Knowledge-Based System for Crop Pests and Diseases Recognition
Abstract
1. Introduction
- A knowledge model representing the crop pest domain in the form of an ontology has been defined as a revision of our previous work [22].
- Natural language processing tools have been used to automatically populate the ontology from unstructured data sources in Spanish. During this process, data about the plant pests and diseases, including their symptoms and recommended treatments, are gathered.
- A novel approach for representing symptoms associated plant diseases is proposed based on the combination of plant parts and observed damage.
- A knowledge-based crop pest recognizer has been built which is capable of identifying multiple overlapping pests. The proposed framework is easily extensible to support new evidentiary inputs such as images.
- Sustainable agricultural practices are fostered by suggesting organic agriculture-compliant treatments. Nonetheless, the proposed framework provides support for both conventional and organic management strategies.
- A dataset has been compiled with symptoms–pests associations for three crops: almond tree, olive tree and grape vine. In total, the dataset contains 212 symptoms declared by means of sentences in Spanish, connected to 75 pests and diseases. This dataset is publicly available at http://agrisemantics.inf.um.es/datasets/ (accessed on 9 April 2021).
2. Related Work
2.1. Pests and Diseases Recognition
2.2. Language Technologies for Knowledge Acquisition
3. Crop Pests and Diseases Identification from Natural Language Text
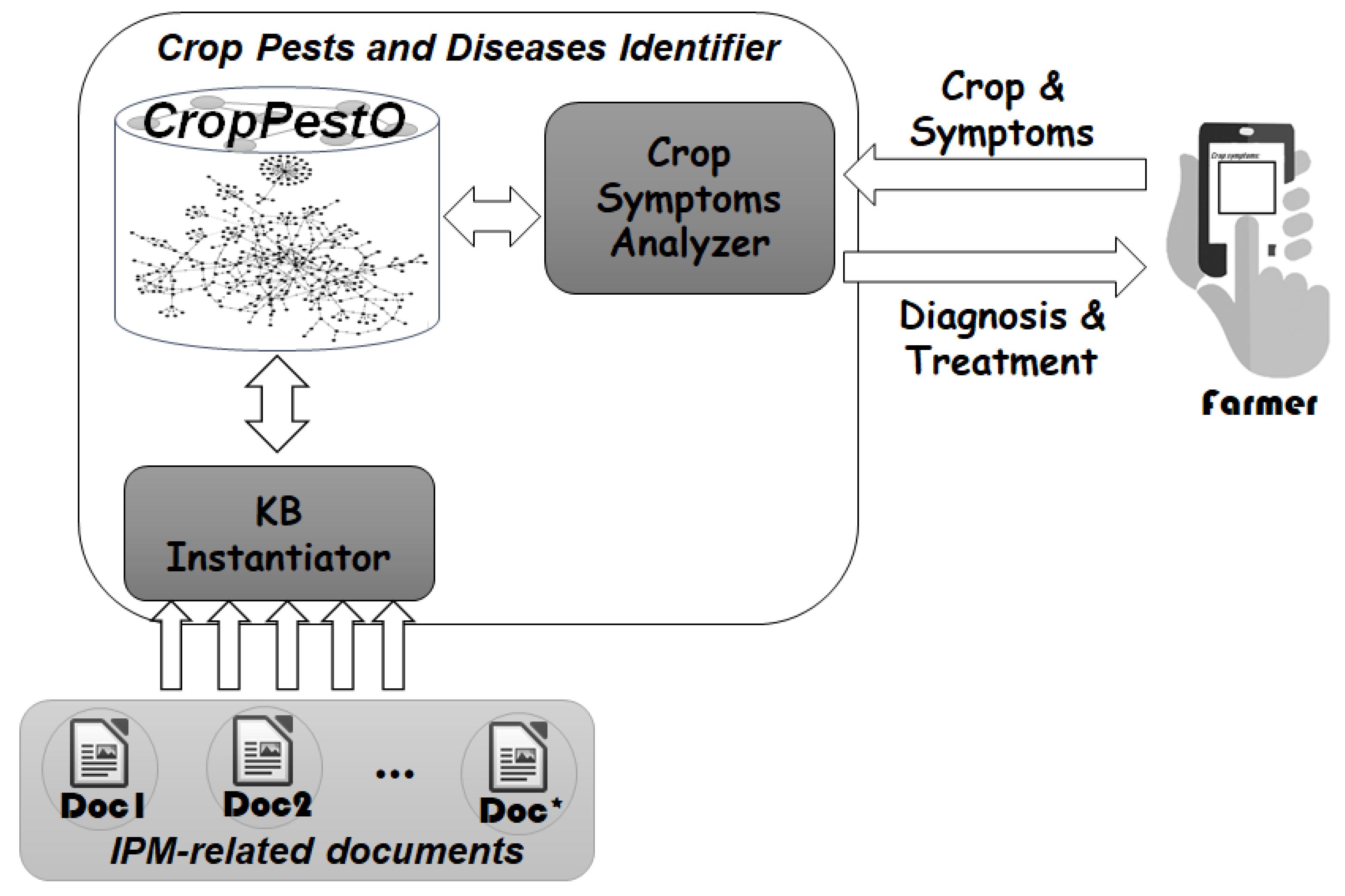
3.1. Proposed Framework
3.2. CropPestO: Pests and Diseases Management Ontology
3.3. KB Instantiator: Knowledge Base Population
| Algorithm 1: SymptomsAnalyser |
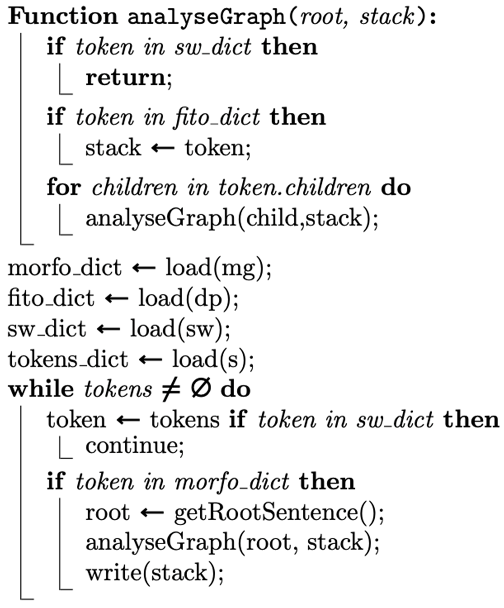 |
3.4. Crop Symptoms Analyzer
4. Evaluation
4.1. Exemplary Usage Scenario
4.2. Dataset
4.3. Evaluation Metrics
4.4. Results
4.5. Discussion
5. Conclusions and Future Work
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Food and Agriculture Organization of the United Nations. In Organically Produced Foods, 3rd ed.; World Health Organization: Rome, Italy, 2007; ISBN 978-92-5-105835-0. [Google Scholar]
- Reganold, J.P.; Wachter, J.M. Organic agriculture in the twenty-first century. Nat. Plants 2016, 2, 15221. [Google Scholar] [CrossRef]
- Tamm, L. The impact of pests and diseases in organic agriculture. In Proceedings of the The BCPC Conference: Pests and Diseases, Brighton, UK, 13–16 November 2000; Volume 1, pp. 159–166. [Google Scholar]
- Deutsch, C.A.; Tewksbury, J.J.; Tigchelaar, M.; Battisti, D.S.; Merrill, S.C.; Huey, R.B.; Naylor, R.L. Increase in crop losses to insect pests in a warming climate. Science 2018, 361, 916–919. [Google Scholar] [CrossRef] [PubMed]
- Velásquez, A.C.; Castroverde, C.D.M.; Yang He, S. Plant-pathogen warfare under changing climate conditions. Curr. Biol. 2018, 28, R619–R634. [Google Scholar] [CrossRef]
- Woodard, J.; Andriessen, M.; Cohen, C.; Cox, C.; Fritz, S.; Johnson, D.; Koo, J.; McLean, M.; See, L.; Speck, T.; et al. ICT in Agriculture (Updated Edition): Connecting Smallholders to Knowledge, Networks, and Institutions; World Bank: Washington, DC, USA, 2017; ISBN 978-1-4648-1002-2. Available online: https://openknowledge.worldbank.org/handle/10986/27526 (accessed on 9 April 2021).
- Unal, Z. Smart farming becomes even smarter with deep learning—A bibliographical analysis. IEEE Access 2020, 8, 105587–105609. [Google Scholar] [CrossRef]
- Lagos-Ortiz, K.; Medina-Moreira, J.; Sinche-Guzmán, A.; Garzón-Goya, M.; Vergara-Lozano, V.; Valencia-García, R. Mobile applications for crops management. In Proceedings of the Technologies and Innovation—4th International Conference, CITI 2018, Guayaquil, Ecuador, 6–9 November 2018; Communications in Computer and Information Science 883. Valencia-García, R., Alcaraz-Mármol, G., del Cioppo-Morstadt, J., Vera-Lucio, N., Bucaram-Leverone, M., Eds.; Springer: Guayaquil, Ecuador, 2018; pp. 57–69. [Google Scholar] [CrossRef]
- Lee, S.H.; Goëau, H.; Bonnet, P.; Joly, A. New perspectives on plant disease characterization based on deep learning. Comput. Electron. Agric. 2020, 170, 105220. [Google Scholar] [CrossRef]
- Loey, M.; ElSawy, A.; Afify, M. Deep learning in plant diseases detection for agricultural crops: A survey. Int. J. Serv. Sci. Manag. Eng. Technol. 2020, 11, 41–58. [Google Scholar] [CrossRef]
- Sinha, A.; Shekhawat, R.S. Review of image processing approaches for detecting plant diseases. IET Image Process. 2020, 14, 1427–1439. [Google Scholar] [CrossRef]
- Lagos-Ortiz, K.; Salas-Zárate, M.D.P.; Paredes-Valverde, M.A.; García-Díaz, J.A.; Valencia-García, R. AgriEnt: A knowledge-based Web platform for managing insect pests of field crops. Appl. Sci. 2020, 10, 1040. [Google Scholar] [CrossRef]
- Bernabé-Díaz, J.A.; del Carmen Legaz-García, M.; García, J.M.; Fernández-Breis, J.T. Efficient, semantics-rich transformation and integration of large datasets. Expert Syst. Appl. 2019, 133, 198–214. [Google Scholar] [CrossRef]
- Asfand-E-Yar, M.; Ali, R. Semantic integration of heterogeneous databases of same domain using ontology. IEEE Access 2020, 8, 77903–77919. [Google Scholar] [CrossRef]
- Jiang, S.; Angarita, R.; Chiky, R.; Cormier, S.; Rousseaux, F. Towards the integration of agricultural data from heterogeneous sources: Perspectives for the French agricultural context using semantic technologies. In Advanced Information Systems Engineering Workshops. CAiSE 2020. Lecture Notes in Business Information Processing; Dupuy-Chessa, S., Proper, H., Eds.; Springer: Cham, Switzerland, 2020; Volume 382, pp. 89–94. [Google Scholar] [CrossRef]
- Shadbolt, N.; Berners-Lee, T.; Hall, W. The Semantic Web revisited. IEEE Intell. Syst. 2006, 21, 96–101. [Google Scholar] [CrossRef]
- Studer, R.; Benjamins, R.; Fensel, D. Knowledge engineering: Principles and methods. Data Knowl. Eng. 1998, 25, 161–197. [Google Scholar] [CrossRef]
- García-Sánchez, F.; Colomo-Palacios, R.; Valencia-García, R. A social-semantic recommender system for advertisements. Inf. Process. Manag. 2020, 57, 102153. [Google Scholar] [CrossRef]
- Shanavas, N.; Wang, H.; Lin, Z.; Hawe, G. Ontology-based enriched concept graphs for medical document classification. Inf. Sci. 2020, 525, 172–181. [Google Scholar] [CrossRef]
- Drury, B.; Fernandes, R.; Moura, M.-F.; de Andrade Lopes, A. A survey of semantic web technology for agriculture. Inf. Process. Agric. 2019, 6, 487–501. [Google Scholar] [CrossRef]
- Hernández-Castillo, C.; Guedea-Noriega, H.H.; Rodríguez-García, M.Á.; García-Sánchez, F. Pest recognition using natural language processing. In Technologies and Innovation. CITI 2019. Communications in Computer and Information Science; Valencia-García, R., Alcaraz-Mármol, G., Del Cioppo-Morstadt, J., Vera-Lucio, N., Bucaram-Leverone, M., Eds.; Springer: Cham, Switzerland, 2019; Volume 1124, pp. 3–16. [Google Scholar] [CrossRef]
- Rodríguez-García, M.Á.; García-Sánchez, F. CropPestO: An ontology model for identifying and managing plant pests and diseases. In Technologies and Innovation. CITI 2020. Communications in Computer and Information Science; Valencia-García, R., Alcaraz-Marmol, G., del Cioppo-Morstadt, J., Vera-Lucio, N., Bucaram-Leverone, M., Eds.; Springer: Cham, Switzerland; Guayaquil, Ecuador, 2020; Volume 1309, pp. 18–29. [Google Scholar] [CrossRef]
- European Commission. Integrated Pest Management (IPM). Available online: https://ec.europa.eu/food/plant/pesticides/sustainable_use_pesticides/ipm_en (accessed on 29 March 2021).
- Sankaran, S.; Mishra, A.; Ehsani, R.; Davis, C. A review of advanced techniques for detecting plant diseases. Comput. Electron. Agric. 2010, 72, 1–13. [Google Scholar] [CrossRef]
- Martinelli, F.; Scalenghe, R.; Davino, S.; Panno, S.; Scuderi, G.; Ruisi, P.; Villa, P.; Stroppiana, D.; Boschetti, M.; Goulart, L.R. Advanced methods of plant disease detection. A review. Agron. Sustain. Dev. 2015, 35, 27–51. [Google Scholar] [CrossRef]
- Vishnoi, V.K.; Kumar, K.; Kumar, B. Plant disease detection using computational intelligence and image processing. J. Plant Dis. Prot. 2020. [Google Scholar] [CrossRef]
- Ngugi, L.C.; Abelwahab, M.; Abo-Zahhad, M. Recent advances in image processing techniques for automated leaf pest and disease recognition—A review. Inf. Process. Agric. 2020. [Google Scholar] [CrossRef]
- Nagaraju, M.; Chawla, P. Systematic review of deep learning techniques in plant disease detection. Int. J. Syst. Assur. Eng. Manag. 2020, 11, 547–560. [Google Scholar] [CrossRef]
- Toda, Y.; Okura, F. How convolutional neural networks diagnose plant disease. Plant Phenomics 2019, 2019, 1–14. [Google Scholar] [CrossRef]
- Too, E.C.; Yujian, L.; Njuki, S.; Yingchun, L. A comparative study of fine-tuning deep learning models for plant disease identification. Comput. Electron. Agric. 2019, 161, 272–279. [Google Scholar] [CrossRef]
- Ferentinos, K.P. Deep learning models for plant disease detection and diagnosis. Comput. Electron. Agric. 2018, 145, 311–318. [Google Scholar] [CrossRef]
- Kartikeyan, P.; Shrivastava, G. Review on emerging trends in detection of plant diseases using image processing with machine learning. Int. J. Comput. Appl. 2021, 174, 39–48. [Google Scholar] [CrossRef]
- Liu, J.; Wang, X. Plant diseases and pests detection based on deep learning: A review. Plant Methods 2021, 17, 22. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-W.; Lin, W.-J.; Cheng, H.-J.; Hung, C.-L.; Lin, C.-Y.; Chen, S.-P. A smartphone-based application for scale pest detection using multiple-object detection methods. Electronics 2021, 10, 372. [Google Scholar] [CrossRef]
- Velásquez, D.; Sánchez, A.; Sarmiento, S.; Toro, M.; Maiza, M.; Sierra, B. A method for detecting coffee leaf rust through wireless sensor networks, remote sensing, and deep learning: Case study of the caturra variety in Colombia. Appl. Sci. 2020, 10, 697. [Google Scholar] [CrossRef]
- Messina, G.; Modica, G. Applications of UAV thermal imagery in precision agriculture: State of the art and future research outlook. Remote Sens. 2020, 12, 1491. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, Y.; Pu, R.; Gonzalez-Moreno, P.; Yuan, L.; Wu, K.; Huang, W. Monitoring plant diseases and pests through remote sensing technology: A review. Comput. Electron. Agric. 2019, 165, 104943. [Google Scholar] [CrossRef]
- Petrellis, N. Plant disease diagnosis for smart phone applications with extensible set of diseases. Appl. Sci. 2019, 9, 1952. [Google Scholar] [CrossRef]
- Cui, S.; Ling, P.; Zhu, H.; Keener, H. Plant pest detection using an artificial nose system: A review. Sensors 2018, 18, 378. [Google Scholar] [CrossRef]
- Hazarika, S.; Choudhury, R.; Montazer, B.; Medhi, S.; Goswami, M.P.; Sarma, U. Detection of Citrus Tristeza virus in mandarin orange using a custom-developed electronic nose system. IEEE Trans. Instrum. Meas. 2020, 69, 9010–9018. [Google Scholar] [CrossRef]
- Ruusunen, O.; Jalli, M.; Jauhiainen, L.; Ruusunen, M.; Leiviskä, K. Advanced data analysis as a tool for net blotch density estimation in spring barley. Agriculture 2020, 10, 179. [Google Scholar] [CrossRef]
- Maneesha, A.; Suresh, C.; Kiranmayee, B.V. Prediction of rice plant diseases based on soil and weather conditions. In Proceedings of the International Conference on Advances in Computer Engineering and Communication Systems. Learning and Analytics in Intelligent Systems, Greater Noida, India, 19–20 February 2021; Volume 20, pp. 155–165. [Google Scholar] [CrossRef]
- Lagos-Ortiz, K.; Medina-Moreira, J.; Paredes-Valverde, M.A.; Espinoza-Morán, W.; Valencia-García, R. An ontology-based decision support system for the diagnosis of plant diseases. J. Inf. Technol. Res. 2017, 10, 42–55. [Google Scholar] [CrossRef]
- Cañadas, J.; del Águila, I.M.; Palma, J. Development of a web tool for action threshold evaluation in table grape pest management. Precis. Agric. 2017, 18, 974–996. [Google Scholar] [CrossRef]
- Toseef, M.; Khan, M.J. An intelligent mobile application for diagnosis of crop diseases in Pakistan using fuzzy inference system. Comput. Electron. Agric. 2018, 153, 1–11. [Google Scholar] [CrossRef]
- Goodridge, W.; Bernard, M.; Jordan, R.; Rampersad, R. Intelligent diagnosis of diseases in plants using a hybrid Multi-Criteria decision making technique. Comput. Electron. Agric. 2017, 133, 80–87. [Google Scholar] [CrossRef]
- Halder, S.; Kumar Singh, S. Knowledge-based expert system for diagnosis of agricultural crops. In Proceedings of International Conference on Frontiers in Computing and Systems. Advances in Intelligent Systems and Computing; Springer: Singapore, 2021; Volume 1255, pp. 351–359. [Google Scholar] [CrossRef]
- Li, Y.; Luo, Z.; Wang, F.; Wang, Y. Hyperspectral leaf image-based cucumber disease recognition using the extended collaborative representation model. Sensors 2020, 20, 4045. [Google Scholar] [CrossRef]
- Deng, X.; Zhu, Z.; Yang, J.; Zheng, Z.; Huang, Z.; Yin, X.; Wei, S.; Lan, Y. Detection of citrus huanglongbing based on multi-input neural network model of UAV hyperspectral remote sensing. Remote Sens. 2020, 12, 2678. [Google Scholar] [CrossRef]
- Li, K.; Lin, J.; Liu, J.; Zhao, Y. Using deep learning for image-based different degrees of ginkgo leaf disease classification. Information 2020, 11, 95. [Google Scholar] [CrossRef]
- Skawsang, S.; Nagai, M.; Tripathi, N.; Soni, P. Predicting rice pest population occurrence with satellite-derived crop phenology, ground meteorological observation, and machine learning: A case study for the central plain of Thailand. Appl. Sci. 2019, 9, 4846. [Google Scholar] [CrossRef]
- Chen, J.; Liu, Q.; Gao, L. Visual tea leaf disease recognition using a convolutional neural network model. Symmetry 2019, 11, 343. [Google Scholar] [CrossRef]
- Anagnostis, A.; Asiminari, G.; Papageorgiou, E.; Bochtis, D. A convolutional neural networks based method for anthracnose infected walnut tree leaves identification. Appl. Sci. 2020, 10, 469. [Google Scholar] [CrossRef]
- Liang, Q.; Xiang, S.; Hu, Y.; Coppola, G.; Zhang, D.; Sun, W. PD2SE-Net: Computer-assisted plant disease diagnosis and severity estimation network. Comput. Electron. Agric. 2019, 157, 518–529. [Google Scholar] [CrossRef]
- Pantazi, X.E.; Moshou, D.; Tamouridou, A.A. Automated leaf disease detection in different crop species through image features analysis and One Class Classifiers. Comput. Electron. Agric. 2019, 156, 96–104. [Google Scholar] [CrossRef]
- Picon, A.; Alvarez-Gila, A.; Seitz, M.; Ortiz-Barredo, A.; Echazarra, J.; Johannes, A. Deep convolutional neural networks for mobile capture device-based crop disease classification in the wild. Comput. Electron. Agric. 2019, 161, 280–290. [Google Scholar] [CrossRef]
- Arsenovic, M.; Karanovic, M.; Sladojevic, S.; Anderla, A.; Stefanovic, D. Solving current limitations of deep learning based approaches for plant disease detection. Symmetry 2019, 11, 939. [Google Scholar] [CrossRef]
- Sun, R.; Zhang, M.; Yang, K.; Liu, J. Data enhancement for plant disease classification using generated lesions. Appl. Sci. 2020, 10, 466. [Google Scholar] [CrossRef]
- Garcerán-Sáez, J.; García-Sánchez, F. SePeRe: Semantically-enhanced system for pest recognition. In ICT for Agriculture and Environment. CITAMA2019 2019. Advances in Intelligent Systems and Computing; Valencia-García, R., Alcaraz-Mármol, G., Cioppo-Morstadt, J., Vera-Lucio, N., Bucaram-Leverone, M., Eds.; Springer: Cham, Switzerland, 2019; Volume 901, pp. 3–11. [Google Scholar] [CrossRef]
- Labaña, F.M.; Ruiz, A.; García-Sánchez, F. PestDetect: Pest recognition using convolutional neural network. In ICT for Agriculture and Environment. CITAMA2019 2019. Advances in Intelligent Systems and Computing; Valencia-García, R., Alcaraz-Mármol, G., Cioppo-Morstadt, J., Vera-Lucio, N., Bucaram-Leverone, M., Eds.; Springer: Cham, Switzerland, 2019; Volume 901, pp. 99–108. [Google Scholar] [CrossRef]
- Colombo-Mendoza, L.O.; Valencia-García, R.; Colomo-Palacios, R.; Alor-Hernández, G. A knowledge-based multi-criteria collaborative filtering approach for discovering services in mobile cloud computing platforms. J. Intell. Inf. Syst. 2020, 54, 179–203. [Google Scholar] [CrossRef]
- Rodríguez-García, M.Á.; Valencia-García, R.; Colomo-Palacios, R.; Gómez-Berbís, J.M. BlindDate recommender: A context-aware ontology-based dating recommendation platform. J. Inf. Sci. 2019, 45, 573–591. [Google Scholar] [CrossRef]
- Rodríguez Iglesias, A.; Egaña Aranguren, M.; Rodríguez González, A.; Wilkinson, M.D. Plant-Pathogen Interactions Ontology (PPIO). In Proceedings of the International Work-Conference on Bioinformatics and Biomedical Engineering IWBBIO 2013, Granada, Spain, 18–20 March 2013; Rojas, I., Ortuño Guzman, F.M., Eds.; Copicentro Editorial: Granada, Spain, 2013; pp. 695–702. [Google Scholar]
- Walls, R.; Smith, B.; Elser, J.; Goldfain, A.; Stevenson, D.W.; Jaiswal, P. A plant disease extension of the Infectious Disease Ontology. In Proceedings of the 3rd International Conference on Biomedical Ontology (ICBO 2012), KR-MED Series, Graz, Austria, 21–25 July 2012; Cornet, R., Stevens, R., Eds.; CEUR-WS.org: Graz, Austria, 2012; pp. 1–5. [Google Scholar]
- Dalvi, P.; Mandave, V.; Gothkhindi, M.; Patil, A.; Kadam, S.; Pawar, S.S. Overview of agriculture domain ontologies. Int. J. Recent Adv. Eng. Technol. 2016, 4, 5–9. [Google Scholar]
- Caracciolo, C.; Stellato, A.; Morshed, A.; Johannsen, G.; Rajbhandari, S.; Jaques, Y.; Keizer, J. The AGROVOC linked dataset. Semant. Web. 2013, 4, 341–348. [Google Scholar] [CrossRef]
- Beck, H.W.; Kim, S.; Hagan, D. A crop-pest ontology for extension publications. In Proceedings of the 2005 EFITA/WCCA Joint Congress on IT in Agriculture, Vila Real, Portugal, 25–28 July 2005; pp. 1169–1176. [Google Scholar]
- Jonquet, C.; Toulet, A.; Arnaud, E.; Aubin, S.; Dzalé Yeumo, E.; Emonet, V.; Graybeal, J.; Laporte, M.-A.; Musen, M.A.; Pesce, V.; et al. AgroPortal: A vocabulary and ontology repository for agronomy. Comput. Electron. Agric. 2018, 144, 126–143. [Google Scholar] [CrossRef]
- Xiaoxue, L.; Xuesong, B.; Longhe, W.; Bingyuan, R.; Shuhan, L.; Lin, L. Review and trend analysis of knowledge graphs for crop pest and diseases. IEEE Access 2019, 7, 62251–62264. [Google Scholar] [CrossRef]
- Lacasta, J.; Lopez-Pellicer, F.J.; Espejo-García, B.; Nogueras-Iso, J.; Zarazaga-Soria, F.J. Agricultural recommendation system for crop protection. Comput. Electron. Agric. 2018, 152, 82–89. [Google Scholar] [CrossRef]
- Titiya, M.D.; Shah, V.A. Ontology based expert system for pests and disease management of cotton crop in India. Int. J. Web. Portals 2018, 10, 32–49. [Google Scholar] [CrossRef]
- Jearanaiwongkul, W.; Anutariya, C.; Andres, F. An ontology-based approach to plant disease identification system. In Proceedings of the Proceedings of the 10th International Conference on Advances in Information Technology—IAIT 2018, Bangkok, Thailand, 10–13 December 2018; ACM Press: New York, NY, USA, 2018; pp. 1–8. [Google Scholar] [CrossRef]
- Somodevilla, M.J.; Vilariño Ayala, D.; Pineda, I. An overview on ontology learning tasks. Comput. Sist. 2018, 22, 137–146. [Google Scholar] [CrossRef]
- Khadir, A.C.; Aliane, H.; Guessoum, A. Ontology learning: Grand tour and challenges. Comput. Sci. Rev. 2021, 39, 100339. [Google Scholar] [CrossRef]
- Xiang, Z.; Zheng, J.; Lin, Y.; He, Y. Ontorat: Automatic generation of new ontology terms, annotations, and axioms based on ontology design patterns. J. Biomed. Semant. 2015, 6, 4. [Google Scholar] [CrossRef]
- Laaz, N.; Wakil, K.; Gotti, S.; Gotti, Z.; Mbarki, S. An automatic generation of domain ontologies based on an MDA approach to support big data analytics. In Advancements in Model-Driven Architecture in Software Engineering; IGI Global: Hershey, PA, USA, 2021; pp. 26–45. [Google Scholar] [CrossRef]
- Lubani, M.; Noah, S.A.M.; Mahmud, R. Ontology population: Approaches and design aspects. J. Inf. Sci. 2019, 45, 502–515. [Google Scholar] [CrossRef]
- Petasis, G.; Karkaletsis, V.; Paliouras, G.; Krithara, A.; Zavitsanos, E. Ontology population and enrichment: State of the art. In Proceedings of the Knowledge-Driven Multimedia Information Extraction and Ontology Evolution; Springer: Berlin/Heidelberg, Germany, 2011; pp. 134–166. [Google Scholar]
- Qiu, J.; Chai, Y.; Liu, Y.; Gu, Z.; Li, S.; Tian, Z. Automatic non-taxonomic relation extraction from big data in smart city. IEEE Access 2018, 6, 74854–74864. [Google Scholar] [CrossRef]
- Reynaud, J.; Toussaint, Y.; Napoli, A. Redescription mining for learning definitions and disjointness axioms in linked open data. In Proceedings of the International Conference on Conceptual Structures, Marburg, Germany, 1–4 July 2019; Springer: Cham, Switzerland; Marburg, Germany, 2019; pp. 175–189. [Google Scholar] [CrossRef]
- Nguyen, T.H.; Tettamanzi, A.G.B. Grammatical evolution to mine OWL disjointness axioms involving complex concept expressions. In Proceedings of the 2020 IEEE Congress on Evolutionary Computation (CEC), Glasgow, UK, 19–24 July 2020; IEEE: Glasgow, UK, 2020; pp. 1–8. [Google Scholar] [CrossRef]
- Ochoa, J.L.; Valencia-García, R.; Perez-Soltero, A.; Barceló-Valenzuela, M. A semantic role labelling-based framework for learning ontologies from Spanish documents. Expert Syst. Appl. 2013, 40, 2058–2068. [Google Scholar] [CrossRef]
- Sad-Houari, N.; Taghezout, N.; Nador, A. A knowledge-based model for managing the ontology evolution: Case study of maintenance in SONATRACH. J. Inf. Sci. 2019, 45, 529–553. [Google Scholar] [CrossRef]
- Abdou, M.; AbdelGaber, S.; Farhan, M. A semi-automated framework for semantically annotating web content. Futur. Gener. Comput. Syst. 2018, 81, 94–102. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, J.; Zhang, L.; Chen, S.; Zhang, Z. A new semantic annotation approach for software vulnerability source code. Int. J. Simul. Process Model. 2021, 16, 1–13. [Google Scholar] [CrossRef]
- Rodríguez-García, M.Á.; Valencia-García, R.; García-Sánchez, F.; Samper-Zapater, J.J. Creating a semantically-enhanced cloud services environment through ontology evolution. Futur. Gener. Comput. Syst. 2014, 32, 295–306. [Google Scholar] [CrossRef]
- Rodríguez-García, M.Á.; Valencia-García, R.; García-Sánchez, F.; Samper-Zapater, J.J. Ontology-based annotation and retrieval of services in the cloud. Knowl.-Based Syst. 2014, 56, 15–25. [Google Scholar] [CrossRef]
- Albukhitan, S.; Helmy, T.; Alnazer, A. Arabic ontology learning using deep learning. In Proceedings of the Proceedings of the International Conference on Web Intelligence—WI ’17; ACM Press: New York, NY, USA, 2017; pp. 1138–1142. [Google Scholar] [CrossRef]
- Noy, N.F.; McGuinness, D.L. Ontology Development 101: A Guide to Creating Your First Ontology. Available online: https://protege.stanford.edu/publications/ontology_development/ontology101.pdf (accessed on 6 March 2021).
- Food and Agriculture Organization (FAO). AGROVOC. Available online: http://aims.fao.org/vest-registry/vocabularies/agrovoc (accessed on 6 March 2021).
- W3C OWL Working Group. OWL 2 Web Ontology Language Document Overview (Second Edition). Available online: https://www.w3.org/TR/owl2-overview/ (accessed on 6 March 2021).
- Ministerio de Agricultura Alimentación y Medio Ambiente. Metodología de la Estadística Sobre Superficies y Producciones Anuales de Cultivos. Available online: https://www.mapa.gob.es/es/estadistica/temas/estadisticas-agrarias/NotasMetodológicasSuperficiesyproduccionesanualesdecultivos_tcm30-122273.pdf (accessed on 6 March 2021).
- Ministerio de Medio Ambiente y Medio Rural y Marino. Patógenos de Plantas Descritos en España, 2nd ed.; Sociedad Española de Fitopatología: Madrid, Spain, 2010; ISBN 978-84-491-0954-6. [Google Scholar]
- Salton, G.; McGill, M.J. Introduction to Modern INFORMATION Retrieval; McGraw-Hill, Inc.: New York, NY, USA, 1983; ISBN 0070544840. [Google Scholar]
- Goldstein, A.; Fink, L.; Ravid, G. A framework for evaluating agricultural ontologies. arXiv 2019, arXiv:1906.10450. [Google Scholar]
- Loizou, E.; Karelakis, C.; Galanopoulos, K.; Mattas, K. The role of agriculture as a development tool for a regional economy. Agric. Syst. 2019, 173, 482–490. [Google Scholar] [CrossRef]
- Shennan, C.; Krupnik, T.J.; Baird, G.; Cohen, H.; Forbush, K.; Lovell, R.J.; Olimpi, E.M. Organic and conventional agriculture: A useful framing? Annu. Rev. Environ. Resour. 2017, 42, 317–346. [Google Scholar] [CrossRef]


| Plant Product | #Pests | |
|---|---|---|
| 1 | “Grapes” | 46 |
| 2 | “Peaches” | 38 |
| 3 | “Apples” | 36 |
| 4 | “Almonds” | 31 |
| 5 | “Cherries” | 23 |
| Pest | #Symptoms | |
|---|---|---|
| 1 | “Cryphonectria parasitica” | 65 |
| 2 | “Armillaria mellea” | 49 |
| 3 | “Phomopsis actinidiae” | 49 |
| 4 | “Taphrina spp” | 39 |
| 5 | “Botryosphaeria dothidea” | 37 |
| Symptom | #Pests | |
|---|---|---|
| 1 | “Hojas podredumbre” | 11 |
| 2 | “Flores caída” | 7 |
| 3 | “Ramas manchas” | 5 |
| 4 | “Tronco anillos” | 3 |
| 5 | “Troncos chancros” | 2 |
| Almond Tree Dataset |
|---|
| ... PEST NAME: Monilia o podredumbre parda (Monilinia sp.) PEST SYMPTOMS AND DAMAGES: las flores secas quedan adheridas al árbol, los frutos adquieren color negro y quedan momificados en las ramas. los chancros en los brotes son de color marrón claro con emisiones de goma que en madera de más edad se abren. ... |
| Almond Tree | Olive Tree | GRAPE VINE | |
|---|---|---|---|
| # of symptoms | 86 | 42 | 102 |
| # of pests | 26 | 25 | 24 |
| Experiment | Accuracy | Precision | Recall | F-Measure |
|---|---|---|---|---|
| Almond tree | 0.993 | 0.846 | 0.885 | 0.859 |
| Olive tree | 0.974 | 0.460 | 0.480 | 0.467 |
| Grape vine | 0.997 | 0.813 | 0.958 | 0.819 |
| Overall | 0.988 | 0.707 | 0.773 | 0.716 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-García, M.Á.; García-Sánchez, F.; Valencia-García, R. Knowledge-Based System for Crop Pests and Diseases Recognition. Electronics 2021, 10, 905. https://doi.org/10.3390/electronics10080905
Rodríguez-García MÁ, García-Sánchez F, Valencia-García R. Knowledge-Based System for Crop Pests and Diseases Recognition. Electronics. 2021; 10(8):905. https://doi.org/10.3390/electronics10080905
Chicago/Turabian StyleRodríguez-García, Miguel Ángel, Francisco García-Sánchez, and Rafael Valencia-García. 2021. "Knowledge-Based System for Crop Pests and Diseases Recognition" Electronics 10, no. 8: 905. https://doi.org/10.3390/electronics10080905
APA StyleRodríguez-García, M. Á., García-Sánchez, F., & Valencia-García, R. (2021). Knowledge-Based System for Crop Pests and Diseases Recognition. Electronics, 10(8), 905. https://doi.org/10.3390/electronics10080905








