Mammographic Classification of Breast Cancer Microcalcifications through Extreme Gradient Boosting
Abstract
1. Introduction
2. Related Work
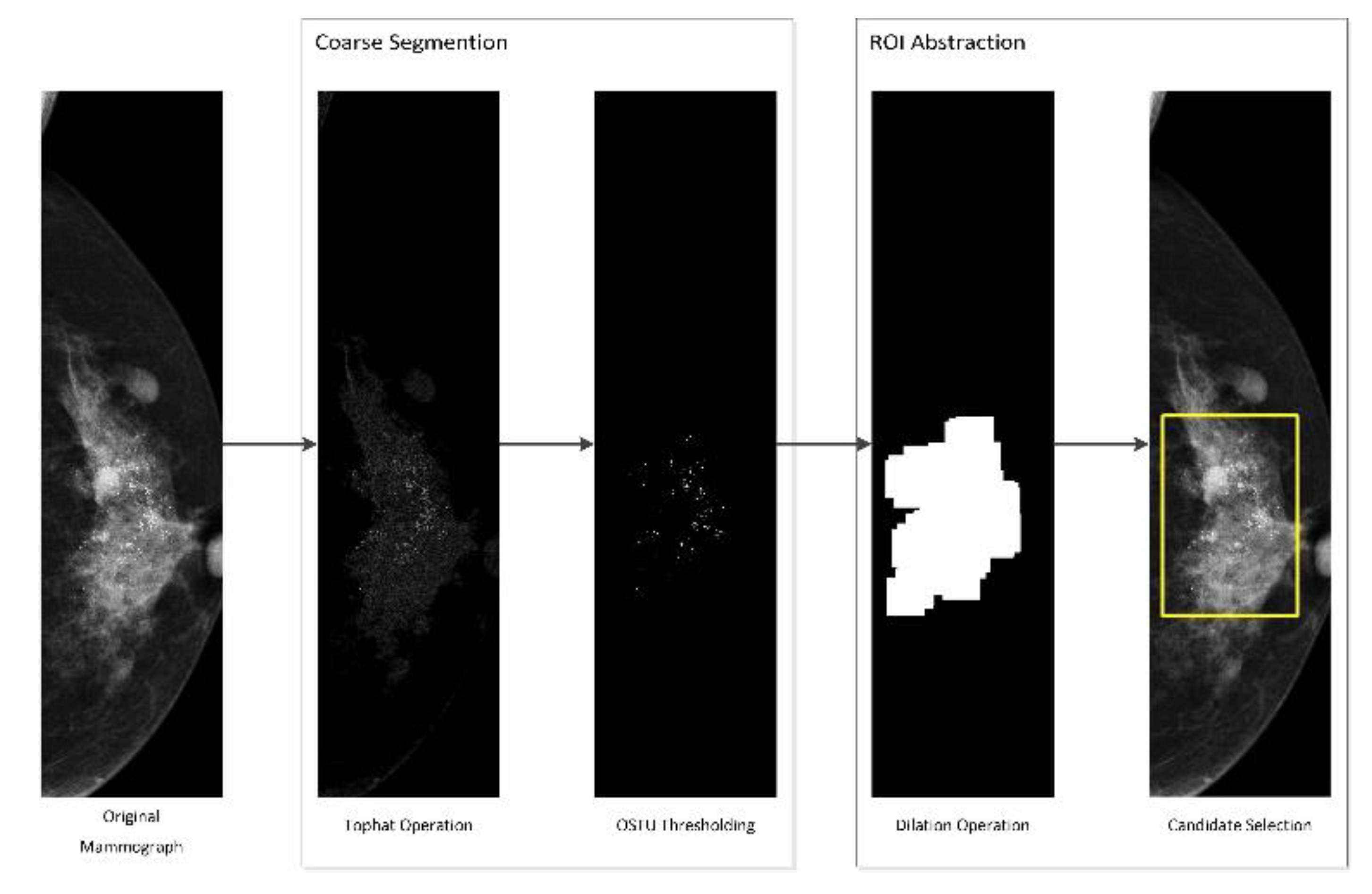
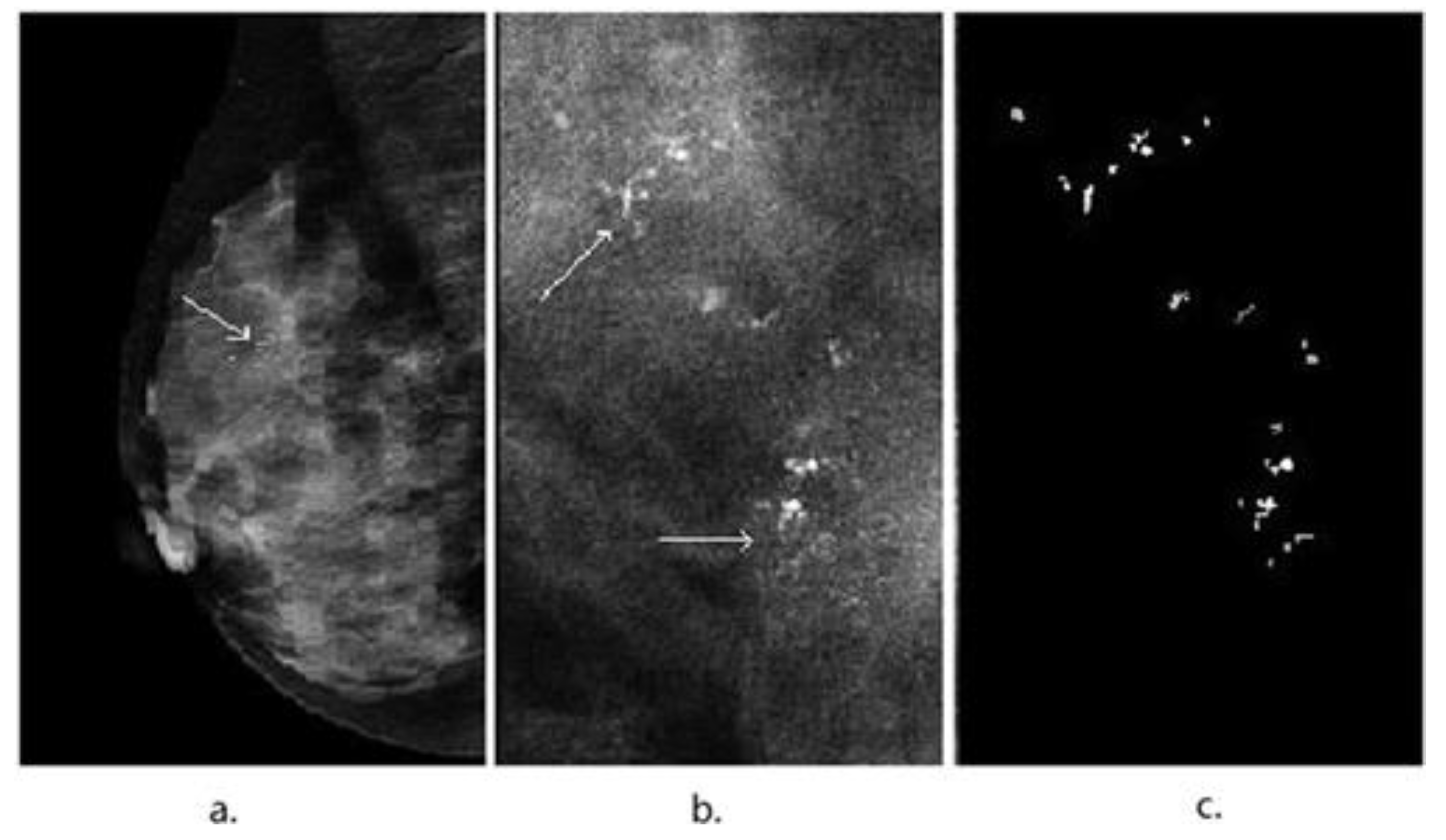
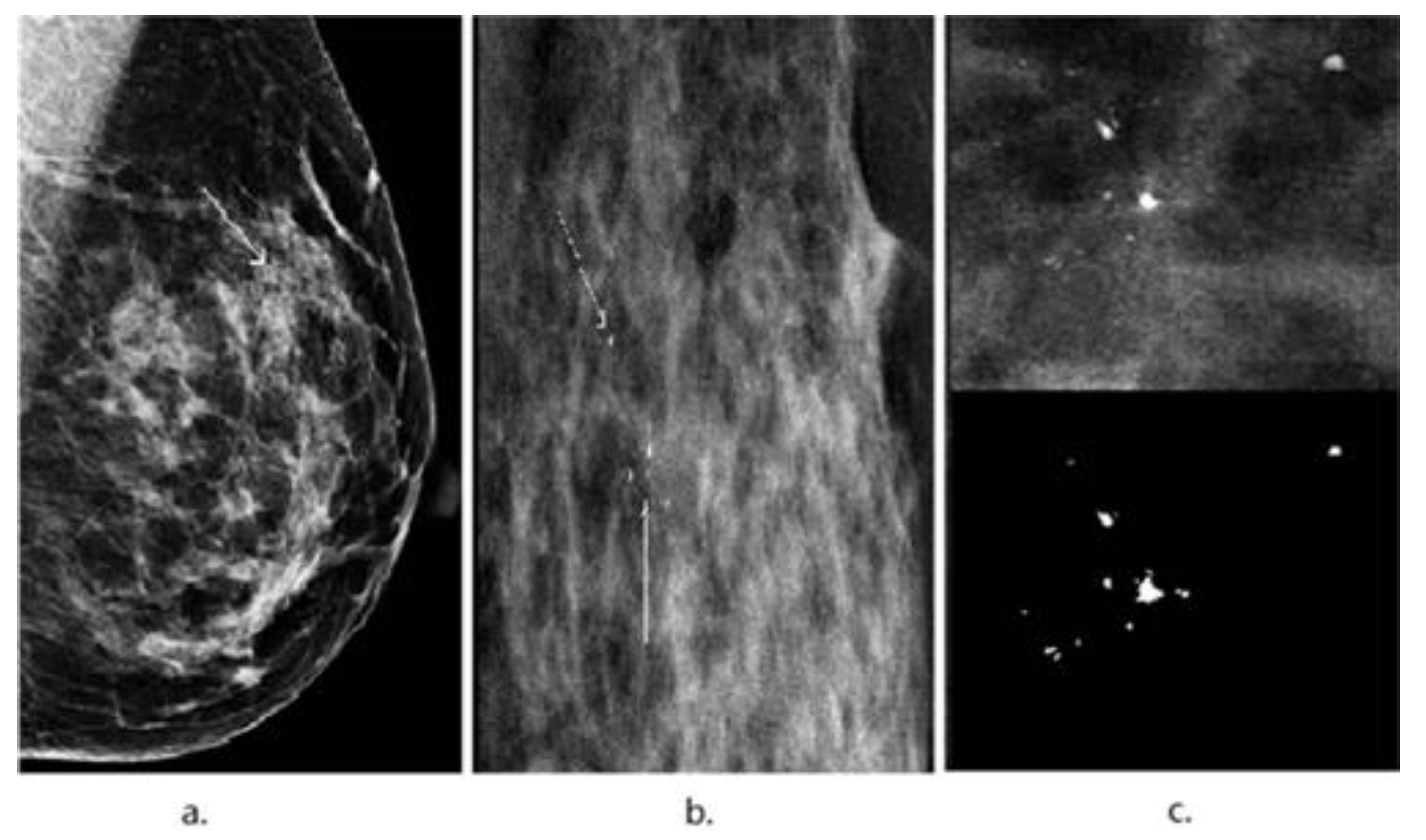
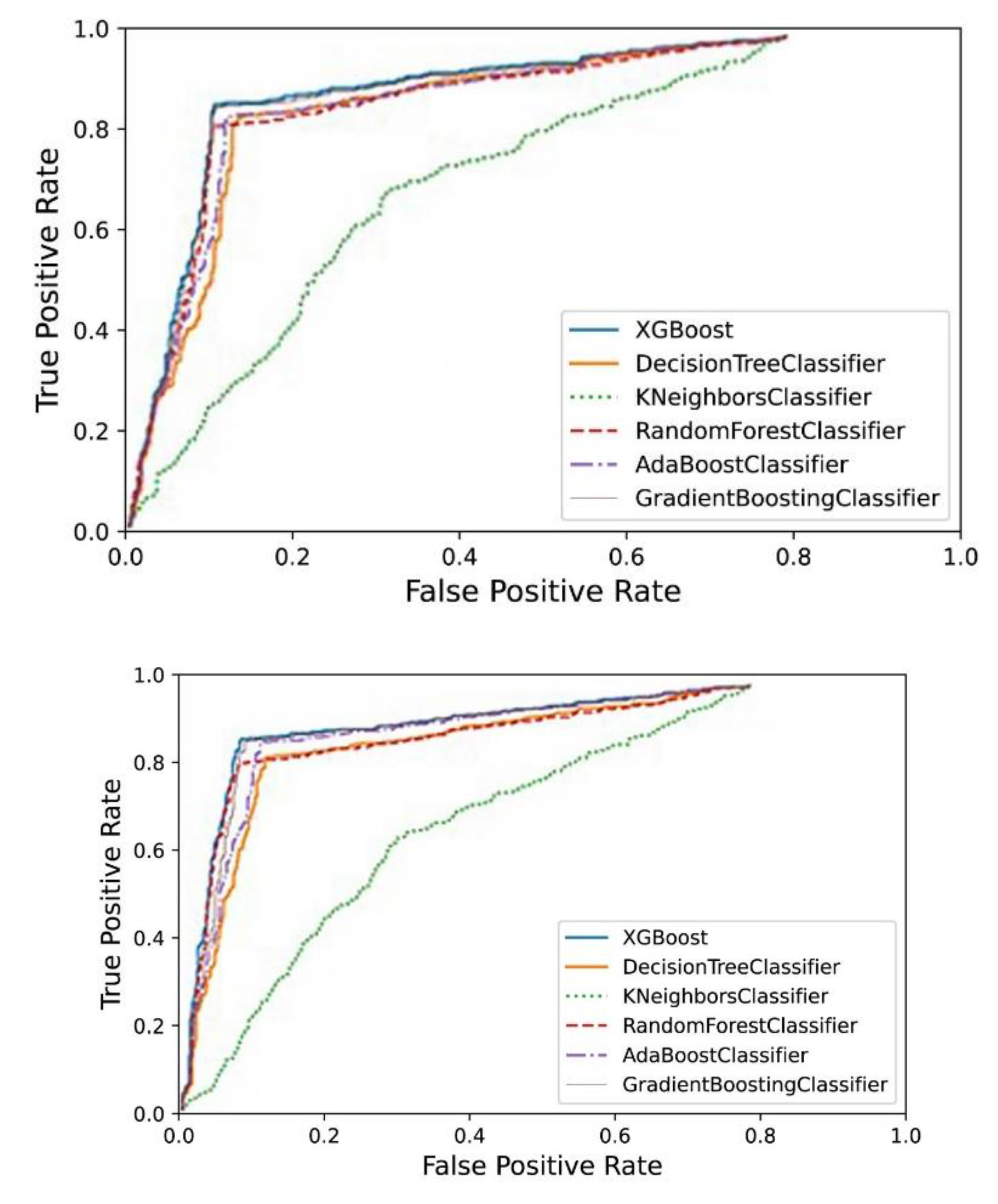
3. Image Segmentation and Feature Selection
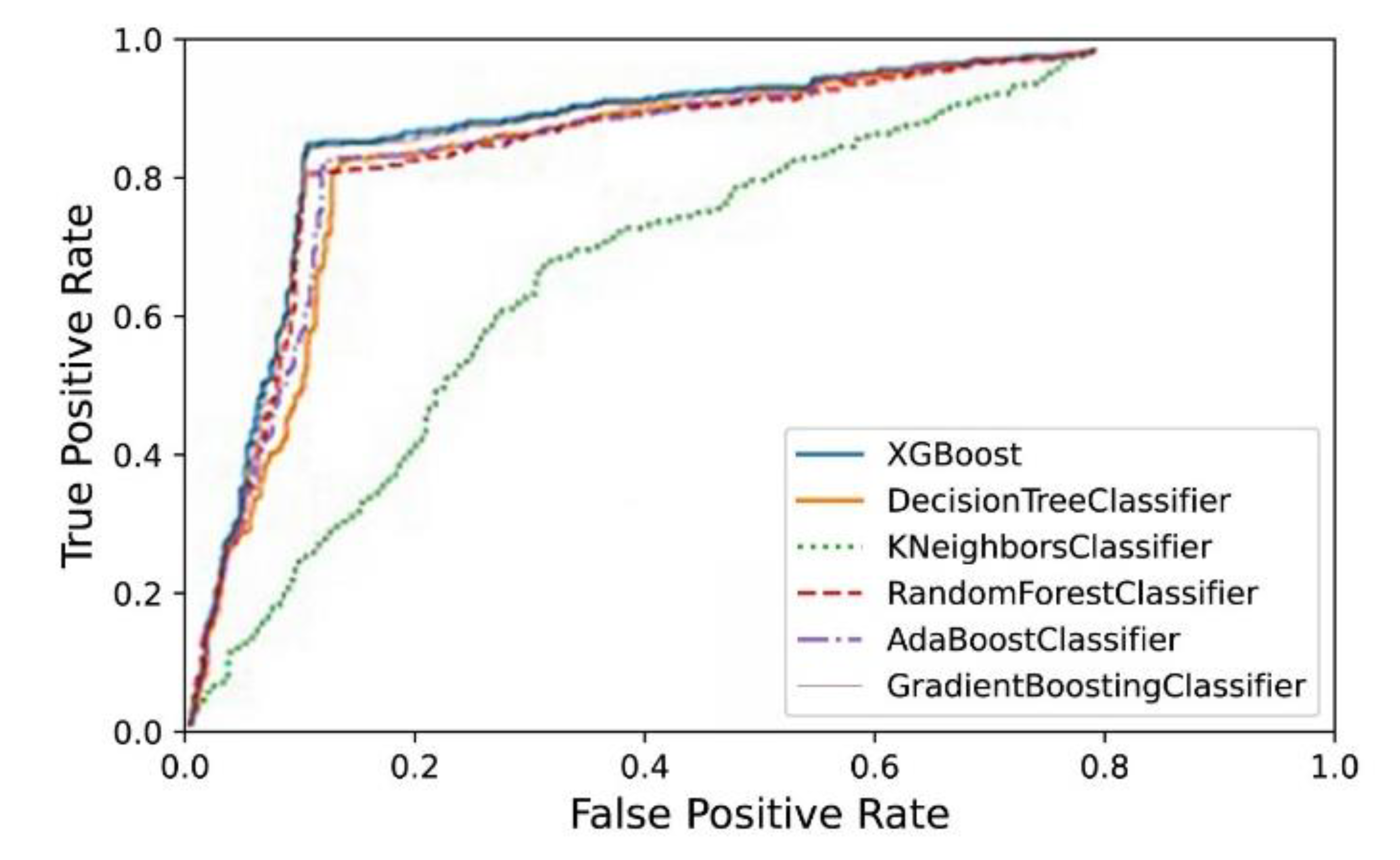
4. Results
5. Discussion
6. Conclusions and Future Work
Author Contributions
Funding
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Soerjomataram, I.; Ervik, M.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. GLOBOCAN 2012 v1. 0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11. 2013; International Agency for Research on Cancer: Lyon, France, 2014. [Google Scholar]
- Chen, W.; Zheng, R.; Baade, P.D.; Zhang, S.; Zeng, H.; Bray, F.; Jemal, A.; Yu, X.Q.; He, J. Cancer statistics in China, 2015. CA A Cancer J. Clin. 2016, 66, 115. [Google Scholar] [CrossRef] [PubMed]
- Specht, J.M.; Mankoff, D.A. Advances in molecular imaging for breast cancer detection and characterization. Breast Cancer Res. 2012, 14, 206. [Google Scholar] [CrossRef] [PubMed]
- Radiology ACo. Breast Imaging Reporting and Data System Atlas (BI-RADS® Atlas); American College of Radiology: Reston, VA, USA, 2003. [Google Scholar]
- Fletcher, S.W.; Elmore, J.G. Mammographic screening for breast cancer. N. Engl. J. Med. 2003, 348, 1672–1680. [Google Scholar] [CrossRef] [PubMed]
- Cady, B.; Chung, M. Mammographic screening: No longer controversial: LWW. Am. J. Clin. Oncol. 2005, 28, 1–4. [Google Scholar] [CrossRef]
- Lehman, C.D.; Lee, A.Y.; Lee, C.I. Imaging management of palpable breast abnormalities. Am. J. Roentgenol. 2014, 203, 1142–1153. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Cai, X.; Chen, X.; Hu, L.; Lou, X. Computer-aided detection and classification of microcalcifications in mammograms: A survey. Pattern Recognit. 2003, 36, 2967–2991. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd Acm Sigkdd International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016. [Google Scholar]
- Sajeev, S.; Bajger, M.; Lee, G. Superpixel texture analysis for classification of breast masses in dense background. IET Comput. Vis. 2018, 12, 779–786. [Google Scholar] [CrossRef]
- Saleck, M.; ElMoutaouakkil, A.; Mouçouf, M. Tumor detection in mammography images using fuzzy C-means and GLCM texture features. In Proceedings of the 2017 14th International Conference on Computer Graphics, Imaging and Visualization, Marrakesh, Morocco, 23–25 May 2017; pp. 122–125. [Google Scholar]
- Mohanty, A.K.; Senapati, M.R.; Lenka, S.K. An improved data mining technique for classification and detection of breast cancer from mammograms. Neural Comput. Appl. 2013, 22, 303–310. [Google Scholar] [CrossRef]
- Zebari, D.A.; Ibrahim, D.A.; Zeebaree, D.Q.; Haron, H.; Salih, M.S.; Damaševičius, R.; Mohammed, M.A. Systematic Review of Computing Approaches for Breast Cancer Detection Based Computer Aided Diagnosis Using Mammogram Images. Appl. Artif. Intell. 2021, 35, 2157–2203. [Google Scholar] [CrossRef]
- Dartois, L.; Gauthier, É.; Heitzmann, J.; Baglietto, L.; Michiels, S.; Mesrine, S.; Boutron-Ruault, M.; Delaloge, S.; Ragusa, S.; Clavel-Chapelon, F.; et al. A comparison between different prediction models for invasive breast cancer occurrence in the French E3N cohort. Breast Cancer Res. Treat. 2015, 150, 415–426. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Peng, Y.; Ou, C.; Chen, M.; Li, L. Diagnosis of breast masses from dynamic contrast-enhanced and diffusion-weighted MR: A machine learning approach. PLoS ONE 2014, 9, e87387. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, M.M.R.; Banerjee, S.; Chakraborty, C.; Chakraborty, C.; Ray, A.K. Statistical analysis of mammographic features and its classification using support vector machine. Expert Syst. Appl. 2010, 37, 470–478. [Google Scholar] [CrossRef]
- Holsbach, N.; Fogliatto, F.S.; Anzanello, M.J. A data mining method for breast cancer identification based on a selection of variables. Cienc. Saude Colet. 2014, 19, 1295–1304. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Şahan, S.; Polat, K.; Kodaz, H.; Güneş, S. A new hybrid method based on fuzzy-artificial immune system and k-nn algorithm for breast cancer diagnosis. Comput. Biol. Med. 2007, 37, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.H. Stochastic gradient boosting. Comput. Stat. Data Anal. 2002, 38, 367–378. [Google Scholar] [CrossRef]
- Friedman, J.H.; Popescu, B.E. Importance sampled learning ensembles. J. Mach. Learn. Res. 2003, 94305, 1–32. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Friedman, J.; Hastie, T.; Tibshirani, R. Additive logistic regression: A statistical view of boosting (with discussion and a rejoinder by the authors). Ann. Stat. 2000, 28, 337–407. [Google Scholar] [CrossRef]
- Moura, D.C.; López, M.A.G. An evaluation of image descriptors combined with clinical data for breast cancer diagnosis. Int. J. Comput. Assist. Radiol. Surg. 2013, 8, 561–574. [Google Scholar] [CrossRef]
- Pérez, N.P.; López, M.A.G.; Silva, A.; Ramos, I. Improving the Mann–Whitney statistical test for feature selection: An approach in breast cancer diagnosis on mammography. Artif. Intell. Med. 2015, 63, 19–31. [Google Scholar] [CrossRef]
- Arevalo, J.; González, F.A.; Ramos-Pollán, R.; Oliveira, J.L.; Lopez, M.A.G. Representation learning for mammography mass lesion classification with convolutional neural networks. Comput. Methods Programs Biomed. 2016, 127, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Pérez, N.; Guevara, M.A.; Silva, A. Improving breast cancer classification with mammography, supported on an appropriate variable selection analysis. In Medical Imaging 2013: Computer-Aided Diagnosis; International Society for Optics and Photonics: Bellingham, WA, USA, 2013; p. 22. [Google Scholar]
- Pérez, N.; Guevara, M.A.; Silva, A.; Ramos, I.; Loureiro, J. Improving the performance of machine learning classifiers for Breast Cancer diagnosis based on feature selection. In Proceedings of the 2014 Federated Conference on Computer Science and Information Systems, Warsaw, Poland, 7–10 September 2014. [Google Scholar]
- Clausi, D.A. An analysis of co-occurrence texture statistics as a function of grey level quantization. Can. J. Remote Sens. 2002, 28, 45–62. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K. Textural features for image classification. IEEE Trans. Syst. Man Cybern. 1973, 3, 610–621. [Google Scholar] [CrossRef]
- Soh, L.-K.; Tsatsoulis, C. Texture analysis of SAR sea ice imagery using gray level co-occurrence matrices. IEEE Trans. Geosci. Remote Sens. 1999, 37, 780–795. [Google Scholar] [CrossRef]
- Wei, X. Gray Level Run Length Matrix Toolbox, v1. 0. Software; Aeronautical Technology Research Center: Beijing, China, 2007. [Google Scholar]
- Chu, A.; Sehgal, C.M.; Greenleaf, J.F. Use of gray value distribution of run lengths for texture analysis. Pattern Recognit. Lett. 1990, 11, 415–419. [Google Scholar] [CrossRef]
- Hassani, H.; Silva, E.S. A Kolmogorov-Smirnov Based Test for Comparing the Predictive Accuracy of Two Sets of Forecasts. Econometrics 2015, 3, 590–609. [Google Scholar] [CrossRef]
- Li, L.; Wu, Z.; Salem, A.; Chen, Z.; Chen, L.; George, F.; Kallergi, M.; Berman, C. Computerized analysis of tissue density effect on missed cancer detection in digital mammography. Comput. Med. Imaging Graph. 2006, 30, 291–297. [Google Scholar] [CrossRef]
- Brem, R.F.; Hoffmeister, J.W.; Rapelyea, J.A.; Zisman, G.; Mohtashemi, K.; Jindal, G.; DiSimio, M.P.; Rogers, S.K. Impact of Breast Density on Computer-Aided Detection for Breast Cancer. Am. J. Roentgenol. 2005, 184, 439–444. [Google Scholar] [CrossRef]
- Malich, A.; Marx, C.; Facius, M.; Boehm, T.; Fleck, M.; Kaiser, W.A. Tumour detection rate of a new commercially available computer-aided detection system. Eur. Radiol. 2001, 11, 2454–2459. [Google Scholar] [CrossRef]
- Barlow, W.E.; Chi, C.; Carney, P.A.; Taplin, S.H.; D’Orsi, C.; Cutter, G.; Hendrick, R.E.; Elmore, J.G. Accuracy of Screening Mammography Interpretation by Characteristics of Radiologists. JNCI J. Natl. Cancer Inst. 2004, 96, 1840–1850. [Google Scholar] [CrossRef]
- Muttarak, M.; Pojchamarnwiputh, S.; Chaiwun, B. Breast carcinomas: Why are they missed? Singap. Med. J. 2006, 47, 851. [Google Scholar]
- Yu, S.; Guan, L. A CAD system for the automatic detection of clustered microcalcifications in digitized mammogram films. IEEE Trans. Med. Imaging 2000, 19, 115–126. [Google Scholar] [PubMed]
- Jiang, Y.; Metz, C.E.; Nishikawa, R.M.; Schmidt, R.A. Comparison of Independent Double Readings and Computer-Aided Diagnosis (CAD) for the Diagnosis of Breast Calcifications. Acad. Radiol. 2006, 13, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Sankar, D.; Thomas, T. Fast fractal coding method for the detection of microcalcification in mammograms. In Proceedings of the 2008 International Conference on Signal Processing, Communications and Networking, Chennai, India, 4–6 January 2008. [Google Scholar]
- Jiang, J.; Yao, B.; Wason, A. A genetic algorithm design for microcalcification detection and classification in digital mammograms. Comput. Med. Imaging Graph. 2007, 31, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Shomona Gracia, J.; Geetha Ramani, R. Efficient classifier for classification of prognostic breast cancer data through data mining techniques. In Proceedings of the World Congress on Engineering and Computer Science, San Francisco, CA, USA, 24–26 October 2012; 1. [Google Scholar]
- Yoav, F.; Schapire, R.E. Experiments with a New Boosting Algorithm; ICML: Baltimore, MA, USA, 1996; pp. 148–156. [Google Scholar]
- Arefan, D.; Mohamed, A.A.; Berg, W.A.; Zuley, M.L.; Sumkin, J.H.; Wu, S. Deep learning modeling using normal mammograms for predicting breast cancer risk. Med. Phys. 2020, 47, 110–118. [Google Scholar] [CrossRef]
- Ai, H. GSEA–SDBE: A gene selection method for breast cancer classification based on GSEA and analyzing differences in performance metrics. PLoS ONE 2022, 17, e0263171. [Google Scholar] [CrossRef]
- Thalor, A.; Joon, H.K.; Singh, G.; Roy, S.; Gupta, D. Machine learning assisted analysis of breast cancer gene expression profiles reveals novel potential prognostic biomarkers for triple-negative breast cancer. Comput. Struct. Biotechnol. J. 2022, 20, 1618–1631. [Google Scholar] [CrossRef]
- Li, Q.; Yang, H.; Wang, P.; Liu, X.; Lv, K.; Ye, M. XGBoost-based and tumor-immune characterized gene signature for the prediction of metastatic status in breast cancer. J. Transl. Med. 2022, 20, 177. [Google Scholar] [CrossRef]
- Jang, J.Y.; Ko, E.Y.; Jung, J.S.; Kang, K.N.; Kim, Y.S.; Kim, C.W. Evaluation of the Value of Multiplex MicroRNA Analysis as a Breast Cancer Screening in Korean Women under 50 Years of Age with a High Proportion of Dense Breasts. J. Cancer Prev. 2021, 26, 258–265. [Google Scholar] [CrossRef]
- Jang, B.-S.; Kim, I.A. Machine-learning algorithms predict breast cancer patient survival from UK Biobank whole-exome sequencing data. Biomark. Med. 2021, 15, 1529–1539. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Das, S.; Kar, D.; Schwenker, F.; Sarkar, R. Computer Aided Breast Cancer Detection Using Ensembling of Texture and Statistical Image Features. Sensors 2021, 21, 3628. [Google Scholar] [CrossRef] [PubMed]
- Chai, H.; Zhou, X.; Zhang, Z.; Rao, J.; Zhao, H.; Yang, Y. Integrating multi-omics data through deep learning for accurate cancer prognosis prediction. Comput. Biol. Med. 2021, 134, 104481. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Hu, Y.; Wang, D.; Sun, M.; Li, H.; Yan, P.; Meng, Y.; Zhang, R.; Li, L.; Yu, D.; et al. Value of CT-Based Radiomics in Predicating the Efficacy of Anti-HER2 Therapy for Patients With Liver Metastases From Breast Cancer. Front. Oncol. 2022, 12, 852809. [Google Scholar] [CrossRef] [PubMed]
- Vamvakas, A.; Tsivaka, D.; Logothetis, A.; Vassiou, K.; Tsougos, I. Breast Cancer Classification on Multiparametric MRI – Increased Performance of Boosting Ensemble Methods. Technol. Cancer Res. Treat. 2022, 21, 15330338221087828. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, Z.; Yin, G.; Sui, C.; Liu, Z.; Li, X.; Chen, W. Prediction of HER2 expression in breast cancer by combining PET/CT radiomic analysis and machine learning. Ann. Nucl. Med. 2022, 36, 172–182. [Google Scholar] [CrossRef]
- Vy, V.P.T.; Yao, M.M.-S.; Le, N.Q.K.; Chan, W.P. Machine Learning Algorithm for Distinguishing Ductal Carcinoma In Situ from Invasive Breast Cancer. Cancers Basel. 2022, 14, 2437. [Google Scholar] [CrossRef]
- Wang, J.; Yang, X.; Cai, H.; Tan, W.; Jin, C.; Li, L. Discrimination of Breast Cancer with Microcalcifications on Mammography by Deep Learning. Sci. Rep. 2016, 6, 27327. [Google Scholar] [CrossRef]
| Algorithms | Accuracy | Recall | Precision | F1-Score | AUC |
|---|---|---|---|---|---|
| kNN | 63.99% | 0.6612 | 0.6263 | 0.6433 | 0.6407 |
| Decision Tree | 84.78% | 0.8511 | 0.8390 | 0.8511 | 0.8501 |
| adaboostM1 | 85.07% | 0.8734 | 0.8451 | 0.8590 | 0.8504 |
| random forest | 85.08% | 0.8526 | 0.8514 | 0.8378 | 0.8501 |
| GBDT | 85.45% | 0.8801 | 0.8595 | 0.8696 | 0.8695 |
| XGBoost | 87.21% | 0.8750 | 0.8597 | 0.8697 | 0.8699 |
| Algorithms | Accuracy | Recall | Precision | F1-Score | AUC |
|---|---|---|---|---|---|
| kNN | 65.06% | 0.6673 | 0.6350 | 0.6500 | 0.6707 |
| Decision Tree | 86.89% | 0.8701 | 0.8514 | 0.8378 | 0.8520 |
| adaboostM1 | 85.27% | 0.8734 | 0.8300 | 0.8590 | 0.8527 |
| random forest | 87.29% | 0.8774 | 0.8672 | 0.8643 | 0.8629 |
| GBDT | 88.74% | 0.8801 | 0.8765 | 0.8758 | 0.8774 |
| XGBoost | 90.24% | 0.8845 | 0.9000 | 0.8952 | 0.8903 |
| Rank | Feature | Remark |
|---|---|---|
| 1 | number of calcification spots | morphologic features |
| 2 | percentage of gravel calcification | morphologic features |
| 3 | sum average | texture features |
| 4 | sum entropy | texture features |
| 5 | average diameter of calcification | morphologic features |
| 6 | percentage of circular degree | morphologic features |
| 7 | number of linear calcification point | morphologic features |
| 8 | circular degree | morphologic features |
| 9 | axis ratio | morphologic features |
| 10 | proportion of calcification | morphologic features |
| 11 | entity | morphologic features |
| 12 | volume rate | morphologic features |
| 13 | difference entropy | texture features |
| 14 | difference variance | texture features |
| 15 | average grey-level | morphologic features |
| Algorithms | Accuracy | Recall | Precision | F1-Score | AUC |
|---|---|---|---|---|---|
| kNN | 64.90% | 0.6566 | 0.6059 | 0.6303 | 0.6408 |
| Decision Tree | 85.04% | 0.8290 | 0.8490 | 0.8429 | 0.8411 |
| adaboostM1 | 85.17% | 0.8367 | 0.8300 | 0.8428 | 0.8426 |
| random forest | 85.77% | 0.8249 | 0.8574 | 0.8456 | 0.8519 |
| GBDT | 85.90% | 0.8670 | 0.8627 | 0.8598 | 0.8591 |
| XGBoost | 88.13% | 0.8713 | 0.8810 | 0.8690 | 0.8792 |
| kNN vs. XGBoost | Decision Tree vs. XGBoost | adaboostM1 vs. XGBoost | Random Forest vs. XGBoost | GBDT vs. XGBoost | |
|---|---|---|---|---|---|
| Two-Sided (p-Value) | 2.2 × 10−16 * | 6.633 × 10−6 * | 0.005486 * | 0.3342 | 0.03193 * |
| One-Sided (p-Value) | 2.2 × 10−16 * | 3.317 × 10−6 * | 0.002743 * | 0.1679 | 0.01597 * |
| kNN vs. XGBoost | Decision Tree vs. XGBoost | adaboostM1 vs. XGBoost | Random Forest vs. XGBoost | GBDT vs. XGBoost | |
|---|---|---|---|---|---|
| Two-Sided (p-Value) | 2.2×10−16 * | 5.228 × 10−5 * | 0.00135 * | 0.03609 * | 0.01286 * |
| One-Sided (p-Value) | 2.2 × 10−16 * | 2.614 × 10−5 * | 0.0006752 * | 0.01805 * | 0.006429 * |
| kNN vs. XGBoost | Decision Tree vs. XGBoost | adaboostM1 vs. XGBoost | Random Forest vs. XGBoost | GBDT vs. XGBoost | |
|---|---|---|---|---|---|
| Two-Sided (p-Value) | 2.2 × 10−16 * | 0.0004882 * | 0.01121 * | 0.4522 | 0.0648 |
| One-Sided (p-Value) | 2.2 × 10−16 * | 0.0002441 * | 0.005604 * | 0.2289 | 0.0324 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, H.; Li, J.; Wu, H.; Li, L.; Zhou, X.; Jiang, X. Mammographic Classification of Breast Cancer Microcalcifications through Extreme Gradient Boosting. Electronics 2022, 11, 2435. https://doi.org/10.3390/electronics11152435
Liang H, Li J, Wu H, Li L, Zhou X, Jiang X. Mammographic Classification of Breast Cancer Microcalcifications through Extreme Gradient Boosting. Electronics. 2022; 11(15):2435. https://doi.org/10.3390/electronics11152435
Chicago/Turabian StyleLiang, Haobang, Jiao Li, Hejun Wu, Li Li, Xinrui Zhou, and Xinhua Jiang. 2022. "Mammographic Classification of Breast Cancer Microcalcifications through Extreme Gradient Boosting" Electronics 11, no. 15: 2435. https://doi.org/10.3390/electronics11152435
APA StyleLiang, H., Li, J., Wu, H., Li, L., Zhou, X., & Jiang, X. (2022). Mammographic Classification of Breast Cancer Microcalcifications through Extreme Gradient Boosting. Electronics, 11(15), 2435. https://doi.org/10.3390/electronics11152435







