Abstract
There is a skin allergy epidemic in the western world, and the rate of deterioration has increased significantly in the past 5–10 years. It is probable that there are many environmental contributing factors, yet some studies have linked it primarily to the rise in the use of synthetic chemical ingredients in modern cosmetics. Our challenge, therefore, was to find a mechanism to determine the effect these substances have on skin health, and whether they really are a primary cause of long term damage to the skin. The first problem is the lack of any definitive way to measure skin health. Motivated by the overwhelming evidence for a link between deficient gut flora and ill health, we decided to look at whether our skin microbiota could similarly be used as an indicator of skin health. Our research illustrates how microbiota diversity alone can predict whether skin is healthy or not, after we revealed a complete lack of conclusive findings linking the presence or abundance of particular species of microbe to skin problems. This phenomenon is replicated throughout nature, where high biodiversity always leads to healthy ecosystems. ‘Caveman’ skin, untouched by modern civilisation, was far different to “western” skin and displayed unprecedented levels of bacterial diversity. The less exposed communities were to western practices, the higher the skin diversity, which is clear evidence of an environmental factor in the developed world damaging skin. For the first time we propose benchmark values of diversity against which we can measure skin to determine how healthy it is. This gives us the ability to be able to predict which people are more likely to be prone to skin ailments, and start to test whether cosmetic ingredients and products are a main cause of the skin allergy epidemic.
1. Skin Microbiota
1.1. Skin Immune System
The intestinal microflora and its links to immune system function and overall health has been the subject of much research and discussion in recent years [1,2]. The idea that commensal microbes in the gut need to be encouraged, not destroyed [3], has led to the topic becoming mainstream and inspiring high street books such as “Let Them Eat Dirt” [4]. There is now a prevailing belief in a causal link between ill-health and unbalanced gut flora [5], and the probability that it is western lifestyles that are affecting the composition and diversity of the gut microbiota leading to many chronic health problems [6]. Yet despite the rapid rise in allergy related skin-health problems in the developed world [7], parallel research into the role of the skin microbiome and the body’s immune system responses on overall health has been very limited.
The skin is the largest organ [8,9] and is host to an enormously diverse array of microbes [10]. Only now is the crucial role that the skin plays in overall health starting to be understood; most early literature on skin microbiota concentrates on the pathogenic roles of different types of microbiota on the skin, yet little research has been done on the influence of resident cutaneous microflora on skin health [11]. The bacterial colonies inhabiting the skin have been found to be crucial for defence of the host [12,13], and are suggested to only rarely become pathogenic, being commensal (harmless and very rarely associated with disease) or symbiotic (good) the majority of the time. They live in peaceful co-existence with the host, sheltered in the specific ecological environment [11,14,15,16,17]. The relationship has been described as “mutualistically symbiotic” instead of commensally symbiotic, as both parties benefit [15,18,19].
Microbes make up part of the skin barrier, which, together with a person’s innate immunity, combine to form a delicate balance needed to maintain healthy skin. If this balance is disturbed, the host becomes more susceptible to inflammatory diseases and cutaneous infections [11,20,21]. A common 21st century problem is the overuse of antibiotics [9,22], which leaves the skin susceptible to pathogens which were previously warded off by the great proportion of resident and mutual bacteria [11]. Studies into the gut have shown that a disrupted exposure to microbes in a person’s early years could lead to allergies and disease [23,24]; however, the same in-depth study should be carried out for the skin. Contrary to previous ideas, it’s suggested the difference between harmless bacteria and a pathogenic agent is due to the ability of the skin as a whole to resist disease and infection, not the built-in properties of the microbe [11,17]. “Host cutaneous defence”, the capacity of the skin to withstand infection and disease, is a combination of a very large amount of systems working together [11]. These include the physical barrier, hostile surface pH, and the “active synthesis of gene-encoded host defence molecules”.
1.2. The Rise of Skin Problems in the Developed World
During the 20th century in the developed world there was an alarming rise in skin ailments and allergic conditions such as eczema, and the rate of this deterioration has increased significantly in the past 5–10 years [7,14,25,26,27,28,29,30]. This has led to a huge increase in healthcare spending [31,32], with some calling it an “allergy epidemic” [33]. During the same period, the developed world as a whole has become wealthier and better fed, coupled with a huge increase in access to medical treatments and technologies [34]. So why with better access to healthcare has skin-health deteriorated? What are the environmental factors in western lifestyles that have caused this epidemic?
1.3. Skin Microbiota and Possible Links to Ill Health
Many studies investigating the causes of skin damage and disease have focused on trying to find a link between specific types of microbe present on the skin and specific skin ailments. However, there is little conclusive evidence that healthy or unhealthy skin is determined by the presence of specific dominating types of microbiota [10]. For example, even after decades of studying Propionibacterium acnes and its contribution to acne pathogenesis, the role it plays is still very unclear, especially as it is a major commensal of normal skin flora [9,35].
Contributing to this uncertainty is the discovery of high intra- and inter-personal variation of bacteria types between subjects, where the microbiota of each individual is “virtually unique” [36,37]. Some studies have shown that among subjects, their bacterial makeup is more similar between sites on their own body than compared to different people [22]; however, others contradict this [38]. Moreover, those from different countries showed high inter-personal variation [38], with studies strongly suggesting that the differing genetic make-up between people could be a defining factor in this variation [28], just like in the gut [39], along with environmental causes. Therefore we concluded it was necessary to search for other means of determining healthy skin.
Our research into the gut uncovered evidence of children in rural Burkina Faso having very different—and most interestingly, far more diverse—microbiota in composition than urban, city-dwelling children in Italy [40,41]. This made us question whether the same was true for the skin: as with the gut, could the diversity of bacterial species present on the skin also be the only reliable indicator of healthy or damaged skin?
However, before investigating this question further, we must firstly take a very brief look at the importance of (bio)diversity throughout the natural world, and the inherent problems of measurement.
1.3.1. The Importance of (Bio)diversity
Researchers across many fields of biology and ecology agree that a high biodiversity corresponds to increased healthiness and functionality within an ecosystem [4,42,43,44,45,46,47,48]. From the plains of Yellowstone Park (macro) right down to a millimetre of tubing in the human gut (micro), a decreased biodiversity negatively impacts on the ecosystem [4,43,49,50] and affects the environment in which it occurs in both linear and non-linear fashions [48]. The same rules apply at the macro-scale environment as the micro.
Three major attributes are used to define an ecosystem’s “health”: organisation, productivity and resilience [51]. Organisation of an ecosystem is to do with how diverse the ecosystem is. As an ecosystem gets more diverse it becomes stronger due to having a larger pool of species to contribute to each role that is vital in the upkeep of the system.
To help explain why increased biodiversity results in a healthy ecosystem, we use the example of the re-introduction of wolves to Yellowstone Park in 1995–6.
The re-introduction of wolves to Yellowstone Park in 1995–6 has largely been linked with dramatic changes in the landscape, but most crucially in ways scientists didn’t even think were possible. The biodiversity was hugely increased [52]. Direct and indirect changes were observed, resulting in rivers changing direction; beaver, grizzly bear, eagle and raven populations increasing [53,54], among many others; increased heights and richness of berry-producing shrubs [55]; and elk populations decreasing.
Wolves were a natural part of Yellowstone Park until the last official killing in 1926 [56,57], and were re-introduced to keep the elk population under control. This had become overly large directly as a result of the absent wolves. While other changes in animal populations put strains on the park’s ecosystem [58] (increasing coyote numbers pushed down pronghorn antelope numbers), the rise of the elks was the biggest contributor to the deteriorating conditions of the park [59]. By 1997, Yellowstone contained “some of the worst overgrazed willow communities in the West” [60,61,62,63,64,65].
This example of the re-introduction of wolves highlights a crucially important concept: the addition or removal of just a small number of species leads to non-linear changes to the ecosystem. The extermination and consequent re-introduction of just a single species (out of many thousands) in Yellowstone caused widespread and deep rooted changes throughout the park. The same “delicate balance”, mentioned in Section 1.1, exists on the skin. When it is upset, organisms not associated with pathogenic behaviour can become damaging to the system/host. The elk, when regulated by wolves, pose a lessened threat, but as soon as they aren’t restricted by wolf predation, they become “pathogenic” to the ecosystem. Their numbers swell and, without wolves to scare them off, overgrazing occurs resulting in more pressure on cottonwood and aspen [66,67,68], among other plants, but most importantly stands of willow, on which beavers depend during the winter [60,69,70]. The beaver communities use tall-willow and aspen for food and dam building materials [66,71]. The loss of beaver dams results in increased erosion [65], and the loss of habitats for many creatures, including fish, amphibians, otters, moose, mink, wading birds and more. Using this one specific example we can relate the role of elk—out of proportion in numbers due to the wolves’ demise—to that of a pathogenic skin microbe, opportunistically becoming so to the host (i.e., the skin) once the very delicate balance is disturbed.
1.3.2. Measuring Diversity
The biodiversity of a certain ecosystem depends on different factors. Simply measuring the amount of species present is not enough. Imagine a vegetable patch with 58 tomatoes, one carrot and one leek. This has the same amount of different species of vegetable (three) than a patch which contains 20 tomatoes, 20 carrots and 20 leeks. But it isn’t as diverse, because tomatoes dominate the ecosystem. The idea of “evenness”—how evenly the total number of species present is divided between each species—then has to be taken into account. The Shannon Diversity Index includes this, with an equation for total biodiversity which is taken from the formula for the Gibbs entropy of a system in physics [72,73], and has been used as the metric for characterising diversity by scientists for decades. It looks at how particles/organisms are distributed in a system and predicts the highest diversity when there is the most “spread” of organisms.
Shannon Diversity equation is below:
- H’ = biodiversity index
- i = species
- pi = ni/N
- ni = total number of organisms of a particular species
- N = total number of organisms of all species.
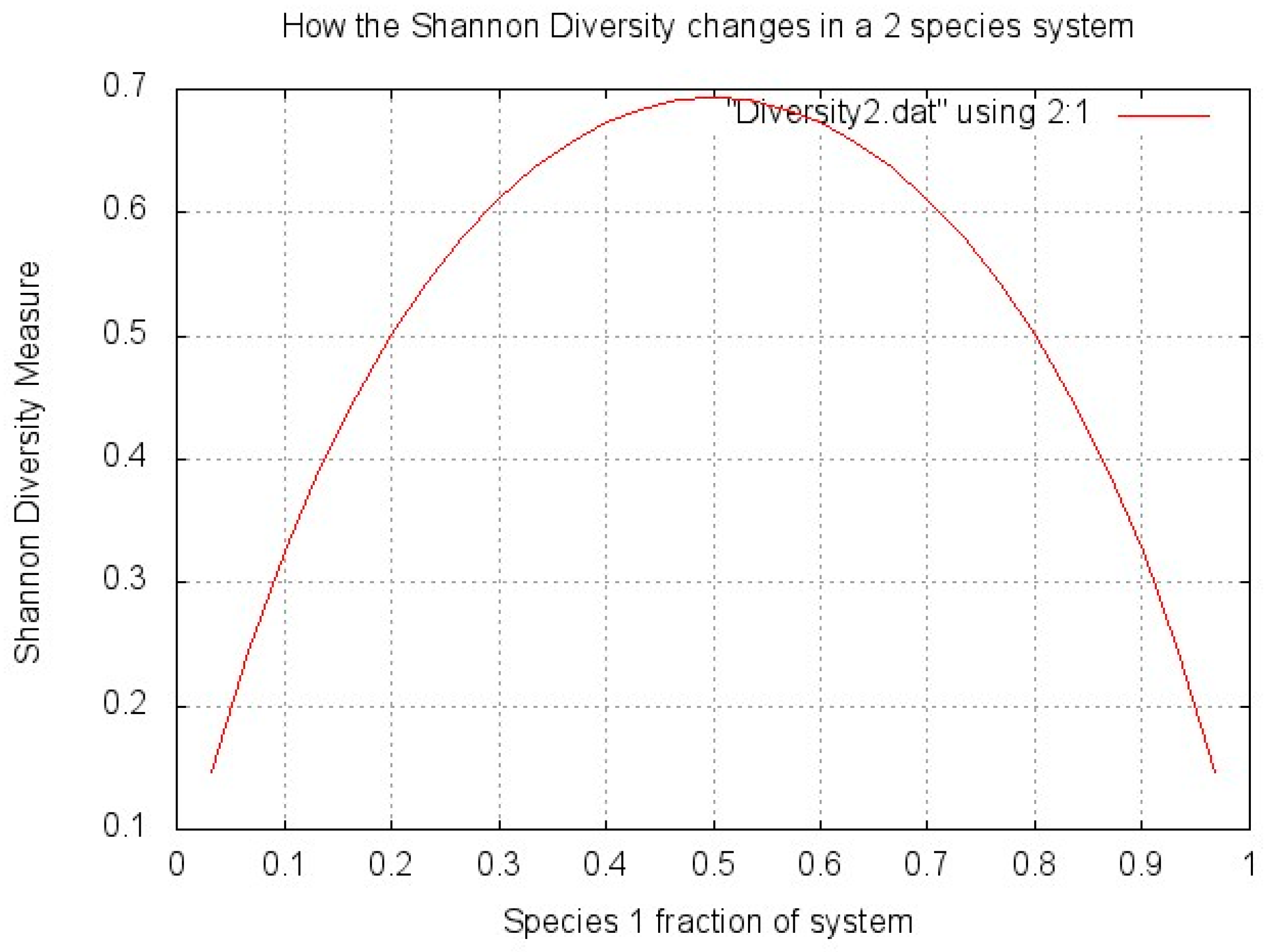
To show how this diversity is affected by the proportions of bacteria, a very simple graph (Figure 1) was made plotting biodiversity (H’) against the percentage split of microbes between two species in a system. The two species system simplifies the problem and shows very clearly that the highest diversity is achieved when the total number of microorganisms is split exactly evenly between the species (two in this case, so 50% are species 1 and 50% are species 2). The diversity score decreases as the microorganisms are split less evenly between species.
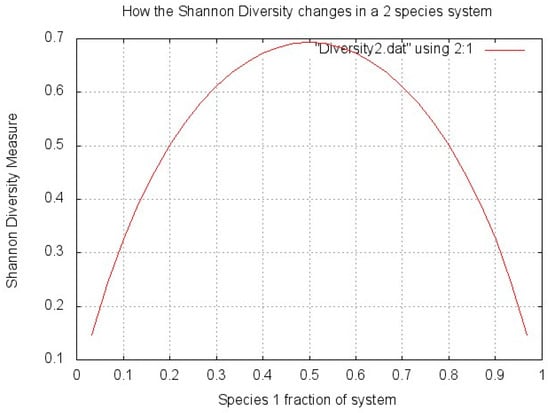
Figure 1.
Shannon Diversity explanation.
The problem is that each species is not equally likely to be seen on the skin, i.e., each species have different natural abundances. Species which are rare will inherently not be as abundant as other more common ones. When using entropy-styled evenness measures like Shannon Diversity (Equation (1)), each species is given the same probability of appearing on the skin. In truth, these systems are non-linear (as are most things in nature) and they depend on which organisms are present, not just the spread of the organisms within it. The presence of a small amount of a very rare species could indicate large diversity, purely for the fact that the conditions allowed it to be present at all.
The non-linearity of the systems may lead researchers to misinterpret the diversity of the communities they are studying, as the “normal” abundance of each species needs to be factored into calculations. Species of organisms within the system also influence each other in ways that can’t be predicted directly.
The Chao1 index is used to try and rectify this; it estimates the total number of species present in a community and is based on the number of rare classes (OTUs) found in a sample.
- Sest = estimated number of species
- Sobs = observed number of species
- f1 = number of singleton taxa (taxa represented by a single read in that community)
- f2 = number of doubleton taxa
- OTU = Operational Taxonomic Unit (the most commonly used units of microbial diversity, refers to clusters of (uncultivated or unknown) organisms, grouped by DNA sequence similarity of a specific taxonomic marker gene)
The idea behind the estimator is that if a community is being sampled, and rare species (“singletons”) are still being discovered, it is likely that there are still more rare species which haven't yet been found; as soon as all species have been recovered at least twice (“doubletons”), it is likely that there are no more species to be found. If a sample contains many singletons, therefore, it is likely that more undetected OTUs exist, and the Chao1 index will estimate greater species richness than it would for a sample without rare OTUs.
However, there are inherent problems with the Chao1 index too: if “singletons” are the basis of predicting diversity, then rare species should be weighted in terms of their rarity/abundance. If not, the presence of every rare species is given the same probability to exist, which probably isn’t the case. If some species are so fragile that they exist only in VERY healthy environments (hugely high diversity), then surely their presence is more significant than another less fragile species? There could also be singletons which are anomalous. These could be commonly occurring species but anomalously be very rare in one certain sample.
However, our investigations on skin diversity will primarily focus on data derived using Chao1 as it is commonly used, and as we will show, the diversity statistics we highlight vary little between measurements formulae used.
2. Field Data and Key Results
2.1. Setting a Baseline for Determining “Healthy” vs. “Damaged” Skin
In order to observe whether the diversity of microbial species on the skin is a reliable indicator of healthy or damaged skin, we first needed to establish a marker for “perfect” skin by looking at the diversity on skin untouched by modern antibiotics, steroids, cosmetics, or civilization.
Several studies were found that provide us with this data. The microbiome of Amerindians was studied in two of the cases. One characterised the skin bacterial microbiome of members of an isolated Yanomami Amerindian village with no documented previous contact with western people [74]. The other sampled Guahibo villagers (Venezuela), not completely isolated like the Yanomami, but who still lived very rurally without some basic amenities [75].
Once we established what “perfect” skin looks like, we needed to set a minimum diversity level below which we could confidently predict that skin was “unhealthy” and liable to disease or damage.
We firstly looked at the diversity level of “healthy” western skin, and then compared this to samples taken from unhealthy subjects from different environments. We use a study of healthy versus localised cutaneous leishmaniasis (LCL) afflicted skin in an agricultural community in Brazil [76], healthy diabetic skin compared to subjects with wounds in the USA [77], and further evidence using other species such as dogs and mice [28,78].
We note the exact skin site on the body where samples were taken in each case, as diversity varies characteristically from site to site. On average, a lower diversity of microbes is found on “sebaceous” areas of the skin (forehead) compared to “dry” (forearm) and “moist” sites [22,38].
2.1.1. Previously Uncontacted Yanomami People
The uncontacted Yanomami people [74] are thought to have been in relative isolation for over 11,000 years since their ancestors arrived in South America. Samples for sequencing were taken from the forearm, which is dry and not sebaceous like the face, so would harbour a characteristically more diverse bacterial community. They had the highest bacterial and functional diversity ever reported in a human group. The complete lack of any antibiotics use and high exposure to the wilderness around them contributed to high bacterial diversity. The difference between this diversity and that of a healthy US resident is very significant. This suggests westernisation severely affects human skin microbiome diversity.
Also observed was the existence of functional antibiotic resistant genes amongst the Yanomami people, even without any exposure to modern antibiotics. Studies have indicated that a natural skin environment possesses innate antibiotic effects via the secretion of substances such as sebum and dermcidin [20,79,80].
2.1.2. Rural Guahibo Settlement
Long running studies tested the skin microbiome of Healthy Amerindians in Platanillal village against healthy residents of Colorado and New York using swab/buffer techniques taken from the volar forearm [75,81,82,83]. They had been nomadic hunter gatherers two to three generations ago and still live in settlements without electricity or running water, meaning their exposure to westernisation was minimal, but still more so than the completely isolated Yanomami people [75].
The forearm skin bacterial community of modern US residents (dominated by Propionibacterium) showed a significant compositional difference to that of Amerindians. Within the Amerindians, there were two very distinct bacterial community membership groups—the first with much higher biodiversity than the US residents, and, more strangely, the second with a biodiversity similar to that of the US residents, but dominated by Staphylococcus and not Propionibacterium.
2.1.3. Agricultural Community in Brazil
Individuals with LCL had their skin microbiome (the full length of the forearm was swabbed) tested against those in the same village who had “healthy” skin [76]. LCL results in chronic, painless ulcers progressing commonly to secondary bacterial infections.
The microbiome of LCL lesion and healthy skin was found to be different. The bacterial diversity present on LCL skin was found to be far lower than healthy skin. The disturbance caused by the LCL lesions is explained by commensal bacteria contaminating the skin opportunistically, leading to pathogens colonising the weakened skin environment. Note that the forearm site where swabbing was conducted is a “dry” area of the skin, resulting in higher bacterial diversity than sites such as the face.
2.1.4. Healthy vs. Diseased or Damaged Skin
This link between ill health and decreased microbial diversity has been further illustrated in several other studies (shown below), including a 2010 report on the bacteriology of diabetic patients [77]: it showed that the intact skin of diabetics was significantly more diverse than the wounded skin of diabetics. However, healthy diabetic skin was still reduced in diversity compared to healthy normal skin.
A significant decrease in bacterial diversity has also been observed in allergic dogs compared to healthy ones [78]. During acute flares and skin lesions, the same was also true. The inflammation of skin even in non-lesional areas on allergic dogs and humans could alter the skin surface enough to influence the diversity of decrease.
This backs up a previous study which reported on the microbial diversity decrease in children during flares of AD (Atopic Dermatitis) [84]. Children with AD had skin dominated by Staphylococcus aureus, which was also observed frequently on the skin of allergic dogs [78]. However, the clinical effectiveness of treatments for AD didn’t depend on the elimination of Staphylococcus aureus, but on the diversification of the bacterial community. When flares of AD are untreated, they display reduced diversity compared to post-flare, baseline and flares being treated intermittently.
Alpha diversity (Chao1 species richness, Faith’s phylogenetic diversity index, the observed number of species, and the Shannon evenness measure) was found to be higher in mice who were healthy than those who had been inflicted with EBA (epidermolysis bullosa acquisita, an autoantibody-induced inflammatory skin disease) [28]. It was suggested that diversity is predictive of disease outcome. The shifts in the composition and diversity of skin bacterial communities observed on healthy mice compared to mice with EBA symptoms were similar to those between humans suffering from AD and psoriatic lesions, and those with healthy skin. Also noted was the significant increase in AD in industrialised countries, but the cause is still unclear.
2.1.5. Summary of Results
Healthy uncontacted Amerindian Yanomami people and rural Guahibo people, with little exposure to western civilisation, showed greater skin diversity than healthy US residents. It is very important to note that even the healthiest of western skin falls far short of the diversity observed in the uncontacted and remote peoples described.
However, the key data that we have observed indicates that, irrespective of background, unhealthy skin has a diversity level below that of any healthy individual. Similar data is reproduced in other species too, and not limited to humans, thus indicating that it is a common characteristic regardless of DNA.
2.2. Using the Data
To summarise, there are two main conclusions to draw from our findings:
(a) Healthy skin untouched by western practices harbours a higher microbial diversity than normal “western” skin;
(b) Damaged skin harbours a reduced diversity of microbial species compared to healthy or normal skin on the same type of subject. This is backed up by previous findings [85,86,87].
If these conclusions are to have a practical use for the testing of healthy skin, then a baseline level of diversity has to be established—what is the level below which, irrespective of culture or environment, we can confidently say that skin is damaged? For instance, does damaged Yanomami skin fall to a similar level to that of western damaged skin? If it only falls slightly (to levels seen in “healthy” western skin), then prediction will be difficult, as an individual’s background and environment will have to be taken into consideration. If, however, there is a lower range below which ALL groups and cultures fall, then an unambiguously reliable mechanism for testing skin health can be proposed.
2.3. Comparing the Data
The problem with the data obtained is that the number of sequences tested tended to be different across each study. We need to be able to match each dataset together, so we created the following formula to approximate the diversity curves from the studies described in Section 2.1.2 [75], Section 2.1.3 [76] and Section 2.1.4 [77]:
This equation is based on the commonly used depreciation formula, but minuses it from a maximum diversity value (which we set using data from the studies in question). Each curve tends to a maximum diversity as the sequence number heads to infinity. In the first sequences, there is a high chance that there will be new species discovered. As more sequences are processed, the chance of finding a new species lessens, and the gradient of the line drops off, i.e., depreciation.
Crucially, this powerful “Diversity Projection Tool” allows us to start comparing all our data by transposing the information onto a single graph with the same axes. It allows us for the first time to illustrate skin health using diversity.
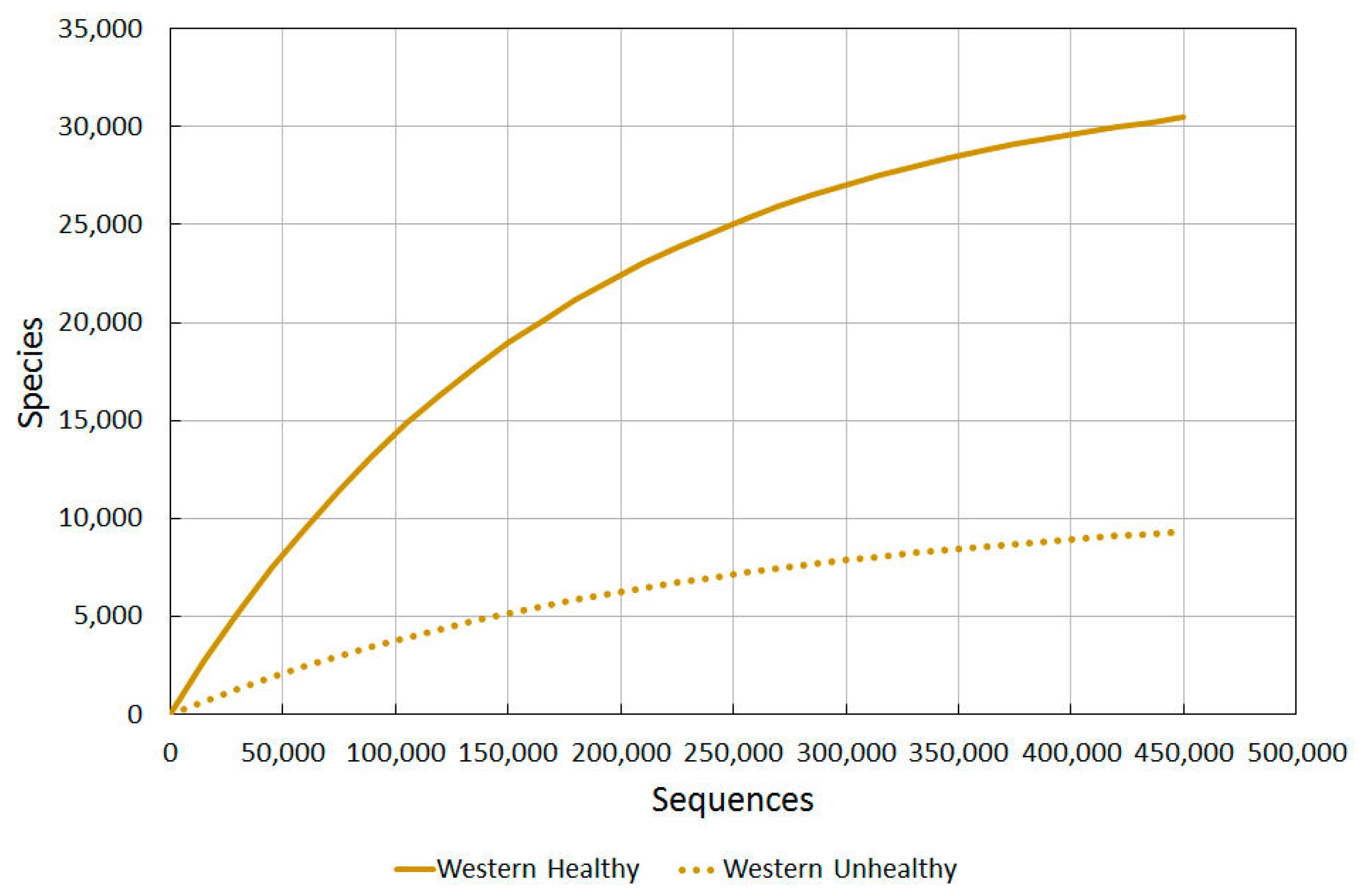
2.3.1. Western Healthy vs. Western Unhealthy
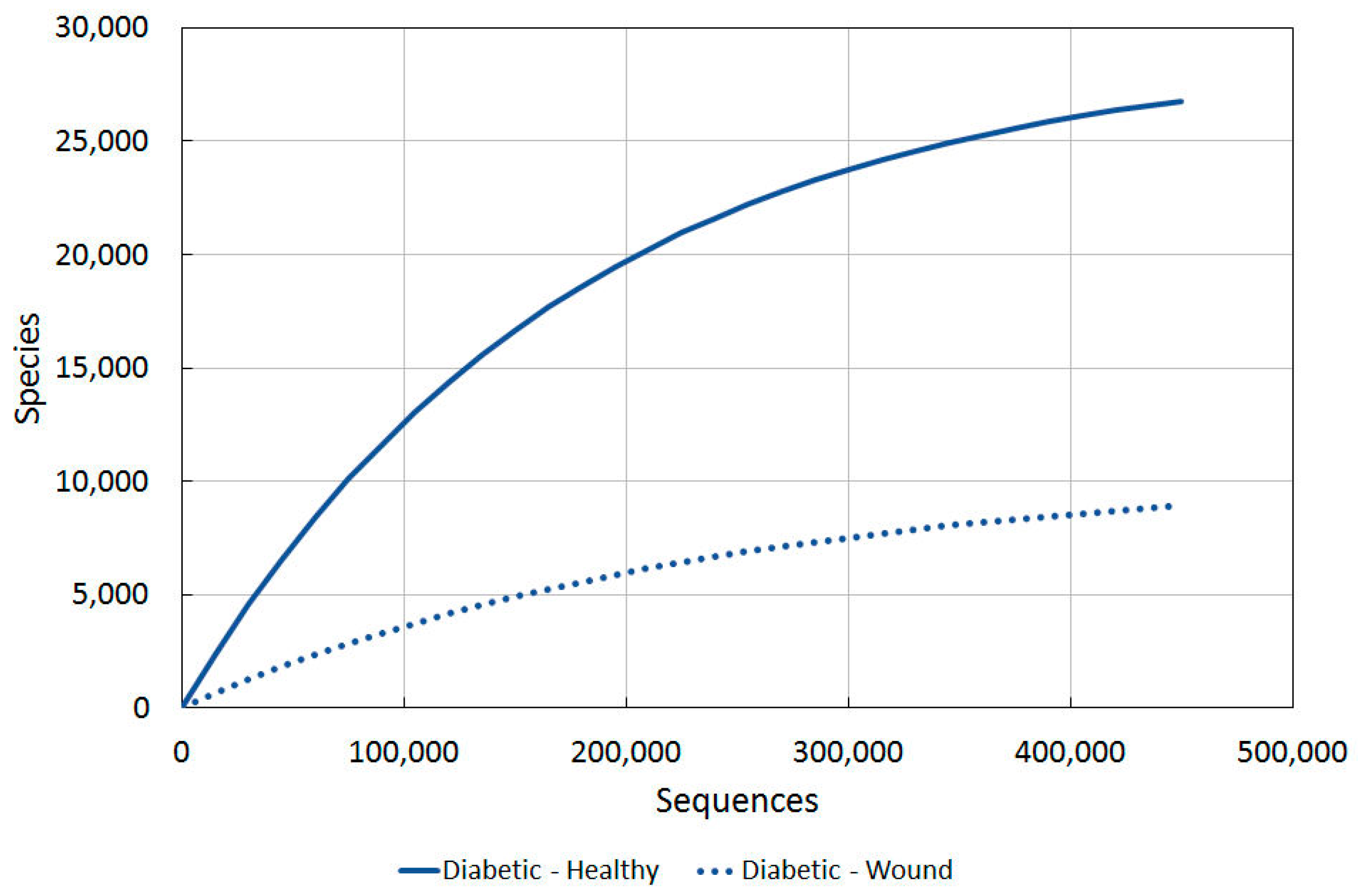
To confidently say that skin is damaged and open to attack from allergies or pathological microbial growth, benchmark diversity values for different levels of healthiness would be needed to be plotted on a graph. Firstly we used skin diversity values of healthy western US residents against those of wounded ones [77]. Different techniques to analyse diversity were used, and varying amounts of sequences per sample. For each type of diversity analysis, a 3% and 5% divergence level was used. The results are seen in Table 3 in Gontcharova et al. [77].
Using our Diversity Projection Tool, we were able to extrapolate diversity projections for up to 450,000 sequences (as shown in Figure 2). We used the Chao1 3% index as this most clearly tied in with the sequencing used in the other studies, thus allowing us to compare them more directly.
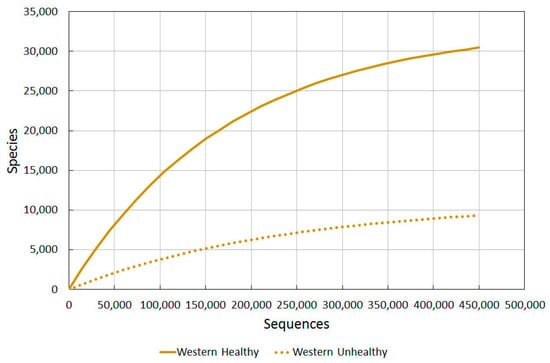
Figure 2.
A comparison of bacterial diversity in healthy and unhealthy western skin. Rarefaction analysis using Chao1 3% index.
The wounded/unhealthy average for western skin is much lower than healthy western skin, as predicted.
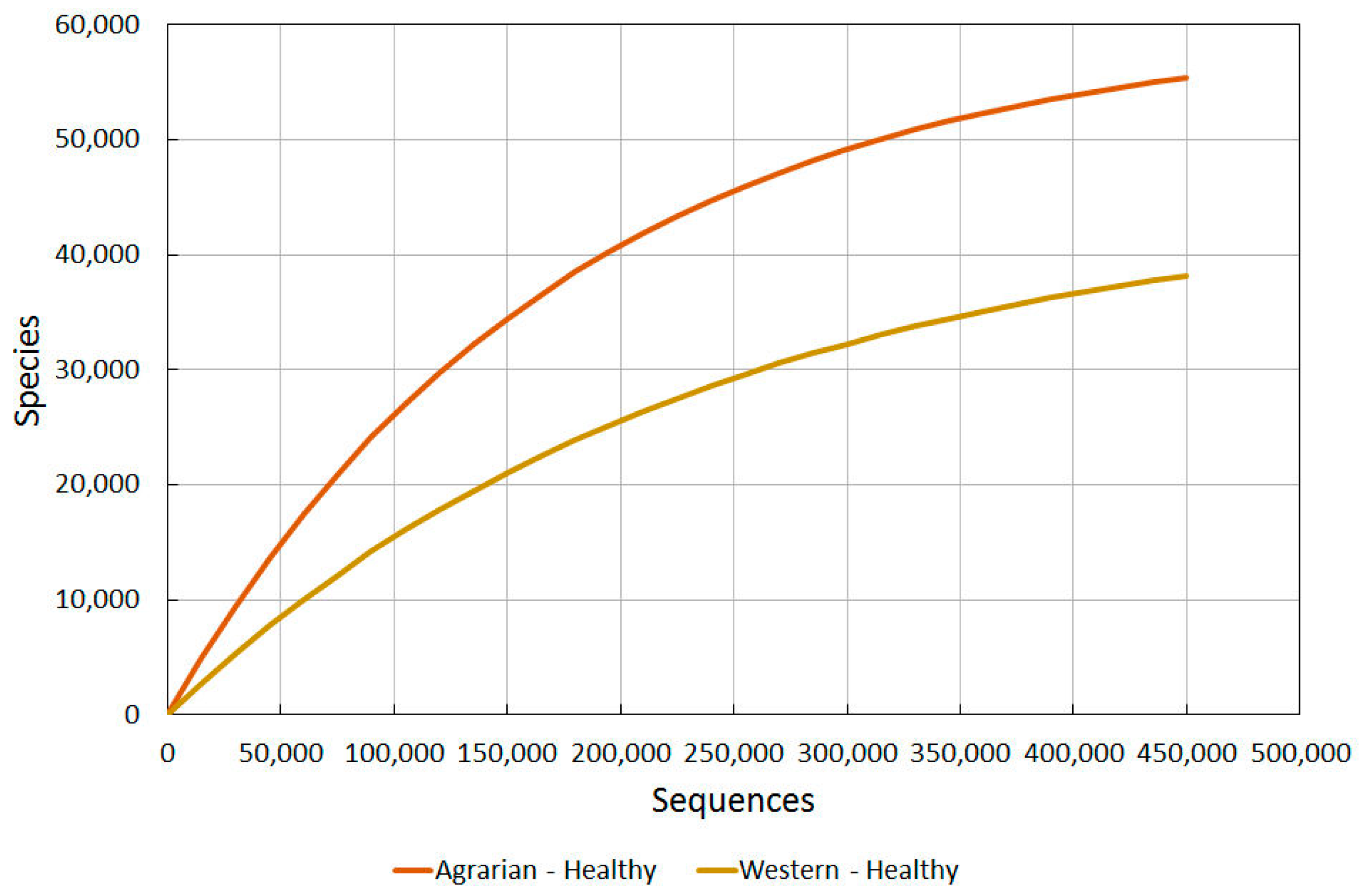
2.3.2. Agrarian Healthy vs. Western Healthy Skin
Using the data for healthy western skin (in Figure 2), we then used our Prediction Tool to extrapolate comparative data from our findings for the Guahibo agrarian culture (see Section 2.1.2) and compare the findings (Figure 3).
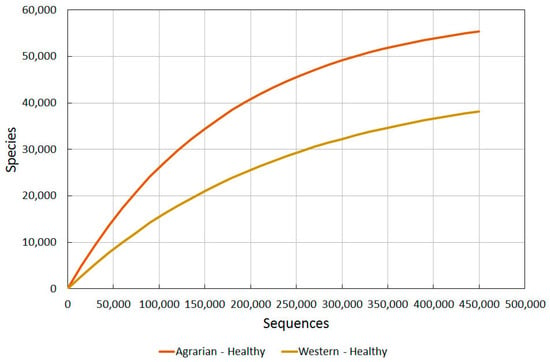
Figure 3.
A comparison of bacterial diversity in healthy agrarian and healthy western skin. Rarefaction analysis using Chao1 3% index.
The difference observed between these different cultures is very revealing, and re-enforces the suggestion that there are major environmental factors causing even healthy western subjects to have a much reduced diversity from that which humans probably all had for most of their existence.
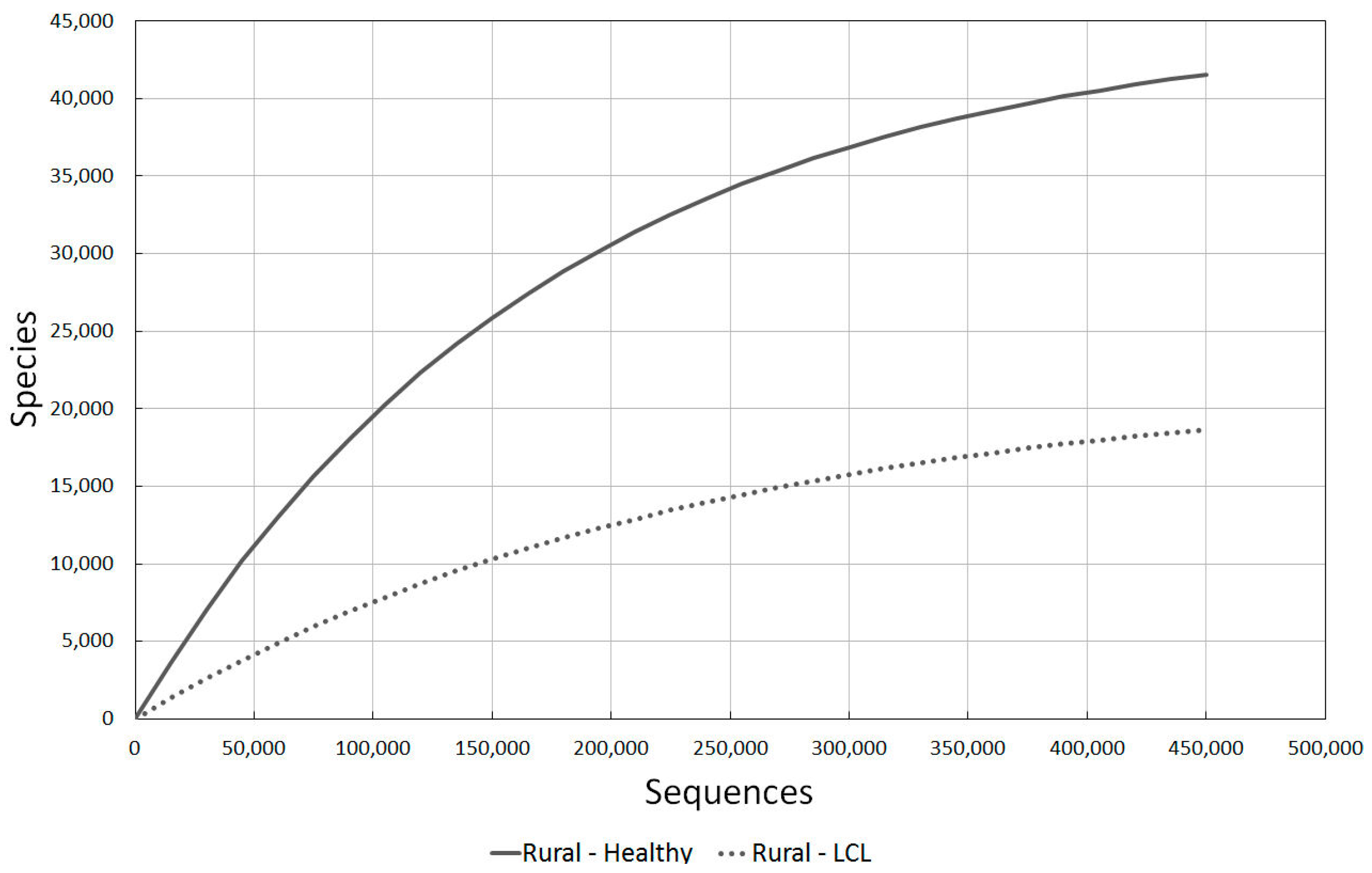
2.3.3. Rural Healthy vs. LCL Lesion Skin
We also need to gain an understanding of how rural cultures’ microbial diversity reduced when skin problems occurred, and compare this to western skin problems.
The data from the agricultural rural community in Brazil was the most useful data source for this information, and the decline in diversity between healthy and non-healthy individuals can be clearly seen in Figure 4.
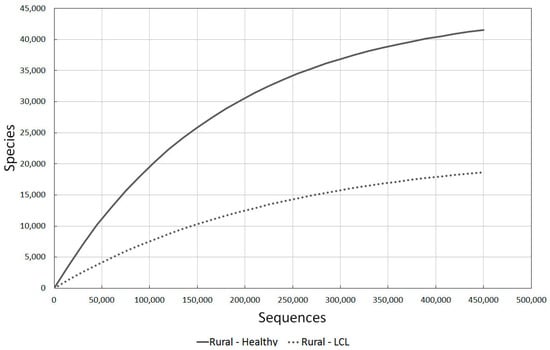
Figure 4.
A comparison of bacterial diversity in healthy rural and localised cutaneous leishmaniasis (LCL) lesioned skin. Rarefaction analysis using Chao1 3% index.
2.3.4. Diabetic vs. Intact Skin
To get an indication of how individuals with chronic problems compared, we used the study of diabetics in the USA. Figure 5 illustrates the marked decline in diversity between healthy and wounded skin.
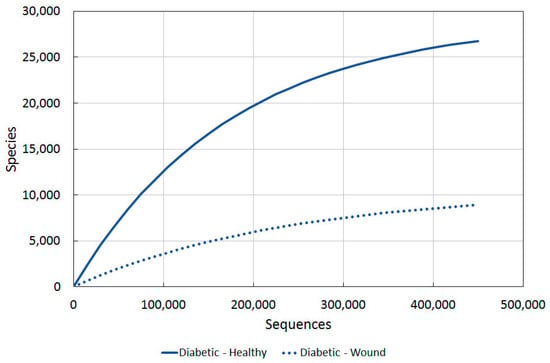
Figure 5.
A comparison of bacterial diversity in diabetic ulcers and contralateral intact skin. Rarefaction analysis using Chao1 3% index.
2.3.5. Combining the Data
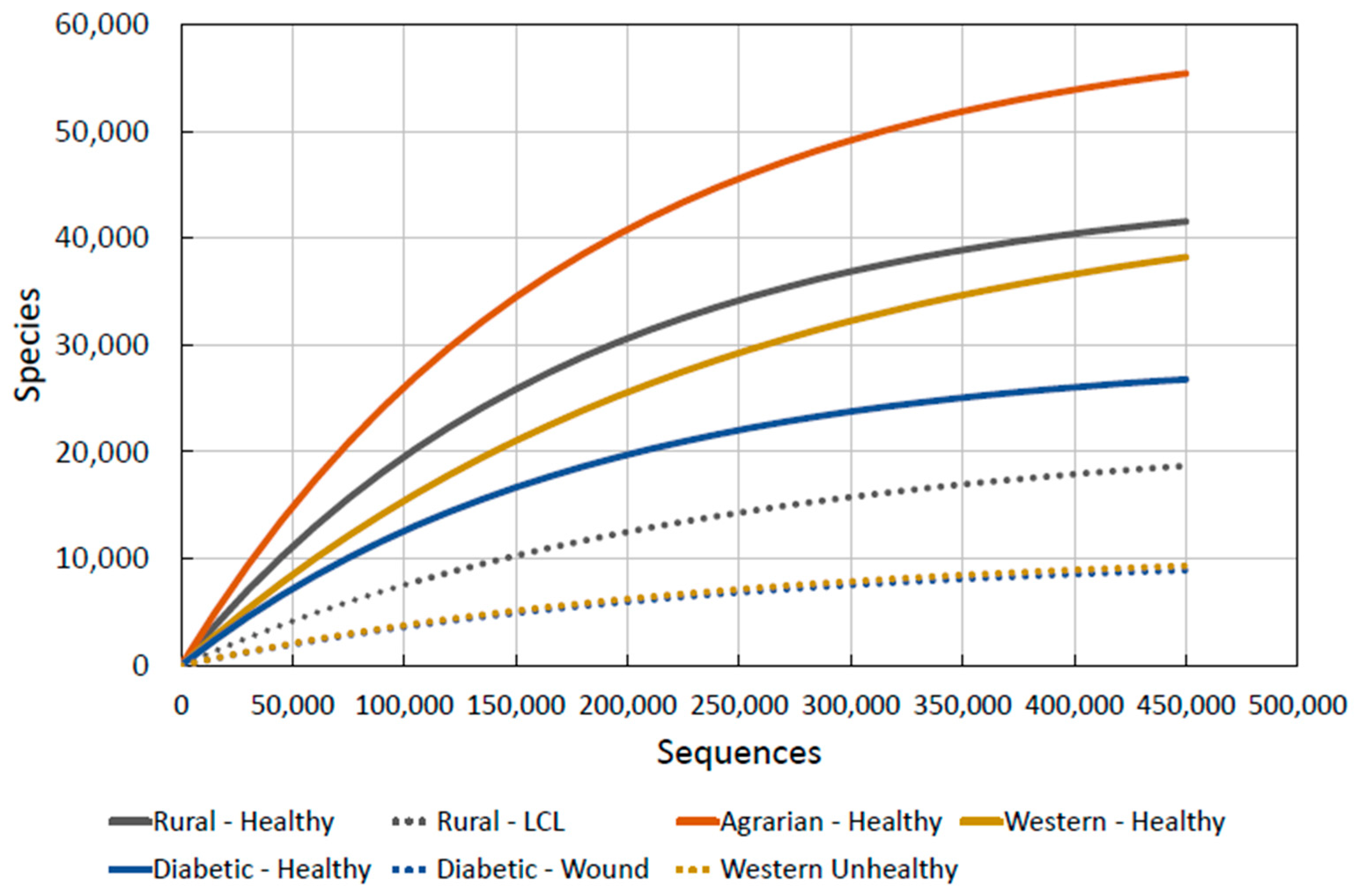
Figure 6 shows the diversity of microbes on the skin is the highest, by far, on the Agrarian Amerindian Guahibo people. They have had little exposure to westernisation. The rural agricultural community in Brazil, more exposed to western practices [75], contains the second highest diversity.
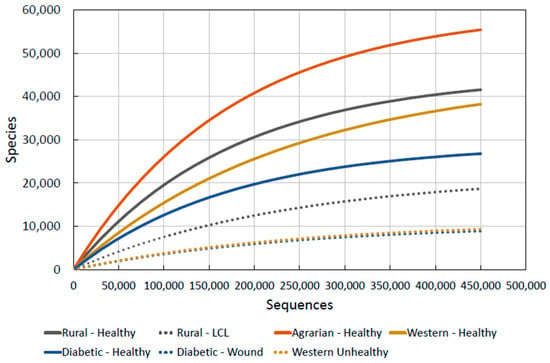
Figure 6.
Combined skin microbiota diversity.
As we now have comparable datasets, we are now able to transpose all of the above graphs onto one:
We note, interestingly, that the diversity of LCL lesioned skin in the Brazilian agricultural communities [76] is well below that of any healthy community, indicating that there may be a minimum diversity level for what could be defined as “healthy skin”. This is a crucial observation for our purposes as it allows us to hypothesise a simple mechanism for determining skin health and thus its likelihood to prevent or promote disease: if human skin microbiota diversity falls anywhere around or under the “unhealthy” lines (marked as dashes in Figure 6), then we can predict that it is damaged and likely to be either currently diseased or certainly open to immune related allergic conditions or attack by pathogens.
To put some figures to this observation using the data, we can now propose numerical statements such as:
“Skin microbial diversity is unhealthy when it drops below 10,000 species (after 150,000 sequences).”
The dotted lines on Figure 6 give the levels of diversity seen in unhealthy skin. By taking samples of skin microbiota from people, their diversity values at certain sequences can be found and compared to this graph. If the values are close to or below the values of “unhealthy” skin given, they can be said to have unhealthy skin. The proposal of 10,000 species at 150,000 sequences is the level we have found from unhealthy skin data and anything below this will also be unhealthy.
This gives us the means, therefore, to propose both a tool for measuring skin health and a means for testing which products damage the skin: if a product is tested on a subject and the diversity drops to unhealthy levels, then it can confidently be stated that the substance is detrimental to skin health.
2.4. Proposal for Using Skin Microbiota Diversity
We conclude that testing for the diversity of skin microbiota gives us a mechanism for determining whether skin is healthy or not. The vital importance of this discovery is that we can now for the first time start to determine which environmental factors are adversely affecting skin health, especially with regard to the skin allergy epidemic in the developed world as mentioned in Section 1.2.
2.4.1 Improving Our Mechanism in Future Work
The “previously uncontacted Yanomami people”, described in Section 2.1.1, were not analysed using the Chao1 index, so we were not able to approximate a curve for their data. Including their values for skin diversity on Figure 6 would be needed in future studies to set an upper benchmark for where “ultimate” skin health would fall.
Secondly, unhealthy or damaged skin diversity curves for both “ultimate” Yanomami skin and Guahibo should be added in future work. These would add weight to our hypothesis of a “threshold” skin diversity, meaning an increase in accuracy for the skin-health determining mechanism we proposed in Section 2.3.5.
Lastly, the exact data in Section 2.1 [28,74,75,76,84] would need to be all plotted on one graph using the “Quantitative Insights Into Microbial Ecology” tool (QIIME)—the standard tool for microbiome analysis of raw DNA sequencing data. This would mean our mechanism would be completely accurate, as the graphs in this study are approximated using the method explained in Section 2.3.
Finally, we acknowledge that the diversity curves for “Agrarian-Healthy”, “Western-Healthy”, “Diabetic-Healthy” and “Diabetic-Wound” in Figure 6 didn’t have raw data which extended up to 400,000 sequences, so were partly projected using our formula. The integrity of this approximated data can only be truly validated through further experimentation.
3. Discussion: Solving the Skin Allergy Epidemic
Our data strongly indicates that healthy skin contains a much more diverse microbial population than unhealthy or diseased skin [28,74,75,76,78,84], and this fits in with several studies that have linked high bacterial diversity with better protection of the host by an immune system which can innovate and adapt [9,12,13].
3.1. Comparisons with Gut Microbiota Studies
As previously discussed, the link between gut health and microbial diversity is also very evident, and it should come as no surprise, therefore, that our studies have revealed that the body’s microbiome as a whole displays the same traits.
3.1.1. Why Microbial Diversity Is Key to Health
In Section 1 we highlighted the crucial role microbes play in regulating the body’s immune system in a “mutualistically symbiotic” relationship, and how there is little conclusive evidence that healthy or unhealthy skin is determined by the presence of specific dominating types of microbiota. We must investigate why microbial diversity appears to be the key to good health.
It is important to note that while there has been much more research done on the gut’s health and microbial diversity than for the skin, the exact reasons for why a higher diversity relates to greater health of a system are still being researched.
Our findings have shown that microbial diversity is important in many ecosystems and encourages stability and equilibrium. On the back of our study, microbiota diversity could be set to become the new indicator of health. The most complex microbial ecosystem was considered to be the gastrointestinal tract: gut microbiota contains a vast number of microorganisms from all three domains of life including bacteria, fungi and viruses, and scientists are at last beginning to characterise the taxonomic abundance and community relationships not only of bacteria, but also the other microbes that inhabit the gut environment [88]. This work is bringing scientists one step closer to understanding the importance of microbial diversity in intestinal health and disease, and gives an understanding of how the multiple groups interact. Thus, while it has been known for some time that the bacteria in the gut play an important role in both health and disease, it is also now becoming clear that the non-bacterial microbiota interacts in a complex way with the bacterial microbiota to contribute to these processes.
Similarly, recent studies have also illustrated how our skin bacteria interact with each other, with other pathogenic microbes, and with human cells. For instance, Staphylococcus epidermidis secretes antimicrobial substances that help fight pathogenic invaders, while Propionibacterium acnes uses the skin’s lipids to generate short-chain fatty acids that can similarly ward off microbial threats.
These and other skin microbes may also be able to alter the behaviour of human immune cells, and impact the local molecular environment. Recent trials have been able to show that adding Staphylococcus epidermidis microbes to sterile skin enabled the immune system to control infection by altering the function of T-cells (a key component of the body’s adaptive immune response) to boost host immunity. Researchers have found that different microbes affect distinct parts of the immune system, and how they communicate with the immune system is highly specific to each microbe.
However, while some microbes are able to control aspects of immunity, others are able to promote inflammation and encourage infection. So, while some microbes are secreting protective antimicrobials, others are inducing our skin cells to release disease triggering substances. People with compromised immune systems, therefore, harbor bacterial and fungal species not normally found on healthy adults. Defects in the immune system allow or permit these otherwise uncommon microbes to be present.
In conclusion, the research into the reasons why microbiota diversity is so key to overall health is still in its infancy, and we are a long way off being able to fully understand the complex processes and interactions involved. All we are now starting to be able to say with some conviction is that microbial diversity is key to health, whether applied to the gut or the skin.
3.1.2. Using Probiotics
The idea of using probiotics to rebuild the gut flora has long been postulated [89], but the problem with the internal system is that it is inaccessible and sealed off from the external environment, even leading to some studies stating that the body’s internal microbiota system is laid down at birth via issues like “vaginal seeding” [90]. Much has also been said about cleanliness issues or drugs such as antibiotics permanently damaging the body’s internal microbiota immune response environment [91,92,93,94]. The problem is to find a mechanism that encourages and replenishes the delicate balance within the gut when it is cut off from its surroundings.
By contrast, our skin is exposed on a daily basis to the external world, and so doesn’t have the problem of access to the required microbes that are essential for the correct running of the skin’s immune defences: “re-stocking” with the correct flora is not an issue. Many of the problems about disease caused by gut imbalances and solutions to rectify the situation should not in theory, therefore, be applicable when it comes to the skin microbiome.
However, although our skin does not have this gut “inaccessibility” problem, our results have shown that unhealthy “low diversity” skin is prevalent across the developed world too. There is something in the environment that is causing these problems, and it is pointless proposing a “pro-biotic” solution to this crisis (i.e., adding “good” microbial communities to the skin) if there is something out there which immediately damages it again.
Left to its own devices, the skin will naturally recreate an environment that will encourage the correct levels of symbiotic microbes to protect the skin and enhance its natural immune defences. For instance, the skin’s natural pH and electrolyte levels will be recreated, and substances such as sebum (that are crucial to the immune symbiosis) produced at the correct levels [19,79,95].
3.2. Why Agrarian Cultures Have a Greater Skin Microbial Diversity
It is almost certain that the reason for less industrially developed agrarian cultures displaying a much higher skin microbial diversity is because their skin has been allowed to look after itself and has not had contact with western cosmetics, cleaners, drugs or pollution, which can strip the skin of its essential oils and bacteria [14]. The skin on the humans in these cultures is probably the closest we can get to what “caveman” skin looked like—i.e., how mankind’s skin microbial diversity has looked for 99% of the c.200,000 years that homo sapiens have inhabited the Earth.
Our findings, which show that other mammalian species exhibit the same microbial diversity decrease on damaged skin, also point to the fact that this “caveman” skin has probably evolved, not just over the 6 million years back to which our direct ancestors can be traced [96,97], but even further back in the evolution of mammalian species tens of millions of years ago [98,99]. Further studies on the skin of birds and reptiles would be another very interesting avenue in determining whether this immune symbiosis possibly pre-dates the rise of the mammals and even the time of the dinosaurs.
3.3. Testing for Skin Health: A New Mechanism Revealed
Our microbial diversity data now allows us to propose a definitive test to measure skin health and hence determine which environmental substances could be contributing to damaged skin. As previously discussed, if the particular substance reduces the skin’s microbial diversity to a value close to that of the “damaged western skin” on Figure 6, we can confidently predict that it is harmful and will almost certainly lead to allergic or pathogenic microbial ill-health.
Our testing mechanism can be used to observe the effect of using particular cosmetic products on the body and if they enhance or degrade the skin’s microbiota diversity.
3.4. Are Cosmetics a Major Contributor to the Skin Allergy Epidemic?
Up until now, the rampant growth of skin allergies and ill-health in the West, and the environmental cause, remains largely unexplained.
An increasing number of studies have linked these health issues to synthetic additives in cosmetics [100,101,102,103,104,105]. The exposure of normal, western skin to twenty-first-century cosmetics, soap, antibiotics, steroids and showering, does appear to have altered the natural microbiota environment of humans, especially in the developed world [14,16]. This alteration, in many cases, has been linked to increased susceptibility to disease and infection [11,12,21]. Healthy “natural” skin keeps the resident bacterial flora attached to the skin, whereas damaged skin promotes their dispersal from the skin and leads to pathogenic bacterial and fungal growth [15,16,19].
A crucial first use for our mechanism will, therefore, be to test whether contact with synthetic chemicals commonly found in cosmetics actually does damage the skin (leading to disease), and which ingredients are the most dangerous. This can be used as a definitive test to find out if claims made by cosmetic manufacturers are true. Regular use of products such as body washes, soaps, shampoos and shower gels should be tested first as individuals are subject to contact with synthetic chemicals on a daily basis with these items.
Although many chemical ingredients are currently allowed under EU and FDA rules, our diversity mechanism could re-write the rule book and lead to the banning of ingredients and products that are proven to harm the skin’s natural environment.
4. Conclusions
Healthy ecosystems, from Yellowstone Park down to bacteria in the human gut, appear to display higher diversity of species, when compared with unhealthy or damaged ones. The environmental and health benefits of biodiversity are found everywhere throughout nature, and our research confirms the consistency of this phenomenon with skin too: our data indicates that microbiota diversity alone can predict whether skin is healthy or not.
“Caveman” skin, untouched by modern civilisation, was far different to “western” skin and displayed unprecedented levels of bacterial diversity. Our research has shown that the more isolated the community that subjects lived in—with consequently less exposure to western practices—the higher the diversity measured on the skin. This is clear evidence of a link between an environmental factor in the developed world and poor microbial diversity on the skin. Consequently:
For the first time, we have been able to propose benchmark values of diversity against which we can measure skin to determine how healthy it is. This gives us the ability to be able to predict which people are more likely to be prone to skin ailments ranging from allergic conditions such as eczema and psoriasis to pathogenic microbial imbalances such as acne, ulcers and lesions.
For the first time we propose a mechanism to determine whether cosmetic ingredients and products are actually the main environmental cause of the skin allergy epidemic in the western world, harming the skin’s natural microbiota and its immune defences and opening it up to allergic or pathogenic microbial attack.
For the first time, we propose a solution to the skin allergy epidemic: the reintroduction of species diversity to the skin through products which enhance and recreate its natural environment—just like in the gut, where a better diet encourages the growth of a wider diversity of microbes. We use the analogy of Yellowstone Park, where the re-introduction of the wolves has been credited by some with returning balance to the ecosystem, allowing other species to flourish and turning “pathogenic” species (e.g., elk) into beneficial ones.
Acknowledgments
This work was supported by Pavane Consultants Ltd.
Author Contributions
Christopher Wallen-Russell was the main contributor. He conceived the idea for the paper, planned, researched, and wrote it. Sam Wallen-Russell analysed the data, created the approximated diversity graphs and fitted them to the real data from previous studies and researched the role of non-linearity in biodiversity along with the problems this caused.
Conflicts of Interest
Christopher Wallen-Russell and Sam Wallen-Russell are employees of research and development company Pavane Consultants Ltd., who commissioned the paper, and directors of JooMo Ltd. Skin health company, JooMo Ltd., holds the exclusive license for the manufacturing, marketing and sale of Pavane Consultant Ltd.’s products. As licence holder for the JooMo Ltd. range of skin health products, Pavane Consultants Ltd. are interested in determining how skin health can be measured and which products in the environment have caused the huge increase in skin allergy problems in the past 75 years.
References
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the human intestinal microbial flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, A.M.; Shanahan, F. The gut flora as a forgotten organ. EMBO Rep. 2006, 7, 688–693. [Google Scholar] [CrossRef] [PubMed]
- Kelly, D.; Conway, S.; Aminov, R. Commensal gut bacteria: Mechanisms of immune modulation. Trends Immunol. 2005, 26, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Finlay, B.B.; Arrieta, M.-C. Let Them Eat Dirt. Saving Your Child from an Oversanitized World; Windmill Books: London, UK, 2016. [Google Scholar]
- Marrs, T.; Flohr, C. The role of skin and gut microbiota in the development of atopic eczema. Br. J. Dermatol. 2016, 175, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Martínez, I.; Stegen, J.C.; Maldonado-Gómez, M.X.; Eren, A.M.; Siba, P.M.; Greenhill, A.R.; Walter, J. The Gut Microbiota of Rural Papua New Guineans: Composition, Diversity Patterns, and Ecological Processes. Cell Rep. 2015, 11, 527–538. [Google Scholar] [CrossRef] [PubMed]
- Taylor, B.; Wadsworth, M.; Wadsworth, J.; Peckham, C. Changes in the reported prevalence of childhood eczema since the 1939–45 war. Lancet 1984, 324, 1255–1257. [Google Scholar] [CrossRef]
- Ribeiro, A.; Estanqueiro, M.; Oliveira, M.; Sousa Lobo, J. Main Benefits and Applicability of Plant Extracts in Skin Care Products. Cosmetics 2015, 2, 48–65. [Google Scholar] [CrossRef]
- Grice, E.A.; Segre, J.A. The skin microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Findley, K.; Grice, E.A. The skin microbiome: A focus on pathogens and their association with skin disease. PLoS Pathog. 2014, 10, e1004436. [Google Scholar] [CrossRef] [PubMed]
- Cogen, A.L.; Nizet, V.; Gallo, R.L. Skin microbiota: A source of disease or defence? Br. J. Dermatol. 2009, 158, 442–455. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A.; Snitkin, E.S.; Yockey, L.J.; Bermudez, D.M.; NISC Comparative Sequencing Program; Liechty, K.W.; Segre, J.A. Longitudinal shift in diabetic wound microbiota correlates with prolonged skin defense response. Proc. Natl. Acad. Sci. USA 2010, 33107, 14799–14804. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.H.; Segre, J.A. Skin microbiome: Looking back to move forward. J. Investig. Dermatol. 2012, 132, 933–939. [Google Scholar] [CrossRef] [PubMed]
- Blaser, M.J.; Falkow, S. What are the consequences of the disappearing human microbiota? Nat. Rev. Microbiol. 2009, 7, 887–894. [Google Scholar] [CrossRef] [PubMed]
- Noble, W.C. Staphylococci on the skin. In The Skin Microflora and Microbial Skin Disease; Noble, W.C., Ed.; Cambridge University Press: London, UK, 2004; pp. 135–152. [Google Scholar]
- Holland, K.T.; Bojar, R.A. Cosmetics: What is their influence on the skin microflora? Am. J. Clin. Dermatol. 2002, 3, 445–449. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A.; Kong, H.H.; Renaud, G.; Young, A.C.; Bouffard, G.G.; Blakesley, R.W.; Wolfsberg, T.G.; Turner, M.L.; Segre, J.A.; NISC, C.S.P. A diversity profile of the human skin microbiota. Genome Res. 2008, 18, 1043–1050. [Google Scholar] [CrossRef] [PubMed]
- Katsuyama, M.; Ichikawa, H.; Ogawa, S.; Ikezawa, Z. A novel method to control the balance of skin microflora: Part 1. Attack on biofilm of Staphylococcus aureus without antibiotics. J. Dermatol. Sci. 2005, 38, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Lambers, H.; Piessens, S.; Bloem, A.; Pronk, H.; Finkel, P. Natural skin surface pH is on average below 5, which is beneficial for its resident flora. Int. J. Cosmet. Sci. 2006, 28, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Brogden, N.K.; Mehalick, L.; Fischer, C.L.; Wertz, P.W.; Brogden, K.A. The emerging role of peptides and lipids as antimicrobial epidermal barriers and modulators of local inflammation. Skin Pharmacol. Physiol. 2012, 25, 167–181. [Google Scholar] [CrossRef] [PubMed]
- Belkaid, Y.; Hand, T.W. Role of the microbiota in immunity and inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; Bouffard, G.G.; Blakesley, R.W.; Murray, P.R.; Green, E.D. Topographical and temporal diversity of the human skin microbiome. Science 2009, 324, 1190–1192. [Google Scholar] [CrossRef] [PubMed]
- Rinne, M.; Kalliomäki, M.; Salminen, S.; Isolauri, E. Probiotic intervention in the first months of life: Short-term effects on gastrointestinal symptoms and long-term effects on gut microbiota. J. Pediatr. Gastroenterol. Nutr. 2006, 43, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Noverr, M.C.; Huffnagle, G.B. The “microflora hypothesis” of allergic diseases. Clin. Exp. Allergy 2005, 35, 1511–1520. [Google Scholar] [CrossRef] [PubMed]
- Simpson, C.R.; Newton, J.; Hippisley-Cox, J.; Sheikh, A. Trends in the epidemiology and prescribing of medication for eczema in England. J. R. Soc. Med. 2009, 102, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Burd, R.M. Psoriasis: A general overview. Br. J. Hosp. Med. 2006, 67, 259–262. [Google Scholar] [CrossRef] [PubMed]
- Prescott, S.L.; Tang, M.L.K. The Australasian Society of Clinical Immunology and Allergy position statement: Summary of allergy prevention in children. Med. J. Aust. 2005, 182, 464–467. [Google Scholar] [PubMed]
- Srinivas, G.; Möller, S.; Wang, J.; Künzel, S.; Zillikens, D.; Baines, J.F.; Ibrahim, S.M. Genome-wide mapping of gene–microbiota interactions in susceptibility to autoimmune skin blistering. Nat. Commun. 2013, 4, 2462. [Google Scholar] [CrossRef] [PubMed]
- Asher, M.I.; Montefort, S.; Björkstén, B.; Lai, C.K.; Strachan, D.P.; Weiland, S.K.; Williams, H.; ISAAC Phase Three Study Group. Worldwide time trends in the prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and eczema in childhood: ISAAC Phases One and Three repeat multicountry cross-sectional surveys. Lancet 2006, 368, 733–743. [Google Scholar] [CrossRef]
- Shaw, T.E.; Currie, G.P.; Koudelka, C.W.; Simpson, E.L. Eczema Prevalence in the United States: Data from the 2003 National Survey of Children’s Health. J. Investig. Dermatol. 2011, 131, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Bickers, D.R.; Lim, H.W.; Margolis, D.; Weinstock, M.A.; Goodman, C.; Faulkner, E.; Gould, C.; Gemmen, E.; Dall, T.; American Academy of Dermatology Association Society for Investigative Dermatology. The burden of skin diseases: 2004. J. Am. Acad. Dermatol. 2006, 55, 490–500. [Google Scholar] [CrossRef] [PubMed]
- Bjarnsholt, T.; Kirketerp-Møller, K.; Jensen, P.Ø.; Madsen, K.G.; Phipps, R.; Krogfelt, K.; Høiby, N.; Givskov, M. Why chronic wounds will not heal: A novel hypothesis. Wound Repair Regen 2008, 16, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, N.G.; Agache, I.; Bavbek, S.; Bilo, B.M.; Braido, F.; Cardona, V.; Custovic, A.; Demoly, P.; Eigenmann, P.; Gayraud, J.; et al. Research needs in allergy: An EAACI position paper, in collaboration with EFA. Clin. Transl. Allergy 2012, 2, 21. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.; Kiyu, A.; Milman, H.M.; Jimenez, J. Improving health and building human capital through an effective primary care system. J. Urban Health 2007, 84 (Suppl. S3), i75–i85. [Google Scholar] [CrossRef] [PubMed]
- Dessinioti, C.; Katsambas, A.D. The role of Propionibacterium acnes in acne pathogenesis: Facts and controversies. Clin. Dermatol. 2010, 28, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Lozupone, C.A.; Hamady, M.; Knight, R.; Gordon, J.I. Worlds within worlds: Evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 2008, 6, 776–788. [Google Scholar] [CrossRef] [PubMed]
- Pei, Z.; Bini, E.J.; Yang, L.; Zhou, M.; Francois, F.; Blaser, M.J. Bacterial biota in the human distal esophagus. Proc. Natl. Acad. Sci. USA 2004, 101, 4250–4255. [Google Scholar] [CrossRef] [PubMed]
- Costello, E.K.; Lauber, C.L.; Hamady, M.; Fierer, N.; Gordon, J.I.; Knight, R. Bacterial Community Variation in Human Body Habitats Across Space and Time. Science 2009, 326, 1694–1697. [Google Scholar] [CrossRef] [PubMed]
- Van de Merwe, J.P.; Stegeman, J.H.; Hazenberg, M.P. The resident faecal flora is determined by genetic characteristics of the host. Implications for Crohn’s disease? Antonie Van Leeuwenhoek 1983, 49, 119–124. [Google Scholar] [CrossRef] [PubMed]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef] [PubMed]
- Sonnenburg, E.D.; Sonnenburg, J.L. Starving our Microbial Self: The Deleterious Consequences of a Diet Deficient in Microbiota-Accessible Carbohydrates. Cell Metab. 2014, 20, 779–786. [Google Scholar] [CrossRef] [PubMed]
- Lefcheck, J.S.; Byrnes, J.E.K.; Isbell, F.; Gamfeldt, L.; Griffin, J.N.; Eisenhauer, N.; Hensel, M.J.; Hector, A.; Cardinale, B.J.; Duffy, J.E. Biodiversity enhances ecosystem multifunctionality across trophic levels and habitats. Nat. Commun. 2015, 6, 6936. [Google Scholar] [CrossRef] [PubMed]
- Balvanera, P.; Pfisterer, A.B.; Buchmann, N.; He, J.-S.; Nakashizuka, T.; Raffaelli, D.; Schmid, B. Quantifying the evidence for biodiversity effects on ecosystem functioning and services. Ecol. Lett. 2006, 9, 1146–1156. [Google Scholar] [CrossRef] [PubMed]
- Gamfeldt, L.; Hillebrand, H.; Jonsson, P.R. Multiple functions increase the importance of biodiversity for overall ecosystem functioning. Ecology 2008, 89, 1223–1231. [Google Scholar] [CrossRef] [PubMed]
- Thrupp, L.A. The importance of biodiversity in agroecosystems. J. Crop Improv. 2004, 12, 315–337. [Google Scholar] [CrossRef]
- Brookfield, H.; Padoch, C. Appreciating Agrodiversity: A Look at the Dynamism and Diversity of Indigenous Farming Practices. Environ. Sci. Policy Sustain. Dev. 1994, 36, 6–45. [Google Scholar] [CrossRef]
- Dudgeon, D.; Arthington, A.H.; Gessner, M.O.; Kawabata, Z.-I.; Knowler, D.J.; Lévêque, C.; Naiman, R.J.; Prieur-Richard, A.H.; Soto, D.; Stiassny, M.L. Freshwater biodiversity: Importance, threats, status and conservation challenges. Biol. Rev. 2006, 81, 163. [Google Scholar] [CrossRef] [PubMed]
- Hooper, D.U.; Chapin, F.S.; Ewel, J.J.; Hector, A.; Inchausti, P.; Lavorel, S.; Lawton, J.H.; Lodge, D.M.; Loreau, M.; Naeem, S.; et al. Effects of biodiversity on ecosystem functioning: A consensus of current knowledge. Ecol. Monogr. 2005, 75, 3–35. [Google Scholar] [CrossRef]
- Cardinale, B.J.; Srivastava, D.S.; Emmett Duffy, J.; Wright, J.P.; Downing, A.L.; Sankaran, M.; et al. Effects of biodiversity on the functioning of trophic groups and ecosystems. Nature 2006, 443, 989–992. [Google Scholar] [CrossRef] [PubMed]
- Worm, B.; Barbier, E.B.; Beaumont, N.; Duffy, J.E.; Folke, C.; Halpern, B.S.; Jackson, J.B.; Lotze, H.K.; Micheli, F.; Palumbi, S.R.; et al. Impacts of Biodiversity Loss on Ocean Ecosystem Services. Science 2006, 314, 787–790. [Google Scholar] [CrossRef] [PubMed]
- Costanza, R.; Mageau, M. What is a healthy ecosystem? In Aquatic Ecology; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1999; pp. 105–115. [Google Scholar]
- Ripple, W.J.; Beschta, R.L. Wolf reintroduction, predation risk, and cottonwood recovery in Yellowstone National Park. For. Ecol. Manag. 2003, 184, 299–313. [Google Scholar] [CrossRef]
- Zahniser, A.; Singh, A. Return of the wolves to Yellowstone National Park, USA: A model of ecosystem restoration. Biodiversity 2004, 5, 3–7. [Google Scholar] [CrossRef]
- Smith, D.W.; Peterson, R.O.; Houston, D.B. Yellowstone after wolves. Bioscience 2003, 53, 330–340. [Google Scholar] [CrossRef]
- Beschta, R.L.; Ripple, W.J. Berry-producing shrub characteristics following wolf reintroduction in Yellowstone National Park. For. Ecol. Manag. 2012, 276, 132–138. [Google Scholar] [CrossRef]
- Haines, A.L. The Yellowstone Story—A History of Our First National Park; University Press of Colorado: Boulder, CO, USA, 1996. [Google Scholar]
- Fischer, H. Wolf Wars: The Remarkable Inside Story of the Restoration of Wolves to Yellowstone; Falcon Press: Nashville, TN, USA, 1995; p. 183. [Google Scholar]
- Berger, K.M.; Gese, E.M.; Berger, J. Indirect effects and traditional trophic cascades: A test involving wolves, coyotes and pronghorn. Ecology 2008, 89, 818–828. [Google Scholar] [CrossRef] [PubMed]
- Stolzenburg, W. Where the Wild Things Were: Life, Death, and Ecological Wreckage in a Land of Vanishing Predators; Bloomsbury: London, UK, 2009; p. 291. ISBN 1596916249. [Google Scholar]
- Kay, C.E. Viewpoint: Ungulate herbivory, willows, and political ecology in yellowstone. J. Range Manag. 1997, 50, 139–145. [Google Scholar] [CrossRef]
- Platts, W.S.; Armour, C.L.; Booth, G.D.; Bryant, M.; Bufford, J.L.; Cuplin, P.; Jensen, S.; Lienkaemper, G.W.; Minshall, G.W.; Monsen, S.T.; et al. Methods for Evaluating Riparian Habitats with Applications to Management; General Technical Report INT-221; USDA Forest Service: Ogden, UT, USA; p. 177.
- Platts, W.S.; Megahan, W.F.; Minshall, G.W. Methods for Evaluating Stream, Riparian, and Biotic Conditions; General Technical Report INT-138; USDA Forest Service: Ogden, UT, USA; p. 70.
- Platts, W.S. Livestock grazing. Am. Fish Soc. Spec. Publ. 1991, 19, 389–424. [Google Scholar]
- Patten, D.T. Herbivore optimization and overcompensation: Does native herbivory on western rangelands support these theories? Ecol. Appl. 1993, 3, 35–36. [Google Scholar] [CrossRef] [PubMed]
- Rosgen, D.L. Stream Classification, Streambank Erosion, and Fluvial Interpretations for the Lamar River and Main Tributaries; Technical Report USDI Park Service; Yellowstone Natl Park: Pagosa Springs, CO, USA, 1993; p. 82. [Google Scholar]
- Kay, C.E. Yellowstone’s Northern Elk Herd: A Critical Evaluation of the “Natural Regulation” Paradigm. Ph.D. Dissertation, Utah State University, Logan, UT, USA, 1990. [Google Scholar]
- Kay, C.E. Browsing by native ungulates: Effects on shrub and seed production in the Greater Yellowstone Ecosystem. In Wild Land Shrub and Arid Land Restoration Symposium: Proceedings; Roundy, B.A., McArthur, E.D., Haley, J.S., Mann, D.K., Eds.; Forest Service General Technical Report INT-GTR-315; U.S. Department of Agriculture: Ogden, UT, USA, 1995; p. 384. [Google Scholar]
- Wagner, F.H.; Sax, J.L. Wildlife Policies in the U.S. National Parks; Island Press: Washington, DC, USA, 1995; p. 242. [Google Scholar]
- Beyer, H.L.; Merrill, E.H.; Varley, N.; Boyce, M.S. Willow on Yellowstone’s northern range: Evidence for a trophic cascade? Ecol. Appl. 2007, 17, 1563–1571. [Google Scholar] [CrossRef] [PubMed]
- Tyers, D.B. The Condition of the Northern Yellowstone Winter Range in Yellowstone National Park—A Discussion of the Controversy. Master’s Thesis, Montana State University, Bozeman, MT, USA, 1981. [Google Scholar]
- Kay, C.E. The impact of native ungulates and beaver on riparian communities in the Intermountain West. Nat. Resour. Environ. Issues 1994, 1, 23–44. [Google Scholar]
- Werndl, C.; Frigg, R. Entropy—A guide for the perplexed. In Probabilities in Phyics; Beisbart, C., Hartmann, S., Eds.; Oxford University Press: Oxford, UK, 2011. [Google Scholar] [CrossRef]
- Jost, L. Entropy and diversity. Oikos 2006, 113, 363–375. [Google Scholar] [CrossRef]
- Clemente, J.C.; Pehrsson, E.C.; Blaser, M.J.; Sandhu, K.; Gao, Z.; Wang, B.; Magris, M.; Hidalgo, G.; Contreras, M.; Noya-Alarcón, Ó.; et al. The microbiome of uncontacted Amerindians. Sci. Adv. 2015, 1, e1500183. [Google Scholar] [CrossRef] [PubMed]
- Blaser, M.J.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Estrada, I.; Gao, Z.; Clemente, J.C.; Costello, E.K.; Knight, R. Distinct cutaneous bacterial assemblages in a sampling of South American Amerindians and US residents. ISME J. 2013, 7, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Salgado, V.R.; de Queiroz, A.T.L.; Sanabani, S.S.; de Oliveira, C.I.; Carvalho, E.M.; Costa, J.M.L.; Barral-Netto, M.; Barral, A. The microbiological signature of human cutaneous leishmaniasis lesions exhibits restricted bacterial diversity compared to healthy skin. Mem. Inst. Oswaldo Cruz 2016, 111, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Gontcharova, V.; Youn, E.; Sun, Y.; Wolcott, R.D.; Dowd, S.E. A comparison of bacterial composition in diabetic ulcers and contralateral intact skin. Open Microbiol. J. 2010, 4, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.R.; Patterson, A.P.; Diesel, A.; Lawhon, S.D.; Ly, H.J.; Stephenson, C.E.; Mansell, J.; Steiner, J.M.; Dowd, S.E.; Olivry, T.; et al. The skin microbiome in healthy and allergic dogs. PLoS ONE 2014, 9, e83197. [Google Scholar] [CrossRef]
- Nakatsuji, T.; Kao, M.C.; Zhang, L.; Zouboulis, C.C.; Gallo, R.L.; Huang, C.-M. Sebum free fatty acids enhance the innate immune defense of human sebocytes by upregulating β-defensin-2 expression. J. Investig. Dermatol. 2010, 130, 985–994. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Weichbrodt, C.; Salnikov, E.S.; Dynowski, M.; Forsberg, B.O.; Bechinger, B.; Steinem, C.; de Groot, B.L.; Zachariae, U.; Zeth, K. Crystal structure and functional mechanism of a human antimicrobial membrane channel. Proc. Natl. Acad. Sci. USA 2013, 110, 4586–4591. [Google Scholar] [CrossRef] [PubMed]
- Ghose, C.; Perez-Perez, G.I.; Dominguez-Bello, M.-G.; Pride, D.T.; Bravi, C.M.; Blaser, M.J. East Asian genotypes of Helicobacter pylori strains in Amerindians provide evidence for its ancient human carriage. Proc. Natl. Acad. Sci. USA 2002, 99, 15107–15111. [Google Scholar] [CrossRef] [PubMed]
- Marini, E.; Maldonado-Contreras, A.L.; Cabras, S.; Hidalgo, G.; Buffa, R.; Marin, A.; Floris, G.; Racugno, W.; Pericchi, L.R.; Castellanos, M.E.; et al. Helicobacter pylori and intestinal parasites are not detrimental to the nutritional status of Amerindians. Am. J. Trop. Med. Hyg. 2007, 76, 534–540. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.H.; Oh, J.; Deming, C.; Conlan, S.; Grice, E.A.; Beatson, M.A.; Nomicos, E.; Polley, E.C.; Komarow, H.D.; Murray, P.R.; Turner, M.L. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012, 22, 850–859. [Google Scholar] [CrossRef] [PubMed]
- Dekio, I.; Hayashi, H.; Sakamoto, M.; Kitahara, M.; Nishikawa, T.; Suematsu, M.; Benno, Y. Detection of potentially novel bacterial components of the human skin microbiota using culture-independent molecular profiling. J. Med. Microbiol. 2005, 54, 1231–1238. [Google Scholar] [CrossRef] [PubMed]
- Bek-Thomsen, M.; Lomholt, H.B.; Kilian, M. Acne is not associated with yet-uncultured bacteria. J. Clin. Microbiol. 2008, 46, 3355–3360. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Tseng, C.; Pei, Z.; Blaser, M.J. Molecular analysis of human forearm superficial skin bacterial biota. Proc. Natl. Acad. Sci. USA 2007, 104, 2927–2932. [Google Scholar] [CrossRef] [PubMed]
- ScienceDaily. More than just Bacteria: Importance of Microbial Diversity in Gut Health, Disease. Available online: www.sciencedaily.com/releases/2014/03/140310090919.htm (accessed on 3 March 2017).
- Rowland, I.R.; Capurso, L.; Collins, K.; Cummings, J.; Delzenne, N.; Goulet, O.; Guarner, F.; Marteau, P.; Meier, R. Current level of consensus on probiotic science—Report of an expert meeting—London, 23 November 2009. Gut Microbes 2010, 1, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Grönlund, M.M.; Lehtonen, O.P.; Eerola, E.; Kero, P. Fecal microflora in healthy infants born by different methods of delivery: Permanent changes in intestinal flora after cesarean delivery. J. Pediatr. Gastroenterol. Nutr. 1999, 28, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Shono, Y.; Docampo, M.D.; Peled, J.U.; Perobelli, S.M.; Velardi, E.; Tsai, J.J.; Slingerland, A.E.; Smith, O.M.; Young, L.F.; Gupta, J.; et al. Increased GVHD-related mortality with broad-spectrum antibiotic use after allogeneic hematopoietic stem cell transplantation in human patients and mice. Sci. Transl. Med. 2016, 8, 339ra71. [Google Scholar] [CrossRef] [PubMed]
- Jernberg, C.; Lofmark, S.; Edlund, C.; Jansson, J.K. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology 2010, 156, 3216–3223. [Google Scholar] [CrossRef] [PubMed]
- Jernberg, C.; Löfmark, S.; Edlund, C.; Jansson, J.K. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 2007, 1, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Dethlefsen, L.; Huse, S.; Sogin, M.L.; Relman, D.A. The Pervasive Effects of an Antibiotic on the Human Gut Microbiota, as Revealed by Deep 16S rRNA Sequencing. PLoS Biol. 2008, 6, e280. [Google Scholar] [CrossRef] [PubMed]
- Fluhr, J.W.; Kao, J.; Ahn, S.K.; Feingold, K.R.; Elias, P.M.; Jain, M. Generation of free fatty acids from phospholipids regulates stratum corneum acidification and integrity. J. Investig. Dermatol. 2001, 117, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Pickford, M.; Senut, B. “Millennium Ancestor”, a 6-million-year-old bipedal hominid from Kenya. S. Afr. J. Sci. 2001, 97, 22. [Google Scholar]
- Kovarovic, K. Human evolution: A very short introduction. J. Hum. Evol. 2007, 52, 467. [Google Scholar] [CrossRef]
- Halliday, T.J.D.; Upchurch, P.; Goswami, A. Resolving the relationships of Paleocene placental mammals. Biol. Rev. 2017, 92, 521–550. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Hamady, M.; Lozupone, C.; Turnbaugh, P.J.; Ramey, R.R.; Bircher, J.S.; Schlegel, M.L.; Tucker, T.A.; Schrenzel, M.D.; Knight, R.; et al. Evolution of mammals and their gut microbes. Science 2008, 320, 1647–1651. [Google Scholar] [CrossRef] [PubMed]
- Goossens, A. Contact-allergic reactions to cosmetics. J. Allergy 2011, 2011, 467071. [Google Scholar] [CrossRef] [PubMed]
- Salverda, J.G.W.; Bragt, P.J.C.; de Wit-Bos, L.; Rustemeyer, T.; Coenraads, P.J.; Tupker, R.A.; Kunkeler, L.; Laheij-de Boer, A.M.; Stenveld, H.J.; van Ginkel, C.J.; et al. Results of a cosmetovigilance survey in The Netherlands. Contact Dermatitis 2013, 68, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Heisterberg, M.V.; Menné, T.; Johansen, J.D. Contact allergy to the 26 specific fragrance ingredients to be declared on cosmetic products in accordance with the EU cosmetics directive. Contact Dermatitis 2011, 65, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Warshaw, E.M.; Buchholz, H.J.; Belsito, D.V.; Maibach, H.I.; Fowler, J.F.; Rietschel, R.L.; Zug, K.A.; Mathias, C.T.; Pratt, M.D.; Sasseville, D.; et al. Allergic patch test reactions associated with cosmetics: Retrospective analysis of cross-sectional data from the North American Contact Dermatitis Group, 2001–2004. J. Am. Acad. Dermatol. 2009, 60, 23–38. [Google Scholar] [CrossRef] [PubMed]
- Berne, B.; Tammela, M.; Färm, G.; Inerot, A.; Lindberg, M. Can the reporting of adverse skin reactions to cosmetics be improved? A prospective clinical study using a structured protocol. Contact Dermatitis 2008, 58, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Berne, B.; Boström, A.; Grahnén, A.F.; Tammela, M. Adverse effects of cosmetics and toiletries reported to the Swedish Medical Products Agency 1989–1994. Contact Dermatitis 1996, 34, 359–362. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).