Impact of Phycosphere-Isolated Marine Bacteria on Nutritional Value, Growth, and Nutrient Uptake of Co-Cultured Chaetoceros calcitrans
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Identification of Phycosphere Bacteria
2.2. Qualitative Characterization of Phycosphere Bacterial Strains
2.2.1. Auxin Production
2.2.2. Siderophore Production
2.2.3. Amylase Activity and Phosphate Solubilization
2.2.4. Specific Biofilm Formation
2.2.5. Bacterial Motility
2.3. Antagonism Assay
2.4. Design of Synthetic Aquaculture Wastewater
2.5. Co-Culture of Chaetoceros Calcitrans with the Bacterial Consortium in SAW
2.6. Statistical Analysis
3. Results
3.1. Identification of Phycosphere-Isolated Bacterial Strains and Selection of the Bacterial Consortium
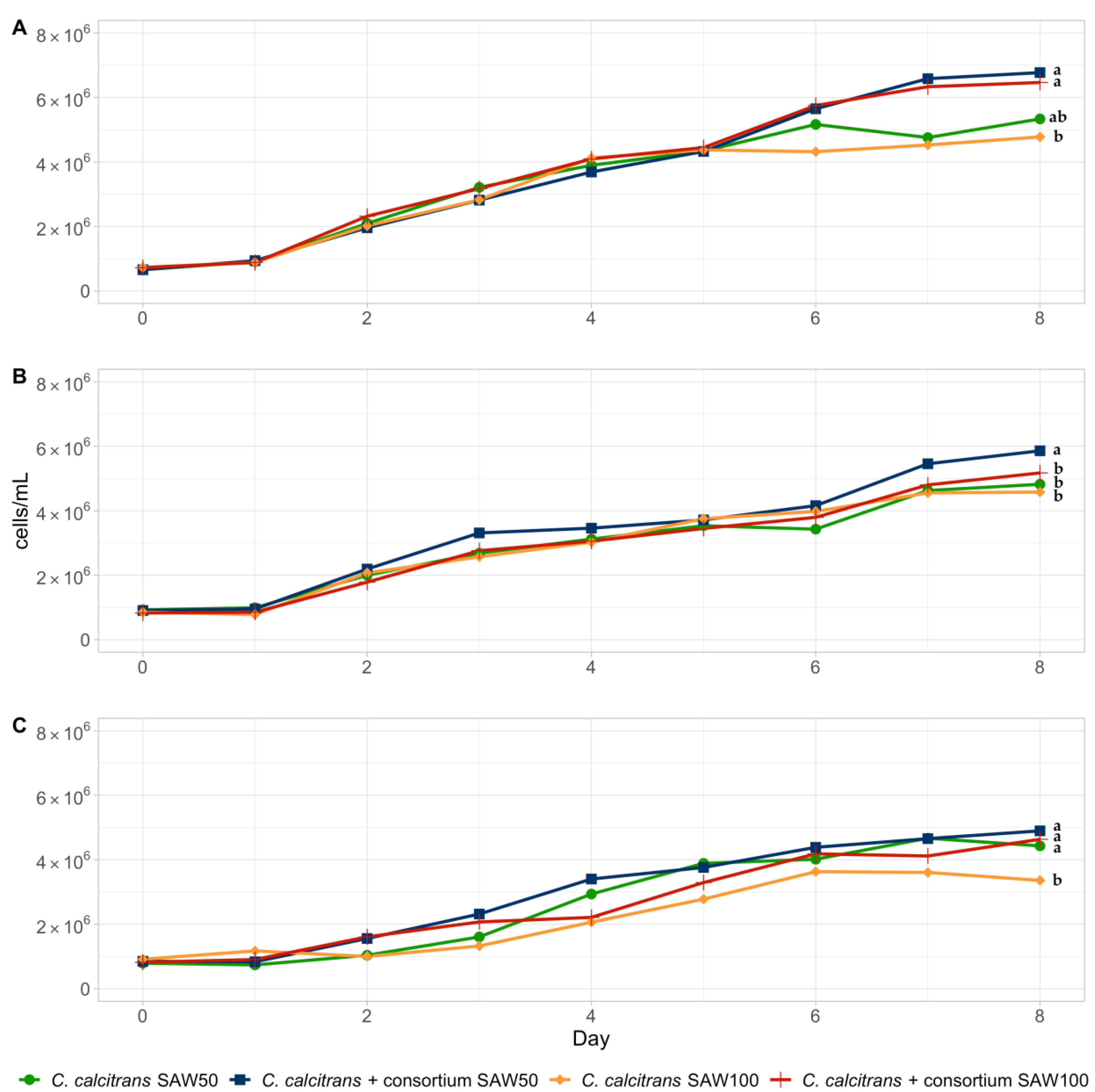
3.2. Growth of Chaetoceros Calcitrans with Bacterial Consortium in SAW
3.3. Composition of Chaetoceros Calcitrans with Bacterial Consortium in SAW
3.4. Bioremediation of Chaetoceros calcitrans with Bacterial Consortium in SAW
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ma, M.; Hu, Q. Microalgae as feed sources and feed additives for sustainable aquaculture: Prospects and challenges. Rev. Aquac. 2024, 16, 818–835. [Google Scholar] [CrossRef]
- Phang, S.M.; Chu, W.L.; Rabiei, R. Phycoremediation. Algae World 2015, 26, 357–389. [Google Scholar]
- Ansari, F.A.; Singh, P.; Guldhe, A.; Bux, F. Microalgal cultivation using aquaculture wastewater: Integrated biomass generation and nutrient remediation. Algal Res. 2017, 21, 169–177. [Google Scholar] [CrossRef]
- Guldhe, A.; Ansari, F.A.; Singh, P.; Bux, F. Heterotrophic cultivation of microalgae using aquaculture wastewater: A biorefinery concept for biomass production and nutrient remediation. Ecol. Eng. 2017, 99, 47–53. [Google Scholar] [CrossRef]
- Khatoon, H.; Banerjee, S.; Syahiran, M.S.; Noordin, N.B.M.; Bolong, A.M.A.; Endut, A. Re-use of aquaculture wastewater in cultivating microalgae as live feed for aquaculture organisms. Desalination Water Treat. 2016, 57, 29295–29302. [Google Scholar] [CrossRef]
- Kanoje, V.V.; Bhatkar, V.R.; Pawase, A.S.; Tibile, R.M.; Sawant, M.S.; Gaud, A.; Chaudhary, B. Effect of different salinities on the growth performance of Isochrysis galbana and Chaetoceros calcitrans at different growth phases. J. Exp. Zool. India 2023, 26, 1863–1869. [Google Scholar] [CrossRef]
- Tantanasarit, C.; Englande, A.J.; Babel, S. Nitrogen, phosphorus and silicon uptake kinetics by marine diatom Chaetoceros calcitrans under high nutrient concentrations. J. Exp. Mar. Biol. Ecol. 2013, 446, 67–75. [Google Scholar] [CrossRef]
- Suantika, G.; Lumbantoruan, G.; Muhammad, H.; Azizah, F.F.N.; Aditiawati, P. Performance of Zero Water Discharge (ZWD) system with nitrifying bacteria and microalgae Chaetoceros calcitrans components in super intensive white shrimp (Litopenaeus vannamei) culture. Aquac. Res. Dev. 2015, 6, 1000359. [Google Scholar]
- Seymour, J.R.; Amin, S.A.; Raina, J.B.; Stocker, R. Zooming in on the phycosphere: The ecological interface for phytoplankton–bacteria relationships. Nat. Microbiol. 2017, 2, 17065. [Google Scholar] [CrossRef]
- Palacios, O.A.; López, B.R.; de-Bashan, L.E. Microalga Growth-Promoting Bacteria (MGPB): A formal term proposed for beneficial bacteria involved in microalgal–bacterial interactions. Algal Res. 2022, 61, 102585. [Google Scholar] [CrossRef]
- Lee, C.; Jeon, M.S.; Kim, J.Y.; Lee, S.H.; Kim, D.G.; Roh, S.W.; Choi, Y.E. Effects of an auxin-producing symbiotic bacterium on cell growth of the microalga Haematococcus pluvialis: Elevation of cell density and prolongation of exponential stage. Algal Res. 2019, 41, 101547. [Google Scholar] [CrossRef]
- Pagnussat, L.A.; Maroniche, G.; Curatti, L.; Creus, C. Auxin-dependent alleviation of oxidative stress and growth promotion of Scenedesmus obliquus C1S by Azospirillum brasilense. Algal Res. 2020, 47, 101839. [Google Scholar] [CrossRef]
- Rajapitamahuni, S.; Bachani, P.; Sardar, R.K.; Mishra, S. Co-cultivation of siderophore-producing bacteria Idiomarina loihiensis RS14 with Chlorella variabilis ATCC 12198, evaluation of micro-algal growth, lipid, and protein content under iron starvation. J. Appl. Phycol. 2019, 31, 29–39. [Google Scholar] [CrossRef]
- Fallahi, A.; Rezvani, F.; Asgharnejad, H.; Nazloo, E.K.; Hajinajaf, N.; Higgins, B. Interactions of microalgae-bacteria consortia for nutrient removal from wastewater: A review. Chemosphere 2021, 272, 129878. [Google Scholar] [CrossRef] [PubMed]
- Shofiyah, S.S.; Yuliani, D.; Widya, N.; Sarian, F.D.; Puspasari, F.; Radjasa, O.K.; Natalia, D. Isolation, expression, and characterization of raw starch degrading α-amylase from a marine lake Bacillus megaterium NL3. Heliyon 2020, 6, e05796. [Google Scholar] [CrossRef]
- Ramanan, R.; Kim, B.H.; Cho, D.H.; Oh, H.M.; Kim, H.S. Algae–bacteria interactions: Evolution, ecology and emerging applications. Biotechnol. Adv. 2016, 34, 14–29. [Google Scholar] [CrossRef]
- Fei, C.; Ochsenkühn, M.A.; Shibl, A.A.; Isaac, A.; Wang, C.; Amin, S.A. Quorum sensing regulates ‘swim-or-stick’ lifestyle in the phycosphere. Environ. Microbiol. 2020, 22, 4761–4778. [Google Scholar] [CrossRef]
- Wei, X.; Shi, F.; Chen, Z.; Feng, J.; Zhu, L. Response of bacterial communities (Marivita, Marinobacter, and Oceanicaulis) in the phycosphere to the growth of Phaeodactylum tricornutum in different inorganic nitrogen sources. Front. Mar. Sci. 2023, 10, 1086166. [Google Scholar] [CrossRef]
- Wirth, R.; Pap, B.; Böjti, T.; Shetty, P.; Lakatos, G.; Bagi, Z.; Kovács, K.L.; Maróti, G. Chlorella vulgaris and its phycosphere in wastewater: Microalgae-bacteria interactions during nutrient removal. Front. Bioeng. Biotechnol. 2020, 8, 557572. [Google Scholar] [CrossRef]
- Paddock, M.B.; Fernández-Bayo, J.D.; VanderGheynst, J.S. The effect of the microalgae-bacteria microbiome on wastewater treatment and biomass production. Appl. Microbiol. Biotechnol. 2020, 104, 893–905. [Google Scholar] [CrossRef]
- ZoBell, C.E. Studies on marine bacteria. I. The cultural requirements of heterotrophic aerobes. J. Mar. Res. 1941, 4, 42–75. [Google Scholar]
- Gravel, V.; Antoun, H.; Tweddell, R.J. Effect of indole-acetic acid (IAA) on the development of symptoms caused by Pythium ultimum on tomato plants. Eur. J. Plant Pathol. 2007, 119, 457–462. [Google Scholar] [CrossRef]
- Charest, M.H.; Beauchamp, C.J.; Antoun, H. Effects of the humic substances of de-inking paper sludge on the antagonism between two compost bacteria and Pythium ultimum. FEMS Microbiol. Ecol. 2005, 52, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Apun, K.; Jong, B.C.; Salleh, M.A. Screening and isolation of a cellulolytic and amylolytic Bacillus from sago pith waste. J. Gen. Appl. Microbiol. 2000, 46, 263–267. [Google Scholar] [CrossRef][Green Version]
- Magallon-Servín, P.; Antoun, H.; Taktek, S.; Bashan, Y.; de-Bashan, L. The maize mycorrhizosphere as a source for isolation of arbuscular mycorrhizae-compatible phosphate rock-solubilizing bacteria. Plant Soil 2020, 451, 169–186. [Google Scholar] [CrossRef]
- Nautiyal, C.S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef]
- Naves, P.; Del Prado, G.; Huelves, L.; Gracia, M.; Ruiz, V.; Blanco, J.; Rodríguez-Cerrato, V.; Ponte, M.C.; Soriano, F. Measurement of biofilm formation by clinical isolates of Escherichia coli is method-dependent. J. Appl. Microbiol. 2008, 105, 585–590. [Google Scholar] [CrossRef]
- Navazo, A.; Barahona, E.; Redondo-Nieto, M.; Martínez-Granero, F.; Rivilla, R.; Martín, M. Three independent signaling pathways repress motility in Pseudomonas fluorescens F113. Microb. Biotechnol. 2009, 2, 489–498. [Google Scholar] [CrossRef]
- de Alva, M.S.; Pabello, V.M.L. Phycoremediation by simulating marine aquaculture effluent using Tetraselmis sp. and the potential use of the resulting biomass. J. Water Process Eng. 2021, 41, 102071. [Google Scholar] [CrossRef]
- Sheets, J.P.; Ge, X.; Park, S.Y.; Li, Y. Effect of outdoor conditions on Nannochloropsis salina cultivation in artificial seawater using nutrients from anaerobic digestion effluent. Bioresour. Technol. 2014, 152, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Guillard, R.R. Culture of phytoplankton for feeding marine invertebrates. In Culture of Marine Invertebrate Animals; Smith, W.L., Chanley, M.H., Eds.; Springer: Boston, MA, USA, 1975; pp. 29–60. [Google Scholar]
- Guillard, R.R.; Ryther, J.H. Studies of marine planktonic diatoms: I. Cyclotella nana Hustedt, and Detonula confervacea (Cleve) Gran. Can. J. Microbiol. 1962, 8, 229–239. [Google Scholar] [CrossRef]
- Arredondo-Vega, B.O.; Voltolina, D.; Zenteno-Savín, T.; Arce-Montoya, M.; Gómez-Anduro, G.A. Métodos y Herramientas Analíticas en la Evaluación de la Biomasa Microalgal, 2nd ed.; Centro de Investigaciones Biológicas: La Paz, Mexico; pp. 1–150.
- Leyva, L.A.; Bashan, Y.; de-Bashan, L.E. Activity of acetyl-CoA carboxylase is not directly linked to accumulation of lipids when Chlorella vulgaris is co-immobilized with Azospirillum brasilense in alginate under autotrophic and heterotrophic conditions. Ann. Microbiol. 2015, 65, 339–349. [Google Scholar] [CrossRef]
- Choix, F.J.; De-Bashan, L.E.; Bashan, Y. Enhanced accumulation of starch and total carbohydrates in alginate-immobilized Chlorella spp. induced by Azospirillum brasilense: II. Heterotrophic conditions. Enzym. Microb. Technol. 2012, 51, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Amavizca, E.; Bashan, Y.; Ryu, C.M.; Farag, M.A.; Bebout, B.M.; de-Bashan, L.E. Enhanced performance of the microalga Chlorella sorokiniana remotely induced by the plant growth-promoting bacteria Azospirillum brasilense and Bacillus pumilus. Sci. Rep. 2017, 7, 41310. [Google Scholar] [CrossRef]
- García-Robledo, E.; Corzo, A.; Papaspyrou, S. A fast and direct spectrophotometric method for the sequential determination of nitrate and nitrite at low concentrations in small volumes. Mar. Chem. 2014, 162, 30–36. [Google Scholar] [CrossRef]
- Fuentes, J.L.; Garbayo, I.; Cuaresma, M.; Montero, Z.; González-del-Valle, M.; Vílchez, C. Impact of microalgae-bacteria interactions on the production of algal biomass and associated compounds. Mar. Drugs 2016, 14, 100. [Google Scholar] [CrossRef]
- Rios Castro, N.; Zavala, J.; Gil-Kodaka, P.; Diringer, B. Evaluation of bacterial strains to improve the productivity of microalgae used in bivalve hatcheries in Peru. J. World Aquac. Soc. 2022, 53, 95–105. [Google Scholar] [CrossRef]
- Cao, J.Y.; Wang, Y.Y.; Wu, M.N.; Kong, Z.Y.; Lin, J.H.; Ling, T.; Xu, S.M.; Ma, S.N.; Zhang, L.; Zhou, C.X.; et al. RNA-seq Insights into the impact of Alteromonas macleodii on Isochrysis galbana. Front. Microbiol. 2021, 12, 711998. [Google Scholar] [CrossRef]
- Lakshmikandan, M.; Wang, S.; Murugesan, A.G.; Saravanakumar, M.; Selvakumar, G. Co-cultivation of Streptomyces and microalgal cells as an efficient system for biodiesel production and bioflocculation formation. Bioresour. Technol. 2021, 332, 125118. [Google Scholar] [CrossRef]
- Hatha, A.A.M.; Mujeeb Rahiman, K.M.; Jasmine, B.; Suresh Kumar, S. Growth enhancement of microalgae, Chaetoceros calcitrans, and Nannochloropsis oculata, using selected bacterial strains. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 352–359. [Google Scholar]
- Pozzobon, V.; Cui, N.; Moreaud, A.; Michiels, E.; Levasseur, W. Nitrate and nitrite as mixed source of nitrogen for Chlorella vulgaris: Growth, nitrogen uptake and pigment contents. Bioresour. Technol. 2021, 330, 124995. [Google Scholar] [CrossRef]
- Aparicio, S.; Robles, Á.; Ferrer, J.; Seco, A.; Falomir, L.B. Assessing and modeling nitrite inhibition in microalgae-bacteria consortia for wastewater treatment by means of photo-respirometric and chlorophyll fluorescence techniques. Sci. Total Environ. 2022, 808, 152128. [Google Scholar] [CrossRef]
- Su, R.; Zhou, L.; Ding, L.; Fu, B.; Fu, H.; Shuang, Y.; Ye, L.; Hu, H.; Ma, H.; Ren, H. How anaerobic sludge microbiome responds to different concentrations of nitrite, nitrate, and ammonium ions: A comparative analysis. Environ. Sci. Pollut. Res. 2023, 30, 49026–49037. [Google Scholar] [CrossRef] [PubMed]
- Palacios, O.A.; León-Vega, R.A.; López, B.R.; de-Bashan, L.E.; Choix, F.J.; Cuevas-Rodríguez, G. Microalga-bacteria interaction mitigates adverse effects on microalga produced by ZnO nanoparticles. Algal Res. 2023, 74, 103198. [Google Scholar] [CrossRef]
- González-Vega, R.I.; Cárdenas-López, J.L.; López-Elías, J.A.; Ruiz-Cruz, S.; Reyes-Díaz, A.; Perez-Perez, L.M.; Cinco-Moroyoqui, F.J.; Robles-Zepeda, R.E.; Borboa-Flores, J.; Del-Toro-Sánchez, C.L. Optimization of growing conditions for pigments production from microalga Navicula incerta using response surface methodology and its antioxidant capacity. Saudi J. Biol. Sci. 2021, 28, 1401–1416. [Google Scholar] [CrossRef] [PubMed]
- Suparmaniam, U.; Lam, M.K.; Lim, J.W.; Yusup, S.; Tan, I.S.; Lau, S.Y.; Kodgire, P.; Kachhwaha, S.S. Influence of environmental stress on microalgae growth and lipid profile: A systematic review. Phytochem. Rev. 2023, 22, 879–901. [Google Scholar] [CrossRef]
- Xue, L.; Shang, H.; Ma, P.; Wang, X.; He, X.; Niu, J.; Wu, J. Analysis of growth and lipid production characteristics of Chlorella vulgaris in artificially constructed consortia with symbiotic bacteria. J. Basic Microbiol. 2018, 58, 358–367. [Google Scholar] [CrossRef]
- Chen, H.; Wang, Q. Microalgae-based nitrogen bioremediation. Algal Res. 2020, 46, 101775. [Google Scholar] [CrossRef]
- Rahayu, S.M.; Damar, A.; Krisanti, M. Variation of nitrate concentration and light level on growth rate of diatom Chaetoceros muelleri. Acta Aquat. Aquat. Sci. J. 2022, 9, 95–100. [Google Scholar] [CrossRef]
- Jia, H.; Yuan, Q. Removal of nitrogen from wastewater using microalgae and microalgae–bacteria consortia. Cogent Environ. Sci. 2016, 2, 1275089. [Google Scholar] [CrossRef]
- Kunhikrishnan, A.; Rahman, M.A.; Lamb, D.; Bolan, N.S.; Saggar, S.; Surapaneni, A.; Chen, C. Rare earth elements (REE) for the removal and recovery of phosphorus: A review. Chemosphere 2022, 286, 131661. [Google Scholar] [CrossRef] [PubMed]
- Carrillo, V.; Fuentes, B.; Gómez, G.; Vidal, G. Characterization and recovery of phosphorus from wastewater by combined technologies. Rev. Environ. Sci. Bio/Technol. 2020, 19, 389–418. [Google Scholar] [CrossRef]
- Dyhrman, S.T. Nutrients and their acquisition: Phosphorus physiology in microalgae. In The Physiology of Microalgae; Borowitzka, M.A., Beardall, J., Raven, J.A., Eds.; Springer: Cham, Switzerland, 2016; pp. 155–183. [Google Scholar]
- Acevedo, S.; Peñuela, G.A.; Pino, N.J. Biomass production of Scenedesmus sp. and removal of nitrogen and phosphorus in domestic wastewater. Ing. Y Compet. 2017, 19, 185–193. [Google Scholar]
- Solovchenko, A.; Verschoor, A.M.; Jablonowski, N.D.; Nedbal, L. Phosphorus from wastewater to crops: An alternative path involving microalgae. Biotechnol. Adv. 2016, 34, 550–564. [Google Scholar] [CrossRef] [PubMed]
| SAWna | SAWni | SAWpho | ||||
|---|---|---|---|---|---|---|
| SAWna50 | SAWna100 | SAWni50 | SAWni100 | SAWpho50 | SAWpho100 | |
| NaNO3 | 303.41 | 606.81 | / | / | 37.50 | 37.50 |
| NaNO2 | / | / | 246.29 | 492.58 | / | / |
| NaH2PO4 H2O | 1.25 | 1.25 | 1.25 | 1.25 | 222.72 | 445.43 |
| NaSiO3 9H2O | 15.00 | 15.00 | 15.00 | 15.00 | 15.00 | 15.00 |
| Metals | 3.90 | 3.90 | 3.90 | 3.90 | 3.90 | 3.90 |
| Vitamins | 3.34 × 10−5 | 3.34 × 10−5 | 3.34 × 10−5 | 3.34 × 10−5 | 3.34 × 10−5 | 3.34 × 10−5 |
| Instant Ocean | 35.79 | 35.79 | 35.79 | 35.79 | 35.79 | 35.79 |
| Isolate ID | Microalgae | Gram Staining | Morphological Characteristics | Closest Match Strain NCBI | Accession Number | Sequence Identity (%) |
|---|---|---|---|---|---|---|
| 40pB | I. galbana | Gram-negative | Rod-shaped | Marinobacter sp. | CP014754.1 | 99.54 |
| 40pA | I. galbana | Gram-negative | Rod-shaped | Marinobacter sp. | CP014754.1 | 100.00 |
| 40g1 | I. galbana | Gram-positive | Round-shaped | Bacillus cereus | CP053997.1 | 99.52 |
| 38 | I. galbana | Gram-negative | Rod-shaped | Marinobacter alkaliphilus | CP115811.1 | 98.84 |
| 37r | I. galbana | Gram-negative | Rod-shaped | Cetobacterium somerae | MN646996.1 | 99.43 |
| 28 | C. calcitrans | Gram-negative | Rod-shaped | Alteromonas macleodii | CP098772.1 | 99.35 |
| 29 | C. calcitrans | Gram-negative | Rod-shaped | Algoriphagus sp. | CP048415.1 | 100.00 |
| Experiments | Control | Strains | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 27a | 28 | 29 | 37b | 37r | 38 | 40g1 | 40pA | 40pB | 41 | 42a | 42b | ||
| Auxin production (μg/mL) | 0.00 ± 0.09 f | 0.00 ± 0.25 f | 0.00 ± 0.40 f | 2.43 ± 0.28 c | 0.00 ± 0.34 f | 1.61 ± 0.48 d | 0.00 ± 0.35 f | 0.00 ± 0.26 f | 5.03 ± 0.38 a | 4.85 ± 0.29 a | 0.15 ± 0.79 e | 4.17 ± 0.46 b | 0.00 ± 0.45 f |
| Siderophore production (%) | 0.00 ± 4.71 g | 6.23 ± 1.64 cd | 10.44 ± 0.46 b | 5.56 ± 1.15 d | 1.10 ± 1.63 f | 10.26 ± 1.70 ab | 9.40 ± 1.28 b | 10.60 ± 1.56 ab | 7.29 ± 0.46 c | 12.54 ± 0.97 a | 11.06 ± 2.08 ab | 2.36 ± 1.89 ef | 3.11 ± 0.18 e |
| Phosphorus solubilization test (cm) | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 1.00 ± 0.00 a | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b |
| Amylolytic activity (cm) | 0.00 ± 0.00 h | 1.85 ± 0.07 d | 2.55 ± 0.07 ab | 0.1 ± 0.00 g | 0.10 ± 0.00 g | 0.10 ± 0.00 g | 2.50 ± 0.00 b | 1.90 ± 0.00 de | 2.50 ± 0.00 b | 2.60 ± 0.00 a | 0.50 ± 0.00 f | 2.50 ± 0.00 b | 2.40 ± 0.14 bc |
| Biofilm formation test (SBF) | 0.00 ± 0.00 b | 5.87 ± 0.52 e | 10.96 ± 0.89 b | 1.37 ± 0.02 h | 1.66 ± 0.14 h | 2.63 ± 0.12 gh | 4.74 ± 0.23 f | 9.16 ± 0.14 c | 18.81 ± 0.67 a | 9.86 ± 0.95 bc | 3.19 ± 0.23 g | 2.59 ± 0.09 gh | 7.37 ± 0.04 d |
| Swimming motility test (cm) | 0.00 ± 0.00 i | 0.75 ± 0.07 fg | 5.65 ± 0.21 b | 0.35 ± 0.07 gh | 0.20 ± 0.00 h | 0.35 ± 0.07 gh | 1.15 ± 0.07 ef | 8.50 ± 0.00 a | 2.30 ± 0.14 c | 1.85 ± 0.21 d | 0.90 ± 0.00 ef | 1.20 ± 0.00 e | 2.00 ± 0.00 cd |
| Antagonism test | / | ++ | ++ | ++ | ++ | ++ | ++ | ++ | + | + | + | + | + |
| Content | C. calcitrans | C. calcitrans | C. calcitrans + consortium | |
|---|---|---|---|---|
| Day 0 | Day 8 | |||
| SAWna50 | Lipids (μg/mL) | 0.00 ± 0.00 b | 304.74 ± 52.98 a | 239.03 ± 61.52 a |
| Carbohydrates (μg/mL) | 0.00 ± 0.00 b | 4.96 ± 0.54 a | 4.87 ± 0.24 a | |
| Chlorophyll a (μg/mL) | 80.09 ± 5.27 c | 170.99 ± 9.52 b | 305.83 ± 9.44 a | |
| Chlorophyll b (μg/mL) | 164.72 ± 4.95 c | 189.83 ± 9.38 b | 270.87 ± 29.63 a | |
| Carotenoids (mg/mL) | 1.04 ± 0.09 c | 2.19 ± 0.18 b | 3.64 ± 0.39 a | |
| SAWna100 | Lipids (μg/mL) | 0.00 ± 0.00 b | 231.65 ± 32.31 a | 313.35 ± 63.98 a |
| Carbohydrates (μg/mL) | 0.00 ± 0.00 b | 5.16 ± 1.02 a | 5.34 ± 0.91 a | |
| Chlorophyll a (μg/mL) | 80.09 ± 5.27 c | 170.67 ± 13.74 b | 297.04 ± 18.50 a | |
| Chlorophyll b (μg/mL) | 164.72 ± 4.95 c | 185.99 ± 6.63 b | 278.57 ± 25.53 a | |
| Carotenoids (mg/mL) | 1.04 ± 0.09 c | 2.23 ± 0.21 b | 3.66 ± 0.19 a | |
| SAWni50 | Lipids (μg/mL) | 0.00 ± 0.00 c | 304.74 ± 52.81 b | 840.10 ± 69.01 a |
| Carbohydrates (μg/mL) | 0.00 ± 0.00 c | 5.35 ± 0.65 a | 2.51 ± 1.08 b | |
| Chlorophyll a (μg/mL) | 94.07 ± 9.91 c | 134.09 ± 12.81 b | 186.50 ± 20.06 a | |
| Chlorophyll b (μg/mL) | 185.70 ± 27.06 a | 184.01 ± 20.65 a | 211.08 ± 13.00 a | |
| Carotenoids (mg/mL) | 1.28 ± 0.20 c | 1.68 ± 0.12 b | 2.39 ± 0.23 a | |
| SAWni100 | Lipids (μg/mL) | 0.00 ± 0.00 b | 233.70 ± 49.93 a | 334.43 ± 26.63 a |
| Carbohydrates (μg/mL) | 0.00 ± 0.00 b | 3.55 ± 0.94 a | 3.25 ± 0.38 a | |
| Chlorophyll a (μg/mL) | 94.07 ± 9.91 c | 135.71 ± 2.50 b | 197.07 ± 10.50 a | |
| Chlorophyll b (μg/mL) | 185.70 ± 27.06 a | 185.70 ± 12.33 a | 211.27 ± 16.54 a | |
| Carotenoids (mg/mL) | 1.28 ± 0.20 c | 1.61 ± 0.16 b | 2.42 ± 0.25 a | |
| SAWpho50 | Lipids (μg/mL) | 0.00 ± 0.00 c | 172.20 ± 8.70 b | 315.57 ± 55.46 a |
| Carbohydrates (μg/mL) | 0.00 ± 0.00 b | 1.56 ± 0.64 a | 2.54 ± 0.55 a | |
| Chlorophyll a (μg/mL) | 102.51 ± 21.50 c | 139.77 ± 12.12 b | 198.34 ± 15.93 a | |
| Chlorophyll b (μg/mL) | 218.58 ± 24.26 b | 233.13 ± 22.64 b | 317.96 ± 31.10 a | |
| Carotenoids (mg/mL) | 1.47 ± 0.26 c | 2.79 ± 0.22 b | 4.04 ± 0.25 a | |
| SAWpho100 | Lipids (μg/mL) | 0.00 ± 0.00 b | 268.96 ± 63.20 a | 328.96 ± 63.73 a |
| Carbohydrates (μg/mL) | 0.00 ± 0.00 b | 0.44 ± 0.58 a | 1.11 ± 0.47 a | |
| Chlorophyll a (μg/mL) | 102.51 ± 21.50 b | 110.64 ± 1.65 b | 135.01 ± 14.46 a | |
| Chlorophyll b (μg/mL) | 218.58 ± 24.26 b | 202.38 ± 7.61 b | 240.78 ± 24.78 a | |
| Carotenoids (mg/mL) | 1.47 ± 0.26 c | 2.06 ± 0.09 b | 2.69 ± 0.28 a | |
| Treatments | Initial Content (mg/L) | Final Content (mg/L) | Removal (%) | |
|---|---|---|---|---|
| SAWna | 50 | 220.92 ± 26.13 a | 199.74 ± 8.69 a | 9.58 |
| 100 | 456.77 ± 13.51 a | 402.91 ± 16.88 b | 11.79 | |
| SAWna + consortium | 50 | 220.92 ± 26.13 a | 226.43 ± 37.18 a | / |
| 100 | 456.77 ± 13.51 a | 424.00 ± 53.73 a | 7.17 | |
| SAWni | 50 | 199.72 ± 3.50 a | 180.39 ± 10.85 b | 9.68 |
| 100 | 288.54 ± 35.00 a | 300.40 ± 28.51 a | / | |
| SAWni + consortium | 50 | 199.72 ± 3.50 a | 179.66 ± 11.17 b | 10.04 |
| 100 | 288.54 ± 35.00 a | 308.54 ± 14.71 a | / | |
| SAWpho | 50 | 51.16 ± 2.87 a | 17.32 ± 1.97 b | 66.14 |
| 100 | 94.06 ± 8.47 a | 67.06 ± 6.22 b | 28.70 | |
| SAWpho + consortium | 50 | 51.16 ± 2.87 a | 15.79 ± 1.03 b | 69.13 |
| 100 | 94.06 ± 8.47 a | 64.48 ± 4.93 b | 31.44 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonçalves, M.A.L.; López-Vela, M.; Palacios-Espinosa, A.; Romero-Bastidas, M.; Rojas-Contreras, M.; Magallón-Servín, P. Impact of Phycosphere-Isolated Marine Bacteria on Nutritional Value, Growth, and Nutrient Uptake of Co-Cultured Chaetoceros calcitrans. Resources 2024, 13, 116. https://doi.org/10.3390/resources13090116
Gonçalves MAL, López-Vela M, Palacios-Espinosa A, Romero-Bastidas M, Rojas-Contreras M, Magallón-Servín P. Impact of Phycosphere-Isolated Marine Bacteria on Nutritional Value, Growth, and Nutrient Uptake of Co-Cultured Chaetoceros calcitrans. Resources. 2024; 13(9):116. https://doi.org/10.3390/resources13090116
Chicago/Turabian StyleGonçalves, Mélissa Angeline Liberia, Melissa López-Vela, Alejandro Palacios-Espinosa, Mirella Romero-Bastidas, Maurilia Rojas-Contreras, and Paola Magallón-Servín. 2024. "Impact of Phycosphere-Isolated Marine Bacteria on Nutritional Value, Growth, and Nutrient Uptake of Co-Cultured Chaetoceros calcitrans" Resources 13, no. 9: 116. https://doi.org/10.3390/resources13090116
APA StyleGonçalves, M. A. L., López-Vela, M., Palacios-Espinosa, A., Romero-Bastidas, M., Rojas-Contreras, M., & Magallón-Servín, P. (2024). Impact of Phycosphere-Isolated Marine Bacteria on Nutritional Value, Growth, and Nutrient Uptake of Co-Cultured Chaetoceros calcitrans. Resources, 13(9), 116. https://doi.org/10.3390/resources13090116





