1584. [Google Scholar] [CrossRef] [PubMed]
- Bharathan, R.; Aggarwal, R.; Darzi, A. Operating room of the future. Best Pract. Res. Clin. Obstet. Gynaecol. 2013, 27, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Cleary, K.; Kinsella, A.; Mun, S.K. OR 2020 workshop report: Operating room of the future. Int. Congr. Ser. 2005, 1281, 832–838. [Google Scholar] [CrossRef]
- Padoy, N.; Blum, T.; Ahmadi, S.-A.; Feussner, H.; Berger, M.-O.; Navab, N. Statistical modeling and recognition of surgical workflow. Med. Image Anal. 2012, 16, 632–641. [Google Scholar] [CrossRef]
- Dergachyova, O.; Bouget, D.; Huaulmé, A.; Morandi, X.; Jannin, P. Automatic data-driven real-time segmentation and recognition of surgical workflow. Int. J. Comput. Assist. Radiol. Surg. 2016, 11, 1081–1089. [Google Scholar] [CrossRef]
- Anteby, R.; Horesh, N.; Soffer, S.; Zager, Y.; Barash, Y.; Amiel, I.; Rosin, D.; Gutman, M.; Klang, E. Deep learning visual analysis in laparoscopic surgery: A systematic review and diagnostic test accuracy meta-analysis. Surg. Endosc. 2021, 35, 1521–1533. [Google Scholar] [CrossRef]
- Maktabi, M.; Neumuth, T. Online time and resource management based on surgical workflow time series analysis. Int. J. Comput. Assist. Radiol. Surg. 2017, 12, 325–338. [Google Scholar] [CrossRef]
- Al Hajj, H.; Lamard, M.; Conze, P.-H.; Cochener, B.; Quellec, G. Monitoring tool usage in surgery videos using boosted convolutional and recurrent neural networks. Med. Image Anal. 2018, 47, 203–218. [Google Scholar] [CrossRef]
- Padoy, N. Machine and deep learning for workflow recognition during surgery. Minim. Invasive Ther. Allied Technol. 2019, 28, 82–90. [Google Scholar] [CrossRef]
- Jin, A.; Yeung, S.; Jopling, J.; Krause, J.; Azagury, D.; Milstein, A.; Fei-Fei, L. Tool detection and operative skill assessment in surgical videos using region-based convolutional neural networks. In Proceedings of the 2018 IEEE Winter Conference on Applications of Computer Vision (WACV), Lake Tahoe, NV, USA, 12–15 March 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 691–699. [Google Scholar]
- Jin, Y.; Li, H.; Dou, Q.; Chen, H.; Qin, J.; Fu, C.-W.; Heng, P.-A. Multi-task recurrent convolutional network with correlation loss for surgical video analysis. Med. Image Anal. 2020, 59, 101572. [Google Scholar] [CrossRef]
- Miyawaki, F.; Tsunoi, T.; Namiki, H.; Yaginuma, T.; Yoshimitsu, K.; Hashimoto, D.; Fukui, Y. Development of automatic acquisition system of surgical-instrument informantion in endoscopic and laparoscopic surgey. In Proceedings of the 2009 4th IEEE Conference on Industrial Electronics and Applications, Xi’an, China, 25–27 May 2009; IEEE: Piscataway, NJ, USA, 2009; pp. 3058–3063. [Google Scholar]
- Doignon, C.; Graebling, P.; De Mathelin, M. Real-time segmentation of surgical instruments inside the abdominal cavity using a joint hue saturation color feature. Real-Time Imaging 2005, 11, 429–442. [Google Scholar] [CrossRef]
- Primus, M.J.; Schoeffmann, K.; Böszörmenyi, L. Temporal segmentation of laparoscopic videos into surgical phases. In Proceedings of the 2016 14th International Workshop on Content-Based Multimedia Indexing (CBMI), Bucharest, Romania, 15–17 June 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 1–6. [Google Scholar]
- Allan, M.; Thompson, S.; Clarkson, M.J.; Ourselin, S.; Hawkes, D.J.; Kelly, J.; Stoyanov, D. 2D-3D pose tracking of rigid instruments in minimally invasive surgery. In Proceedings of the International Conference on Information Processing in Computer-assisted Interventions, Fukuoka, Japan, 28 June 2014; Springer: Berlin/Heidelberg, Germany, 2014; pp. 1–10. [Google Scholar]
- Primus, M.J.; Schoeffmann, K.; Böszörmenyi, L. Instrument classification in laparoscopic videos. In Proceedings of the 2015 13th International Workshop on Content-Based Multimedia Indexing (CBMI), Prague, Czech Republic, 10–12 June 2015; IEEE: Piscataway, NJ, USA, 2015; pp. 1–6. [Google Scholar]
- Twinanda, A.P.; Shehata, S.; Mutter, D.; Marescaux, J.; De Mathelin, M.; Padoy, N. Endonet: A deep architecture for recognition tasks on laparoscopic videos. IEEE Trans. Med. Imaging 2016, 36, 86–97. [Google Scholar] [CrossRef]
- Sahu, M.; Mukhopadhyay, A.; Szengel, A.; Zachow, S. Addressing multi-label imbalance problem of surgical tool detection using CNN. Int. J. Comput. Assist. Radiol. Surg. 2017, 12, 1013–1020. [Google Scholar] [CrossRef]
- Alshirbaji, T.A.; Jalal, N.A.; Möller, K. Surgical tool classification in laparoscopic videos using convolutional neural network. Curr. Dir. Biomed. Eng. 2018, 4, 407–410. [Google Scholar] [CrossRef]
- Mishra, K.; Sathish, R.; Sheet, D. Learning latent temporal connectionism of deep residual visual abstractions for identifying surgical tools in laparoscopy procedures. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Honolulu, HI, USA, 21–26 July 2017; pp. 58–65. [Google Scholar]
- Alshirbaji, T.A.; Jalal, N.A.; Docherty, P.D.; Neumuth, T.; Möller, K. A deep learning spatial-temporal framework for detecting surgical tools in laparoscopic videos. Biomed. Signal Process. Control 2021, 68, 102801. [Google Scholar] [CrossRef]
- Jalal, N.A.; Abdulbaki Alshirbaji, T.; Docherty, P.D.; Neumuth, T.; Möller, K. Surgical Tool Detection in Laparoscopic Videos by Modeling Temporal Dependencies Between Adjacent Frames. In Proceedings of the European Medical and Biological Engineering Conference, Portorož, Slovenia, 29 November–3 December 2020; Springer: Berlin/Heidelberg, Germany, 2020; pp. 1045–1052. [Google Scholar]
- Nwoye, C.I.; Mutter, D.; Marescaux, J.; Padoy, N. Weakly supervised convolutional LSTM approach for tool tracking in laparoscopic videos. Int. J. Comput. Assist. Radiol. Surg. 2019, 14, 1059–1067. [Google Scholar] [CrossRef]
- Namazi, B.; Sankaranarayanan, G.; Devarajan, V. A contextual detector of surgical tools in laparoscopic videos using deep learning. Surg. Endosc. 2022, 36, 679–688. [Google Scholar] [CrossRef]
- Wang, S.; Xu, Z.; Yan, C.; Huang, J. Graph convolutional nets for tool presence detection in surgical videos. In Proceedings of the International Conference on Information Processing in Medical Imaging, Hong Kong, China, 2–7 June 2019; Springer: Berlin/Heidelberg, Germany, 2019; pp. 467–478. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; IEEE: Piscataway, NJ, USA, 2009; pp. 248–255. [Google Scholar]
- Ross, T.; Zimmerer, D.; Vemuri, A.; Isensee, F.; Wiesenfarth, M.; Bodenstedt, S.; Both, F.; Kessler, P.; Wagner, M.; Müller, B. Exploiting the potential of unlabeled endoscopic video data with self-supervised learning. Int. J. Comput. Assist. Radiol. Surg. 2018, 13, 925–933. [Google Scholar] [CrossRef]
- Ross, T.; Reinke, A.; Full, P.M.; Wagner, M.; Kenngott, H.; Apitz, M.; Hempe, H.; Filimon, D.M.; Scholz, P.; Tran, T.N. Robust medical instrument segmentation challenge 2019. arXiv 2020. [Google Scholar] [CrossRef]
- Bar, O.; Neimark, D.; Zohar, M.; Hager, G.D.; Girshick, R.; Fried, G.M.; Wolf, T.; Asselmann, D. Impact of data on generalization of AI for surgical intelligence applications. Sci. Rep. 2020, 10, 22208. [Google Scholar] [CrossRef]
- Abdulbaki Alshirbaji, T.; Jalal, N.A.; Docherty, P.D.; Neumuth, T.; Möller, K. Cross-dataset evaluation of a CNN-based approach for surgical tool detection. In Proceedings of the AUTOMED 2021-Automatisierung in der Medizintechnik, 15. Interdisziplinäres Symposium, Basel, Switzerland, 8–9 June 2021. [Google Scholar]
- Alshirbaji, T.A.; Jalal, N.A.; Docherty, P.D.; Neumuth, T.; Moeller, K. Assessing Generalisation Capabilities of CNN Models for Surgical Tool Classification. Curr. Dir. Biomed. Eng. 2021, 7, 476–479. [Google Scholar] [CrossRef]
- Wagner, M.; Müller-Stich, B.-P.; Kisilenko, A.; Tran, D.; Heger, P.; Mündermann, L.; Lubotsky, D.M.; Müller, B.; Davitashvili, T.; Capek, M. Comparative validation of machine learning algorithms for surgical workflow and skill analysis with the HeiChole benchmark. arXiv 2021. [Google Scholar] [CrossRef]
- Alshirbaji, T.A.; Jalal, N.A.; Möller, K. Data Recording Framework for Physiological and Surgical Data in Operating Theatres. Curr. Dir. Biomed. Eng. 2020, 6, 364–367. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; PMLR. pp. 6105–6114. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE international Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).