Abstract
Existing nuclei segmentation methods have obtained limited results with multi-center and multi-organ whole-slide images (WSIs) due to the use of different stains, scanners, overlapping, clumped nuclei, and the ambiguous boundary between adjacent cell nuclei. In an attempt to address these problems, we propose an efficient stain-aware nuclei segmentation method based on deep learning for multi-center WSIs. Unlike all related works that exploit a single-stain template from the dataset to normalize WSIs, we propose an efficient algorithm to select a set of stain templates based on stain clustering. Individual deep learning models are trained based on each stain template, and then, an aggregation function based on the Choquet integral is employed to combine the segmentation masks of the individual models. With a challenging multi-center multi-organ WSIs dataset, the experimental results demonstrate that the proposed method outperforms the state-of-art nuclei segmentation methods with aggregated Jaccard index (AJI) and F1-scores of and , respectively, while achieving a lower number of parameters.
1. Introduction
The digital pathology revolution has begun by digitizing glass slides using a whole-slide image (WSI) scanner. Afterward, pathologists analyze the WSIs manually on a monitor using an image viewer [1]. Over the last few years, digital pathology has been employed in research, academic, and medical institutions in collecting data, managing databases, as well as designing robust studies for millions of specimens. However, the manual analysis performed by pathologists (i.e., the visual assessment of WSIs) is time consuming, especially in tasks like nuclei cell segmentation and counting [2,3].
Thanks to the vast developments in deep learning-based techniques supported by the recent improvements of hardware (e.g., graphics processing unit), researchers have been attracted to exploring the applicability and utility of deep learning in digital pathology [4]. For instance, deep learning techniques have been used to segment nuclei cells, count cancer cells, and detect cancer metastasis in lymph nodes.
Efficient automated nuclei cell segmentation methods, including deep learning-based methods, are required for realizing several tasks such as cell counting, feature extraction, classification of cancer type, cancer sub-type recognition, and cancer grading. Generally speaking, nuclei cell segmentation can be performed by classifying each pixel in a WSI as nuclei or non-nuclei. In the literature, deep convolutional neural networks (DCNNs) have been employed to segment and detect nuclei [5,6]. Deep learning-based semantic segmentation methods such as the fully convolutional network architecture (FCN) [7], UNet [8], self-correction [9], and SegNet [10] have been used to segment nuclei in WSIs, to name but a few. Besides, task-specific models like CIA-Net [11] and AS-UNet [12] have been introduced to accurately segment nuclei in WSIs.
Over the last decade, the automatic segmentation of nuclei in histopathological images has been well studied. Due to the dominating performance of deep learning in the same era, deep learning-based approaches are becoming increasingly popular in histopathological image analysis including nuclei segmentation. For instance, Cui et al. [13] proposed an automatic end-to-end DCNN algorithm for the segmentation of individual nuclei. They also presented a nuclei boundary model to predict the inner nuclear instance, the nuclear contour, and the background simultaneously in WSIs. Furthermore, they employed a customized weighted loss function based on the relative position of pixels within the image to improve and stabilize the inner nuclei and contour prediction. With the MoNuSegdataset [14], their model obtained an F1-score of . Naylor et al. [15] proposed a deep learning-based nuclei segmentation model called DIST, where they formulated the problem of overlapped nuclei segmentation in WSIs as a regression task. They achieved an F1-score of with the MoNuSeg dataset.
Furthermore, Wang et al. [16] proposed a multi-path dilated residual network for nuclei segmentation and detection. The network included the following key elements: (1) multi-scale feature extraction based on the ResNet model and the feature pyramid network (FPN), (2) the candidate region network, and (3) a final network for the detection and segmentation of nuclei. With the MoNuSeg dataset, they obtained an F1-score of . Zhou et al. [17] introduced the UNet++ model for semantic and instance segmentation. They improved the performance of the standard medical image segmentation model, UNet, by combining UNets of varying depths, redesigning the skip connections, and employing an architecture pruning scheme to accelerate the inference speed while maintaining the performance. Although UNet++ could improve segmentation accuracy over the standard UNet model, the computation cost increased. UNet++ achieved an F1-score of on the MoNuSeg dataset. Zeng et al. [18] introduced the RIC-UNet model for nuclei segmentation. RIC-UNet was constructed by adding residual blocks, multi-scale, and a channel attention mechanism to the UNet model. RIC-UNet obtained an F1-score of with the MoNuSeg dataset.
Additionally, Graham et al. [19] introduced a stain-aware multi-scale convolutional network, so-called SAMS-Net, for segmenting nuclei in WSIs. The architecture of SAMS-Net was inspired by the fully convolutional network (FCN) architecture [7]. In particular, SAMS-Net employs a set of pre-defined weight maps aware of the intensity of hematoxylin in each histopathological image. SAMS-Net extracts deep contextual features in the down-sampling path and localizes nuclei in the up-sampling path. The performance of SAMS-Net was evaluated on the nuclei segmentation dataset [20]—achieving a Dice score of . Xie et al. [21] proposed a deep interval-marker-aware network called DIMAN for nuclei segmentation. DIMAN combines the convolutional neural networks with the marker-controlled watershed. DIMAN has been trained to segment the foreground, marker, and interval of nuclei simultaneously. The performance of DIMAN was assessed on three public histopathology image datasets, achieving an aggregated Jaccard index (AJI) score of with the MoNuSeg dataset.
Existing automated nuclei cell segmentation methods still face several challenges when segmenting nuclei in WSIs, most importantly the color variation in WSIs due to the use of different stains, as well as the intrinsic variation in cell types and sizes. To better explain these challenges, in Figure 1, we present four examples of WSIs collected from multiple organs and multiple centers that use different stains. As one can see, the WSIs include variations like cellular structure (red boxes), overlapping (yellow circles), and clumped nuclei (green boxes). Such variations could limit the accuracy of automated nuclei cell segmentation methods significantly. In an attempt to develop an efficient nuclei segmentation model while tackling the challenges mentioned above, this article proposes an efficient stain-aware nuclei segmentation deep learning framework for multi-center WSIs. In contrast to existing methods that employ a single stain template from the dataset to normalize all images, we propose an efficient algorithm to select a set of stain templates based on stain clustering. Then, a set of individual deep learning-based nuclei segmentation models is trained based on each stain template, and finally, an aggregation function based on the Choquet integral is proposed to fuse the segmentation masks of the individual models.
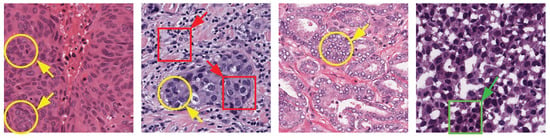
Figure 1.
Whole-slide images (WSIs) collected from multiple organs and many centers employing different stains. Cell overlapping, shape variation, and clumped nuclei are marked by yellow circles, red boxes, and green boxes, respectively.
2. Methodology
Figure 2 presents an overview of the proposed stain-aware nuclei segmentation deep learning method for multi-center WSIs. The key elements of the proposed method are an automatic stain template selection based on stain clustering, stain normalization, the construction of individual nuclei segmentation models, and an aggregation function based on the Choquet integral that produces the final nuclei segmentation masks. It should be noted that most related works randomly selected a stain template from the WSI dataset to normalize all images. Differently, we propose a stain template selection algorithm to select images from the WSI dataset to serve as stain templates. A stain-normalization method is used to transform all WSIs to the color space of each stain template. This normalization process generates WSI datasets. Afterward, individual nuclei segmentation models are built, where each normalized dataset is used to train an individual nuclei segmentation model to predict coarse nuclei segmentation masks (i.e., coarse labels). Ultimately, an aggregation function based on the Choquet integral and the gray wolf optimizer is employed to fuse the coarse nuclei segmentation masks into a single segmentation mask. Such an aggregation process can correctly join the information given in the segmentation masks of the individual models. Below, we describe the proposed method in detail.
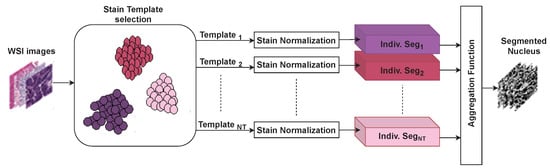
Figure 2.
Key elements of the proposed stain-aware nuclei segmentation method for multi-center WSI images. Indiv. Seg. refers to an individual segmentation model.
2.1. Selection of Stain Templates
To select a set of stain templates (), WSIs are clustered as groups . Here, we employ the K-means clustering algorithm to group WSIs having quite similar stain colors. The K-means clustering algorithm is a centroid-based algorithm, where each cluster is associated with a centroid. The clustering process can be formulated as follows:
Here, refers to the mean of the WSIs of the group and .
It should be noted that the number of selected stain templates () equals the number of centroids (K). Given the value of and the WSIs, the K-means clustering algorithm clusters the WSIs into K groups with K centroids. To define the stain templates, we compute the distance map D between the WSIs and the K centroid as shown below:
Here, refers to the Euclidean distance between the ith image and the jth centroid. After we compute the distance map D, we determine the NT images that achieve the minimum RMSE values with the K centroids. In other words, we determine the index of the minimum of each column in D. The resulting NT images are then employed as stain templates. The value of K (i.e., the number of stain templates—NT) is tuned experimentally. Algorithm 1 presents the steps of the stain template selection algorithm.
| Algorithm 1 Pseudocode for the stain template selection algorithm. |
|
2.2. Stain Normalization
Stain normalization techniques have been widely used in histopathological image assessment tools. It is a fundamental step to reduce color variations among WSIs and obtain a more desirable color consistency between the images before assessing them. In this study, we employed the stain normalization method introduced in [22], in which all WSIs are transformed to the color space of a pre-defined target image (i.e., stain template image). In [22], the stain normalization was formulated as an optimization problem as follows:
Here, H is the stain concentration matrix and W is the stain color matrix. The columns of W represent the RGB color of each stain. V is the relative optical density, and is the sparsity regularization parameter. The size of H is , where n is the number of pixels. The size of W is , where r is the number of stains and m is the three RGB channels. The optimization problem is solved by alternating W and H, meaning it optimizes one of them while fixing the other.
2.3. Constructing the Individual Nuclei Segmentation Models
Different semantic segmentation models based on deep convolution networks have been proposed in the literature, such as the fully connected convolution neural network (FCN) [7], UNet [8], SegNet [10], and fully connected densely connected convolutional networks (FCDenseNets) [23]. Many semantic segmentation models have been adapted in the last few years for the nuclei segmentation problem. In [24], we adapted several efficient semantic segmentation models to the nuclei segmentation problem and compared their results in terms of segmentation accuracy and the number of parameters. The study found that the FCDenseNet model [23] achieved promising nuclei segmentation results with a low number of parameters (i.e., number of weights). Accordingly, we employed the FCDenseNet model in this study to build the individual nuclei segmentation models (NSMs). Specifically, we employed the FCDenseNet103 architecture to construct the NSMs. It should be noted that each NSM is trained after normalizing all images using one of the stain templates selected by the stain template selection algorithm described in Section 2.1.
Figure 3 shows that each NSM includes encoder (top branch) and decoder (bottom branch) networks. There are 11 dense blocks (DBs) with five DBs in the encoder network, one DB in the BottleNeck(in between the encoder and the decoder networks), and 5 DBs in the decoder network. In total, there are 103 convolutional layers. As depicted in Figure 4a, each DB contains a set of layers. Each layer is composed of a batch normalization (BN) layer, a rectified linear unit (ReLU) layer, a convolutional layer, and dropout (Figure 4b). Each layer of the DB is connected to all other layers in a feedforward fashion [25], allowing reusing features and permitting the gradient to flow directly to earlier layers. Assume that is the output of the nth layer. is computed by applying a non-linear transformation to the output of the previous layer in the case of conventional DCNNs:
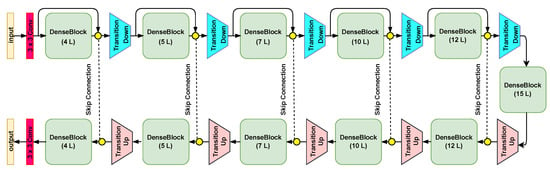
Figure 3.
The architecture of the individual nuclei segmentation model (NSM).
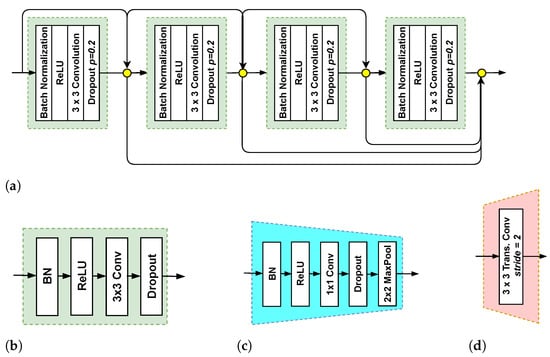
Figure 4.
Key components of the NSM. (a) dense block (DB) with four layers, (b) a layer, (c) transition down (TD), and (d) transition up (TU).
Here, H represents a convolution followed by a ReLU and often dropout. DB concatenates all feature outputs as follows:
In this expression, Z refers to a BN layer followed by ReLU, a convolution, and dropout, and represents the concatenation operation.
In the encoder network (top branch), each DB is followed by a transition down (TD) transformation. As one can see in Figure 4c, TD contains a BN layer, ReLU, a convolutional layer, and a max pooling operation. In the decoder network (bottom branch), each DB is followed by a transition up (TU) transformation. TU includes a transposed convolution with a stride of 2 (Figure 4d). The transposed convolution layers upsample the preceding feature maps. The outputs of TU layers are concatenated with the corresponding feature maps acquired by the skip connections. The concatenated feature maps represent the input of the next DB. To produce the segmentation masks, a convolutional layer followed by a softmax is employed at the end of the decoder network.
2.4. Constructing Aggregation Function Based on the Choquet Integral
An aggregation operator is a function that fuses a set of real numbers () into a real number :
It should be noted that the function must satisfy the following properties to be said aggregation operator:
- Identity: .
- Boundary conditions: and .
- Monotonicity: If , then ≤.
The Choquet integral is a widely-used and powerful aggregation operator that depends on the calculation of fuzzy measures. The Choquet integral-based aggregation function, which satisfies the above-mentioned properties, can represent different combinations among the set of values to be fused through a fuzzy measure [26,27]. Given n-tuple real numbers (), the Choquet integral with a fuzzy measure is expressed as follows:
It should be noted that increasing permutation should be applied on the elements of () to calculate . The increasing permutation on produces a new vector , where the number at the underscore is an index referring to the order of the element, , and .
As one can see in Equation (7), the computation of requires a fuzzy measure. Assume that we trained a set of NSMs, ; a real-valued function is considered as a fuzzy measure if and only if it fulfills the properties mentioned below:
- (boundary condition).
- (normalized).
- Assume that Q is a reference set, If , then (monotonicity).
There are non-empty subsets in the set S. Accordingly, each measure element in the fuzzy measure should correspond to a non-empty subset of S. It should be noted that has a total of elements; the measure element that corresponds to the whole set S must equal to one, whereas the rest of measure elements can hold any real value in the range [0, 1] while satisfying the monotonicity property. In this study, we use the gray wolf optimizer (GWO) [28] to compute the fuzzy measure elements that correspond to all subsets optimally. GWO is inspired by the social hierarchy and the hunting behavior strategy of gray wolves. The main steps of the hunting behavior of the gray wolves are encircling the prey and hunting.
Figure 5 presents the steps to find the fuzzy measure elements using the GWO algorithm. First, we initialize the population of fuzzy measure elements in the range . The GWO algorithm employs N search agents to find the optimal solution, where each search agent (SA) contains candidate values for the fuzzy measure elements . Below, we present the population of the GWO, where each row in the population represents a search agent.
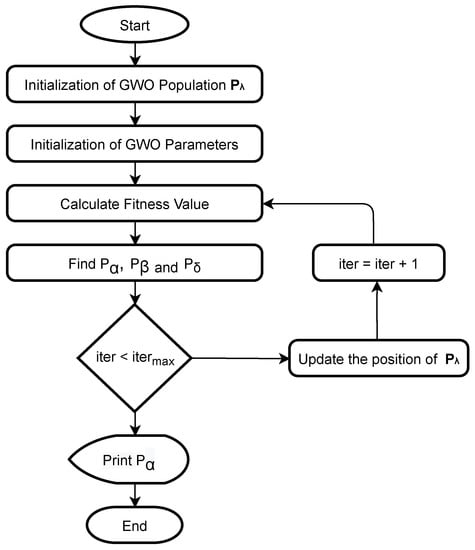
Figure 5.
Finding fuzzy measure elements using the GWO algorithm. , and are the best three search agents [28].
For each search agent in the population , we utilize the Choquet integral-based aggregation function (Equation (7)) to combine the predictions of the NSMs (pixel-level aggregation). Afterward, the result of the aggregation (a nuclei segmentation mask) and the corresponding ground-truth are used to compute the fitness of the search agent. Here, we employ the aggregated Jaccard index (AJI) as a fitness function:
Here, is the ith ground-truth mask of nuclei pixels, is the predicted nuclei segmentation mask, is the connected component from the predicted mask that maximizes the Jaccard index, and is the list of indices of pixels that do not belong to any element in the ground-truth.
After calculating the fitness of all search agents, the population is updated. GWO repeats this process till it reaches the maximum number of iterations or optimizer convergence. The number of search agents and the number of iterations were set to 100 and 50, respectively.
2.5. Implementation of the Proposed Method
In Algorithm 2, we present the steps to implement the proposed method. The WSI dataset was split into training and testing. In the training phase, we employed the proposed stain template selection algorithm (Section 2.1) to select stain templates (). Given a stain template , we normalized all training images (as described in Section 2.2) and then trained a nuclei segmentation model . This process was repeated with all stain templates, resulting in nuclei segmentation models . For trained models, we obtained nuclei segmentation masks by inputting the input WSI into the learned mapping functions of the nuclei segmentation models:
In the testing phase, the trained nuclei segmentation models were used to predict the segmentation masks R that corresponded to the test WSIs. Afterward, the masks of R were fused using Choquet integral expressed in Equation (7).
| Algorithm 2 Implementation of the proposed method. |
|
In this study, we used the F1-score (F1) and aggregated Jaccard Index (AJI) [14] to evaluate the proposed method. The F1-score represents the harmonic average of the precision (the ratio of segmented nuclei in label images) and recall (the percentage of the total number of nuclear pixels in the label image correctly segmented). The F1-score can be expressed as follows:
Here, (true positive) is the number of correctly segmented nuclei cell pixels, (true negative) is the number of correctly segmented non-nuclei cell pixels, (false positive) is the number of non-nuclei cell pixels wrongly segmented as nuclei, and (false negative) is the number of nuclei cell pixels wrongly segmented as non-nuclei.
The AJI is expressed in Equation (9). It is an enhanced version of the Jaccard index, which has a more powerful ability to measure the strength of nuclei segmentation [14].
3. Results and Discussion
3.1. Multi-Center Multi-Organ WSI Image Dataset
To assess the efficacy of the proposed method, we used a well-known and very challenging multi-center multi-organ WSI dataset called MoNuSeg [14], which was made available by the Indian Institute of Technology Guwahati. The MoNuSeg dataset contains 30 WSIs having a size of with a total of 21,000 manually annotated nuclei. The MoNuSeg dataset is tremendously difficult as it has WSIs of seven different organs (breast, kidney, colon, stomach, prostate, liver, and bladder). Besides, the WSIs were collected from many medical centers that employed different stains. It should be noted that in our experiments, we used 23 WSIs for training the proposed method and kept one WSI for each organ for testing (seven WSIs hand-picked from the dataset) to take into consideration the diversity of organs and stain colors. We used the same test set to evaluate all segmentation models.
The input image size to the NSM models was . During training, we re-scaled each WSI to , and then, we divided it into four non-overlapping sub-images of size . Data augmentation was also employed by cropping 200 patches of size from each WSI randomly, then flipping them vertically and horizontally, resulting in a total of 14,076 patches. During the test phase, each WSI was re-scaled to and then divided into four non-overlapping sub-images of size . It should be noted that the proposed segmentation classified each pixel in the input WSI as a nucleus or non-nucleus pixel. Thus, even if the splitting line crossed a nucleus, the proposed method segmented pixels of each part of the cropped nucleus. Afterward, the patches belonging to one input WSI were stacked while keeping their spatial order to form the final segmentation mask.
3.2. Analyzing the Performance of the Proposed Method
First, we analyzed the effect of the stain template selection algorithm on the performance of the proposed method. Figure 6 shows the distance maps of the stain template selection algorithm with different numbers of clusters: K = 2, 3, and 5. In these maps, the darkest color indicates the index of the closest WSI to the cluster centroid. The stain template selection algorithm selects the best stain templates based on these distance maps as explained in Section 2.1. Figure 6a shows the distance map of the stain selection algorithm with K = 2 (two clusters). As shown, the closest WSI to Centroid 1 (WSI # 15) is very far from Centroid 2. Figure 6b shows the distance map of the stain template selection algorithm with K = 3. As we can see, WSI Number 15 is the closest image to the centroid of the first cluster, which obtains a Euclidean distance of 0.31 (the minimum value on the map). WSI Number 5 is the closest image to the centroid of the second cluster (Euclidean distance of 0.33). In turn, WSI Number 1 is the closest image to the centroid of the third cluster (Euclidean distance of 0.42). Figure 6c shows the distance map of the stain selection algorithm with K = 5 (five clusters). As one can see, Centroid 3 and Centroid 5 cannot yield distinct stain templates because most of the images are far from these two clusters. Therefore, the stain selection algorithm with five clusters (K = 5) may yield limited segmentation results. However, the use of three clusters (K = 3) would lead to a more accurate nuclei segmentation.
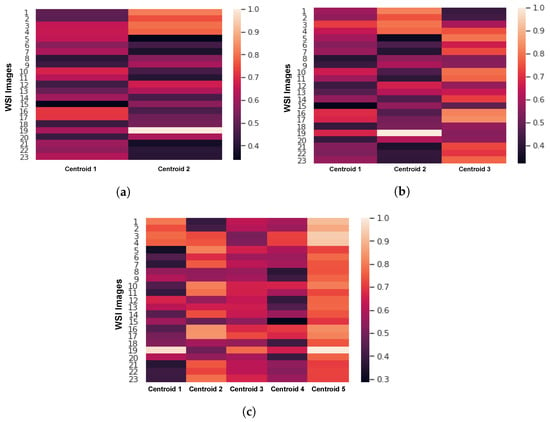
Figure 6.
The distance maps of the stain selection algorithm with (a) K = 2, (b) K = 3, and (c) K = 5.
Table 1 shows the performance of the stain template selection algorithm with different numbers of clusters , and 5 in terms of the F1 and AJI-scores. It is worth noting that means that we train one model after normalizing the data using one stain template, means that we train two models after normalizing the data using two stain templates, and means that we train five models after normalizing the data using five stain templates. As one can see in Table 1, the proposed method obtains the lowest F1- and AJI-scores with one stain template. A noticeable improvement in the segmentation results ( approximately) can be seen when using multiple stain templates. The proposed method achieved the highest F1-score (), AJI-score (), precision , and recall when setting the number of stain templates k to three.

Table 1.
Effect of the stain template selection algorithm on the performance of the proposed method. The best results are in bold. AJI, aggregated Jaccard index.
Second, we evaluated the performance of other variants of the proposed method by replacing the Choquet integral-based aggregation function with other aggregation operators: median, mean, and max aggregation operators. Specifically, in this experiment, we fixed the number of templates to three (, meaning we used three NSMs) while replacing the proposed Choquet integral-based aggregation function with other operators. As presented in Table 2, the mean operator achieved F1- and AJI-scores of and , which were better than the maximum and median operators.

Table 2.
Evaluating other variants of the proposed method by replacing the Choquet integral-based aggregation function with other aggregation operators: median, mean, and max aggregation operators. The best results are in bold.
Although the mean aggregation function gave an acceptable performance, the proposed Choquet integral-based aggregation achieved slightly better results. It is worth noting that one of the main advantages of the Choquet integral is that it considers the interaction between the components to be combined through the fuzzy measure. In turn, other operators like the maximum operator disregard the relationship between the components and ignore inherent information. Differently, the Choquet integral is a generalization of various aggregation operators, such as the arithmetic mean, weighted sum, ordered weighted average, minimum, and maximum [27,29,30].
Figure 7 shows the segmentation results of the proposed method with four different organs: liver, breast, colon, and kidney. Despite the color variation in the four WSIs, the proposed method achieved precise segmentation masks. As shown, the proposed method accurately segmented nuclei in the WSIs of kidney organs with an AJI-score of . Furthermore, it could segment the liver WSI with an AJI-score of , noting that this WSI was comprised of many overlapped nuclei having different shapes.
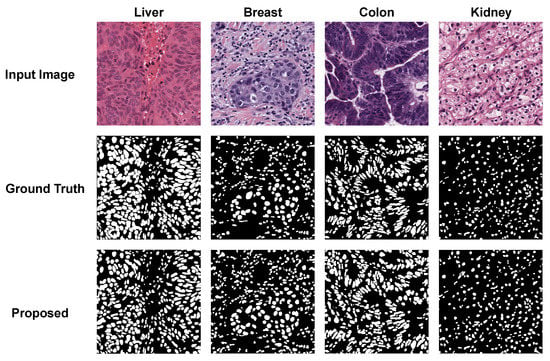
Figure 7.
Segmentation results of the proposed method with different organs (liver, breast, colon, and kidney).
Table 3 compares the performance of the proposed method with recently published nuclei segmentation methods [15,18,31]. Table 3 also compares the proposed method with two popular medical image segmentation models (UNet [8] and UNet++ [17]), as well as efficient semantic segmentation models adopted for the nuclei segmentation task [7,10,23]. It should be noted that all these models were trained using the MoNuSeg dataset. Besides, in this experiment, we used the same data augmentation strategy used to train our model, and we tried different stain templates to get the best possible performance for all compared models. As one can see, the proposed method outperformed all compared methods. Both FCN8sand UNet++ achieved a competitive performance over the other methods; however, they obtained AJI-scores of and , respectively, which were lower than the proposed method. The NSM-based FCDenseNet103 (one of the key components of the proposed model that was trained) achieved F1- and AJI-scores higher than the other compared methods. Besides, we compared the performance of the proposed method with a popular segmentation software called ImageFIJI [32], finding that the proposed method obtained superior results. It should be noted that the recently published nuclei segmentation methods achieved improvements of 1–2% over the state-of-the-art models—for instance, [33]. As one can see in Table 3, the AJI-score of the proposed method was better than the ones of FCN8s, UNet++, and FCDenseNet103 with approximately 3, 3, and 2 points, respectively. Thus, it is clear that the proposed method achieved good enhancements over the existing methods when looking at the range of enhancement achieved with the state-of-the-art models and considering the difficulty of the problem.

Table 3.
Comparing the performance of the proposed method with recently published nuclei segmentation methods, popular medical image segmentation models (UNet [8] and UNet++ [17]), as well as efficient semantic segmentation models adopted for the nuclei segmentation task ([7,10,23]). The best results are in bold.
Figure 8 shows the segmentation masks of the proposed method, FCN8s, UNet++, and ImageFIJI for the WSIs of bladder, prostate, and stomach organs. The ImageFIJI software had the worst segmentation results with all organ images; in particular, there was a serious over-segmentation for the WSIs of bladder and stomach, which have dense nuclei. In the case of the prostate organ WSI, Figure 8 shows that the proposed method can highly recover the mis-segmentation of the other methods. With the prostate WSI, the proposed method achieved an AJI-score of , which was better than the high-performance method UNet++. Figure 9 presents the boxplots of the F1- and AJI-scores of the proposed method, as well as different compared methods (UNet, SegNet, UNet++, and ImageFIJI). Given the F1- and AJI-scores of the test images with a particular model, a boxplot can be displayed based on a five-number summary: the minimum, the maximum, the sample median, and the first and third quartiles. As shown in Figure 9, the proposed method achieved the highest median AJI- and F1-scores without outliers.
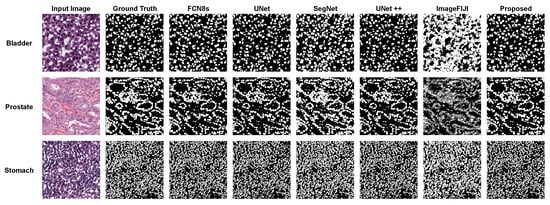
Figure 8.
The nuclei segmentation masks of the proposed method, FCN8s, UNet++, and ImageFIJI for the WSIs of bladder, prostate, and stomach organs.
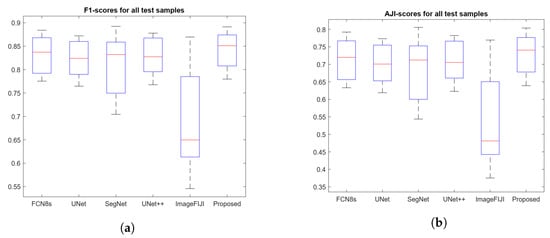
Figure 9.
Boxplots of the F1- and AJI-scores of the proposed method, as well as different compared methods: UNet, SegNet, UNet++, and ImageFIJI. (a) Boxplot of F1-scores and (b) Boxplot of AJI-scores.
To demonstrate the efficiency of the proposed method, in Figure 10, we plot the AJI-scores and the number of parameters of the proposed method, FCN8s, UNet, SegNet, UNet++, and FC-DenseNet103. As shown, the nuclei segmentation models had a noticeable variation in the number of parameters. On the one hand, UNet++ achieved an AJI-score higher than ; however, it is a bit heavy model with >35 million trainable parameters (high computational cost). On the other hand, FCDenseNet achieved an AJI-score higher than ; it had <10 million trainable parameters (light architecture), which made it the primary choice to construct the NSMs. It is worth noting that despite the number of trainable parameters of the proposed method (25 million approximately) being higher than NSMs (FCDenseNet), it achieved state-of-the-art results (AJI >73%). All in all, the proposed method obtained the highest segmentation results while maintaining the number of parameters lower than the other methods having close results.
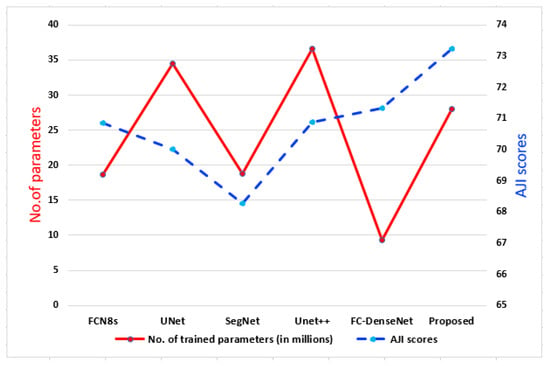
Figure 10.
Plot of the number of parameters vs. the AJI-score.
3.3. Evaluating the Proposed Method on a Second Multi-Organs Dataset
To further prove the efficiency of the proposed method, we tested it on another WSI dataset. Specifically, we assessed the performance of the proposed method with the recently published CryoNuSegdataset [33]. CryoNuSeg includes WSIs from ten human organs, noting that these organs have not been covered in other publicly available datasets, including the MoNuSeg dataset. The ten organs covered in CryoNuSeg are adrenal gland, larynx, lymph node, mediastinum, pancreas, pleura, skin, testis, thymus, and thyroid gland. CryoNuSeg contained 30 WSIs having a size of (). In this experiment, we used the 30 images of the CryoNuSeg dataset as a testing set. Then, we evaluated the performance of the nuclei segmentation models trained on the MoNuSeg dataset using this testing set (as recommended by the authors of the CryoNuSeg dataset [33].
Table 4 presents the segmentation results of the proposed method, FCDenseNet103, UNet++, UNet, FCN8s, and SegNet on the testing set of the CryoNuSeg dataset. Although we did not train the proposed method on the WSIs of the CryoNuSeg dataset, it still outperformed the compared methods. It achieved precision and recall scores of more than 85% with the 30 WSIs of 10 different organs (the models were been trained on the WSIs of these organs). As one can see, the proposed method outperformed the UNet and UNet++, which were two baseline biomedical image segmentation models. The proposed method attained an AJI-score of 64.51%, which was two points higher than the AJI-score of the FCDenseNet103 model. This analysis confirmed that our method could give accurate nuclei segmentation even with unseen WSI datasets and also with images of unseen organs (i.e., the models were been trained on images of the organs included in the testing set).

Table 4.
Evaluating the proposed method on the testing set of the CryoNuSegdataset (30 WSIs of 10 organs not included in the MoNuSegdataset). The best results are in bold.
3.4. Evaluating the Proposed Method on a Single-Organ Dataset
Here, we present an additional evaluation for the proposed method on the BNSdataset [34]. The BNS dataset contained 33 WSI patches having a size of () collected from the breasts of seven patients (a total of 2754 annotated cells). Following the evaluation setting mentioned in Section 3.3, we used all 33 images of the BNS dataset as the testing set to evaluate the performance of the nuclei segmentation models trained on the MoNuSeg dataset.
Table 5 presents the segmentation results of the proposed method, FCDenseNet103, UNet++, UNet, FCN8s, and SegNet on the BNS dataset. The proposed method beat the compared methods. It obtained an AJI-score of 65.76%, which was three points higher than the AJI-score of the FCDenseNet103 model. Furthermore, the AJI-score of the proposed method was much better than the UNet and UNet++ models by eight and six points, respectively. Based on the analysis presented here and in Section 3.2 and Section 3.3, it is clear that the proposed method can achieve accurate nuclei segmentation with multi-organ, single-organ, and multi-center WSIs.

Table 5.
Evaluating the proposed method on the testing set of the BNSdataset (33 WSIs of human breasts). The best results are in bold.
4. Conclusions
This article presented an efficient stain-aware nuclei segmentation deep learning method for multi-center WSIs. The key elements of the proposed method were the stain template selection algorithm based on the K-means clustering technique, stain normalization, construction of individual nuclei segmentation models, and the aggregation function based on the Choquet integral that produced the final nuclei segmentation masks. Notably, most related works randomly selected a single-stain template from the WSI dataset to normalize the images. Differently, in this article, we proposed a stain template selection algorithm to select a set of WSIs from the multi-center WSI dataset to serve as stain target templates.
To demonstrate the performance of the nuclei segmentation models, we used a well-known and very challenging multi-organ WSI dataset, which included WSIs collected from different organs and scanners. We comprehensively compared the performance of the proposed method with several nuclei segmentation models and exiting software in terms of the F1-score and the object-level AJI metric, as well as the number of trainable parameters. Experimental results demonstrated that the proposed method surpassed the state-of-the-art nuclei segmentation methods with an AJI-score of while keeping the number of parameters (25 million approximately) lower than the related methods that obtained close results.
The future work will be focused on the use of different aggregation strategies to combine the individual nuclei segmentation models and the incorporation of stain normalization techniques into the deep learning framework. Cost-sensitive learning will be employed in our future work to give different attention to misclassification errors.
Author Contributions
Conceptualization, L.H., M.A.-N., and A.S.; methodology, L.H., M.A.-N., and A.S.; software, L.H. and A.S.; validation, L.H., M.A.-N., and A.S.; formal analysis, L.H., M.A.-N., and A.S.; investigation, L.H., M.A.-N., and A.S.; resources, L.H., M.A.-N., and A.S.; data curation, L.H., M.A.-N., and A.S.; writing—original draft preparation, L.H., M.A.-N., and A.S.; writing—review and editing, L.H., M.A.-N., A.S., and O.A.O.; visualization, L.H., M.A.-N., and A.S.; supervision, M.A.-N., D.P., A.S., and O.A.O.; project administration, M.A.-N. and D.P.; funding acquisition, M.A.-N. and D.P. All authors read and agreed to the published version of the manuscript.
Funding
The Spanish Government partly supported this research through Project PID2019-105789RB-I00.
Data Availability Statement
In our study, we used MoNuSeg dataset [14], CryoNuSeg dataset [33] and BNS dataset [34], which are publicly available.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pantanowitz, L. Digital images and the future of digital pathology. J. Pathol. Inform. 2010, 1. [Google Scholar] [CrossRef]
- Elazab, N.; Soliman, H.; El-Sappagh, S.; Islam, S.; Elmogy, M. Objective Diagnosis for Histopathological Images Based on Machine Learning Techniques: Classical Approaches and New Trends. Mathematics 2020, 8, 1863. [Google Scholar] [CrossRef]
- Pocevičiūtė, M.; Eilertsen, G.; Lundström, C. Survey of XAI in digital pathology. In Artificial Intelligence and Machine Learning for Digital Pathology; Springer: Berlin/Heidelberg, Germany, 2020; pp. 56–88. [Google Scholar]
- Guo, Z.; Liu, H.; Ni, H.; Wang, X.; Su, M.; Guo, W.; Wang, K.; Jiang, T.; Qian, Y. A fast and refined cancer regions segmentation framework in whole-slide breast pathological images. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Irshad, H.; Veillard, A.; Roux, L.; Racoceanu, D. Methods for Nuclei Detection, Segmentation, and Classification in Digital Histopathology: A Review—Current Status and Future Potential. IEEE Rev. Biomed. Eng. 2014, 7, 97–114. [Google Scholar] [CrossRef] [PubMed]
- Alom, M.Z.; Yakopcic, C.; Taha, T.M.; Asari, V.K. Microscopic Nuclei Classification, Segmentation and Detection with improved Deep Convolutional Neural Network (DCNN) Approaches. arXiv 2018, arXiv:1811.03447. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Abdel-Nasser, M.; Saleh, A.; Puig, D. Channel-wise Aggregation with Self-correction Mechanism for Multi-center Multi-Organ Nuclei Segmentation in Whole Slide Imaging. In Proceedings of the 15th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications—Volume 4: VISAPP, Valletta, Malta, 27–29 February 2020. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Zhou, Y.; Onder, O.F.; Dou, Q.; Tsougenis, E.; Chen, H.; Heng, P.A. CIA-Net: Robust Nuclei Instance Segmentation with Contour-Aware Information Aggregation. In Information Processing in Medical Imaging; Chung, A.C.S., Gee, J.C., Yushkevich, P.A., Bao, S., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 682–693. [Google Scholar]
- Pan, X.; Li, L.; Yang, D.; He, Y.; Liu, Z.; Yang, H. An Accurate Nuclei Segmentation Algorithm in Pathological Image Based on Deep Semantic Network. IEEE Access 2019, 7, 110674–110686. [Google Scholar] [CrossRef]
- Cui, Y.; Zhang, G.; Liu, Z.; Xiong, Z.; Hu, J. A deep learning algorithm for one-step contour aware nuclei segmentation of histopathology images. Med. Biol. Eng. Comput. 2019, 57, 2027–2043. [Google Scholar] [CrossRef]
- Kumar, N.; Verma, R.; Sharma, S.; Bhargava, S.; Vahadane, A.; Sethi, A. A Dataset and a Technique for Generalized Nuclear Segmentation for Computational Pathology. IEEE Trans. Med. Imaging 2017, 36, 1550–1560. [Google Scholar] [CrossRef]
- Naylor, P.; Laé, M.; Reyal, F.; Walter, T. Segmentation of Nuclei in Histopathology Images by Deep Regression of the Distance Map. IEEE Trans. Med. Imaging 2019, 38, 448–459. [Google Scholar] [CrossRef]
- Wang, E.K.; Zhang, X.; Pan, L.; Cheng, C.; Dimitrakopoulou-Strauss, A.; Li, Y.; Zhe, N. Multi-Path Dilated Residual Network for Nuclei Segmentation and Detection. Cells 2019, 8, 499. [Google Scholar] [CrossRef]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. UNet++: Redesigning Skip Connections to Exploit Multiscale Features in Image Segmentation. IEEE Trans. Med. Imaging 2020, 39, 1856–1867. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Xie, W.; Zhang, Y.; Lu, Y. RIC-UNet: An Improved Neural Network Based on UNet for Nuclei Segmentation in Histology Images. IEEE Access 2019, 7, 21420–21428. [Google Scholar] [CrossRef]
- Graham, S.; Rajpoot, N.M. SAMS-NET: Stain-aware multi-scale network for instance-based nuclei segmentation in histology images. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 590–594. [Google Scholar] [CrossRef]
- Computational Precision Medicine Nuclei Segmentation Challenge Website. Available online: http://miccai.cloudapp.net/competitions/57 (accessed on 15 October 2017).
- Xie, L.; Qi, J.; Pan, L.; Wali, S. Integrating deep convolutional neural networks with marker-controlled watershed for overlapping nuclei segmentation in histopathology images. Neurocomputing 2020, 376, 166–179. [Google Scholar] [CrossRef]
- Vahadane, A.; Peng, T.; Albarqouni, S.; Baust, M.; Steiger, K.; Schlitter, A.M.; Sethi, A.; Esposito, I.; Navab, N. Structure-preserved color normalization for histological images. In Proceedings of the 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), Brooklyn Bridge, NY, USA, 16–19 April 2015; pp. 1012–1015. [Google Scholar] [CrossRef]
- Jégou, S.; Drozdzal, M.; Vazquez, D.; Romero, A.; Bengio, Y. The One Hundred Layers Tiramisu: Fully Convolutional DenseNets for Semantic Segmentation. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Honolulu, HI, USA, 21–26 July 2017; pp. 1175–1183. [Google Scholar] [CrossRef]
- Hassan, L.; Saleh, A.; Abdel-Nasser, M.; Omer, O.A.; Puig, D. Promising Deep Semantic Nuclei Segmentation Models for Multi-Institutional Histopathology Images of Different Organs. Int. J. Interact. Multimed. Artif. Intell. 2020, 1–11. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar] [CrossRef]
- Bueno, J.C.S.; Dias, C.A.; Dimuro, G.P.; Santos, H.; Borges, E.N.; Lucca, G.; Bustince, H. Aggregation functions based on the Choquet integral applied to image resizing. In Proceedings of the 11th Conference of the European Society for Fuzzy Logic and Technology (EUSFLAT 2019), Prague, Czech Republic, 9–13 September 2019; Atlantis Press: Paris, France, 2019. [Google Scholar]
- Abdel-Nasser, M.; Mahmoud, K.; Lehtonen, M. Reliable Solar Irradiance Forecasting Approach Based on Choquet Integral and Deep LSTMs. IEEE Trans. Ind. Inform. 2020, 17, 1873–1881. [Google Scholar] [CrossRef]
- Mirjalili, S.; Mirjalili, S.M.; Lewis, A. Grey wolf optimizer. Adv. Eng. Softw. 2014, 69, 46–61. [Google Scholar] [CrossRef]
- Llamazares, B. Constructing Choquet integral-based operators that generalize weighted means and OWA operators. Inf. Fusion 2015, 23, 131–138. [Google Scholar] [CrossRef]
- Detyniecki, M. PART 1: Definition and Properties 1. 1 Definition. 2001. Available online: http://www-poleia.lip6.fr/~marcin/web/wp-content/papercite-data/pdf/Detyniecki_AGOP_2001.pdf (accessed on 14 March 2021).
- Mahmood, F.; Borders, D.; Chen, R.J.; Mckay, G.N.; Salimian, K.J.; Baras, A.; Durr, N.J. Deep Adversarial Training for Multi-Organ Nuclei Segmentation in Histopathology Images. IEEE Trans. Med Imaging 2020, 39, 3257–3267. [Google Scholar] [CrossRef]
- Dong, F.; Irshad, H.; Oh, E.Y.; Lerwill, M.F.; Brachtel, E.F.; Jones, N.C.; Knoblauch, N.W.; Montaser-Kouhsari, L.; Johnson, N.B.; Rao, L.K.F.; et al. Computational Pathology to Discriminate Benign from Malignant Intraductal Proliferations of the Breast. PLoS ONE 2014, 9, e114885. [Google Scholar] [CrossRef]
- Mahbod, A.; Schaefer, G.; Bancher, B.; Löw, C.; Dorffner, G.; Ecker, R.; Ellinger, I. CryoNuSeg: A dataset for nuclei instance segmentation of cryosectioned H&E-stained histological images. Comput. Biol. Med. 2021, 132, 104349. [Google Scholar] [CrossRef] [PubMed]
- Naylor, P.; Laé, M.; Reyal, F.; Walter, T. Nuclei segmentation in histopathology images using deep neural networks. In Proceedings of the 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), Melbourne, Australia, 18–21 April 2017; pp. 933–936. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).