Characterization of Insulin-Like Growth Factor Binding Protein-5 (IGFBP-5) Gene and Its Potential Roles in Ontogenesis in the Pacific Abalone, Haliotis discus hannai
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals and Sample Collection
2.2. RNA Isolation and cDNA Synthesis
2.3. Cloning of IGFBP-5 from Pacific Abalone
2.4. Sequence Analysis
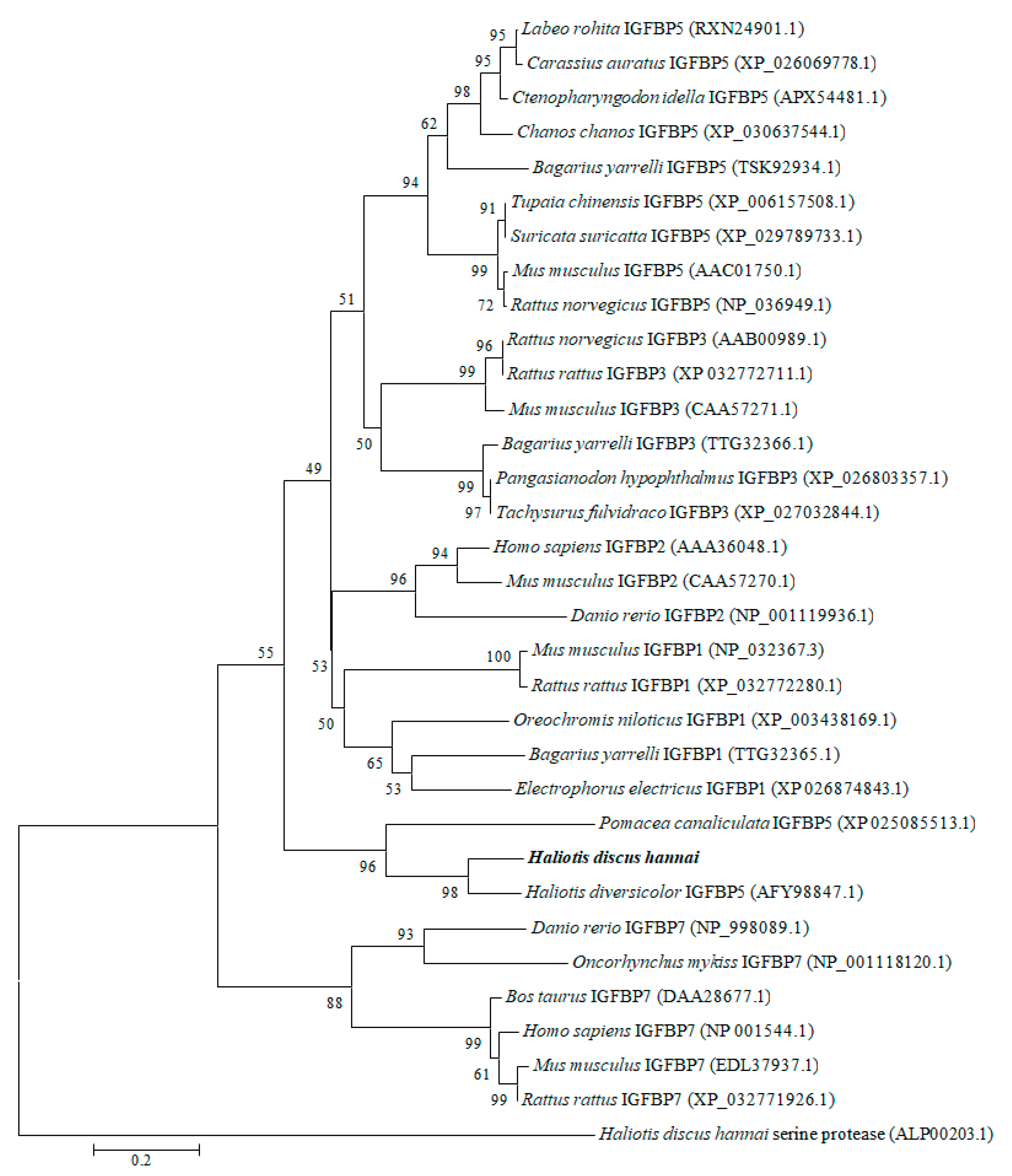
2.5. Phylogenetic Analysis
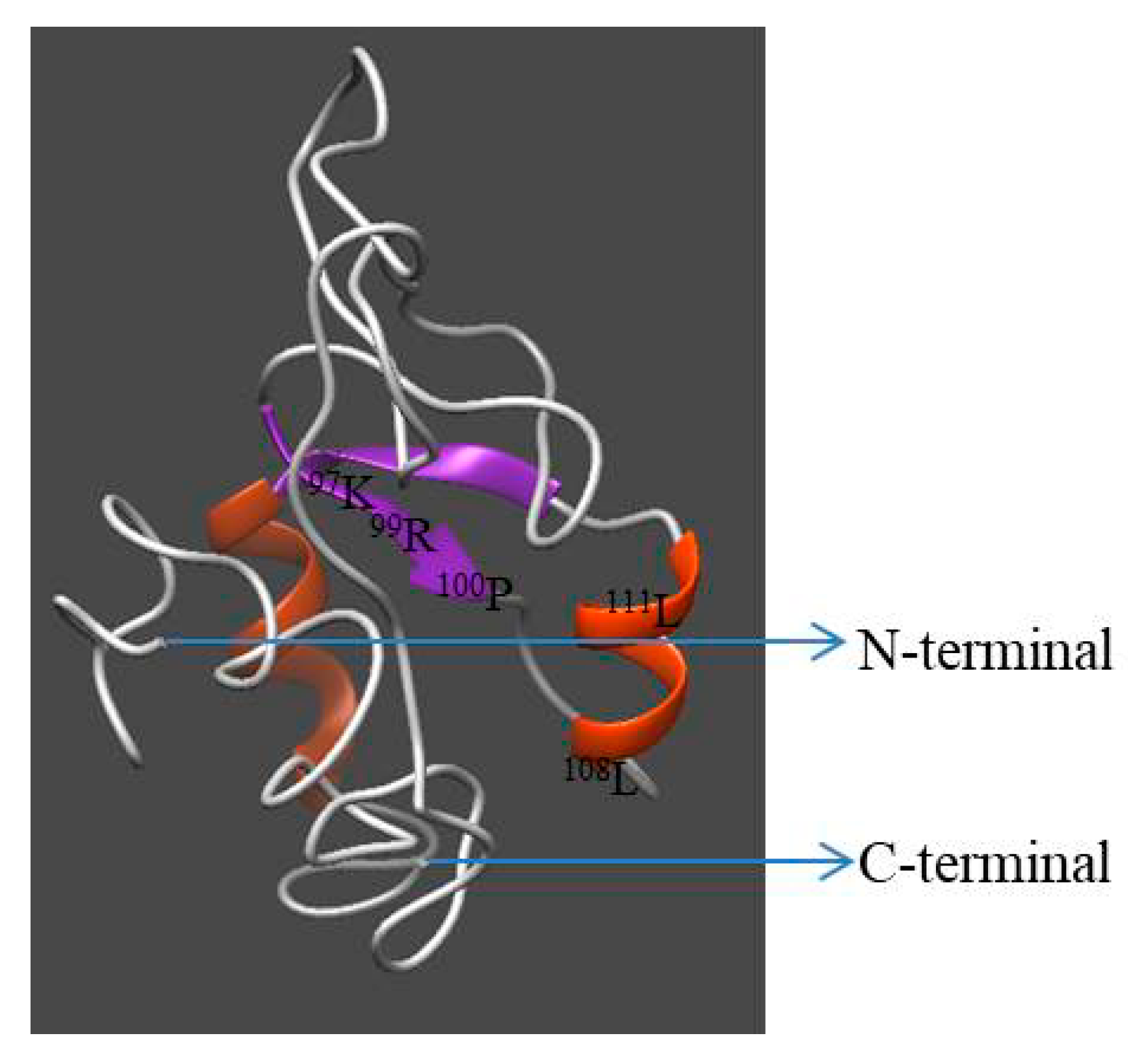
2.6. Homology Modeling of IGFBP-5 in Pacific Abalone
2.7. Semi-Quantitative Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
2.8. Quantitative Real-Time PCR (qRT-PCR) Analysis
2.9. In Situ Hybridization (ISH)
2.10. Nuclear Fast Red Counterstain
2.11. Statistical Analysis
3. Results
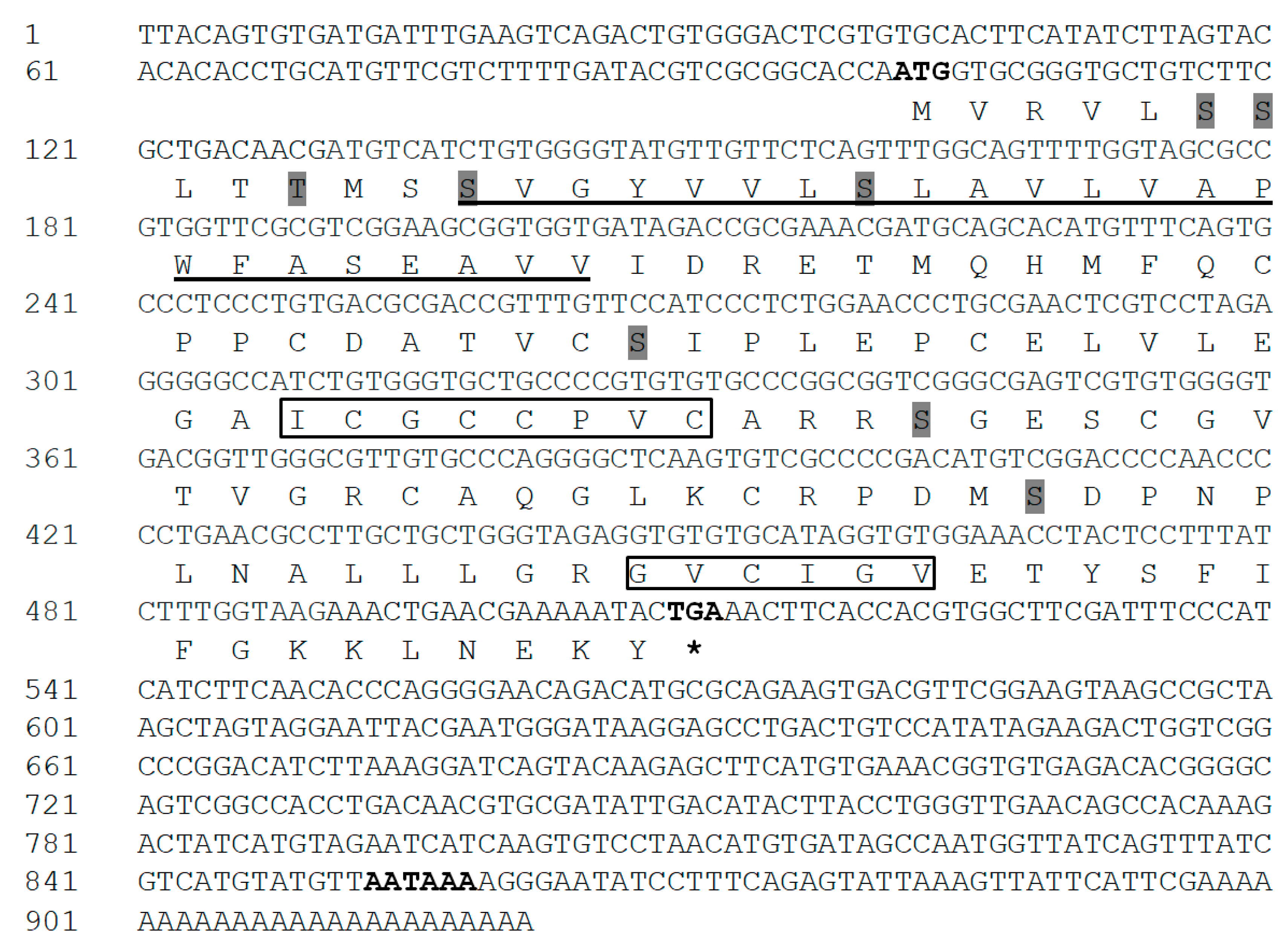
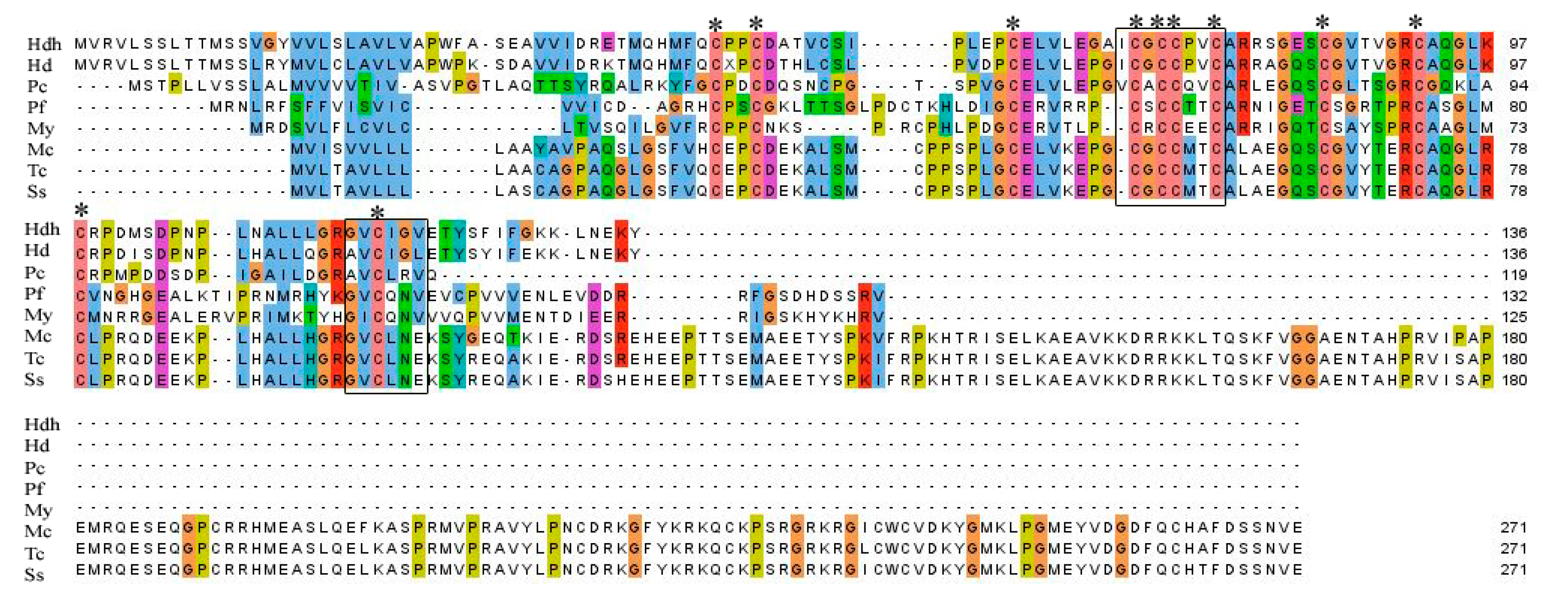
3.1. Cloning and Characterization of IGFBP-5
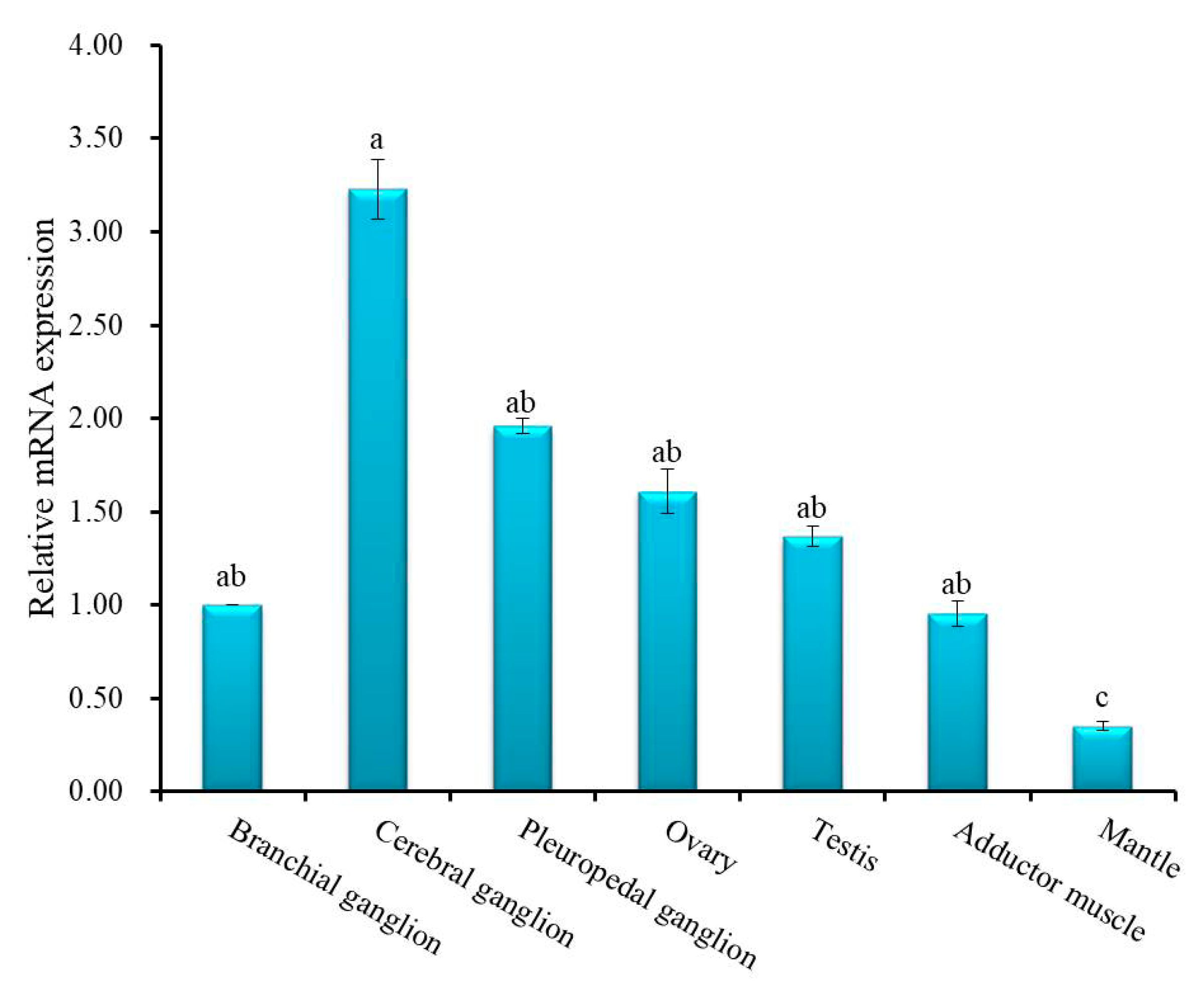
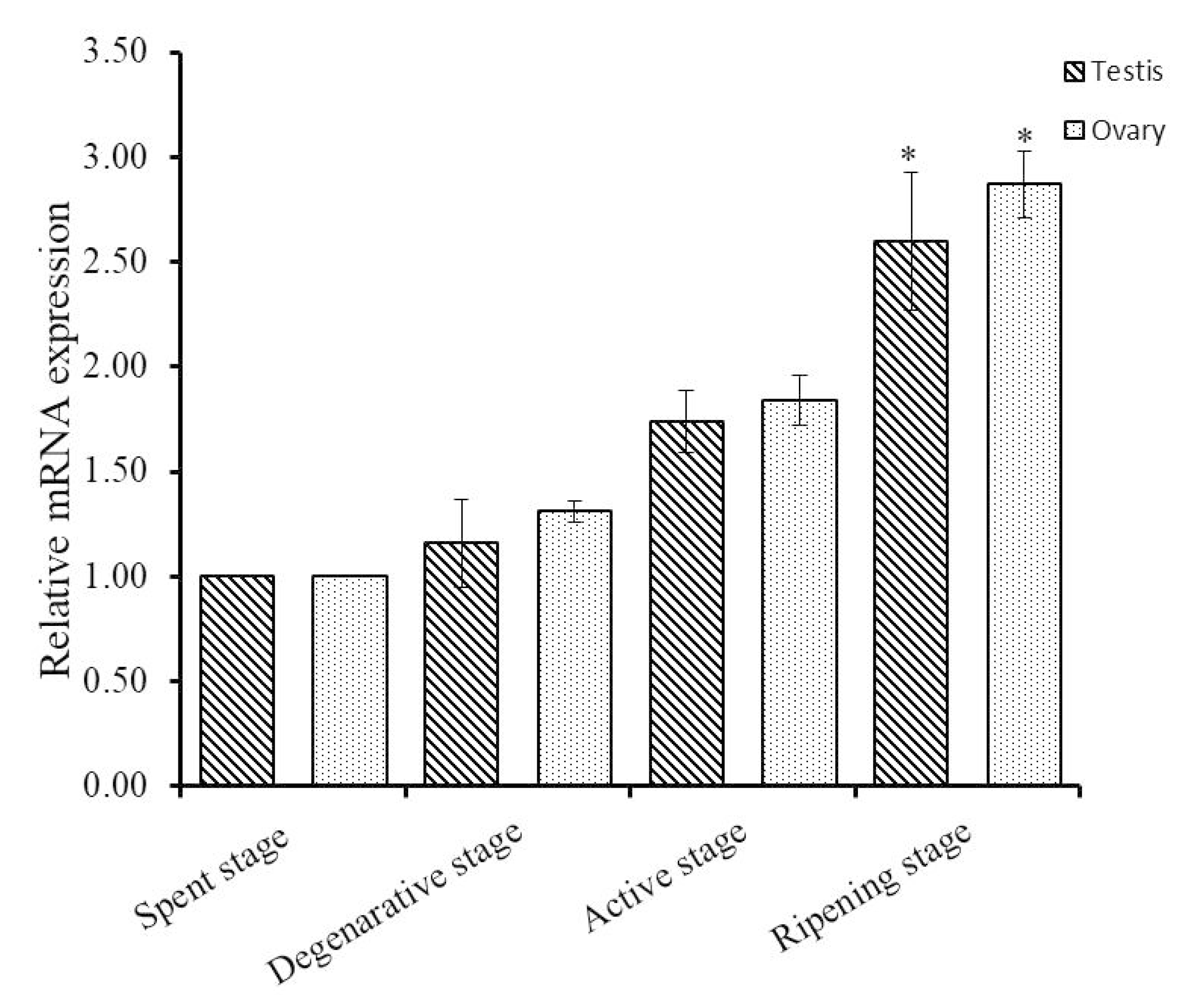
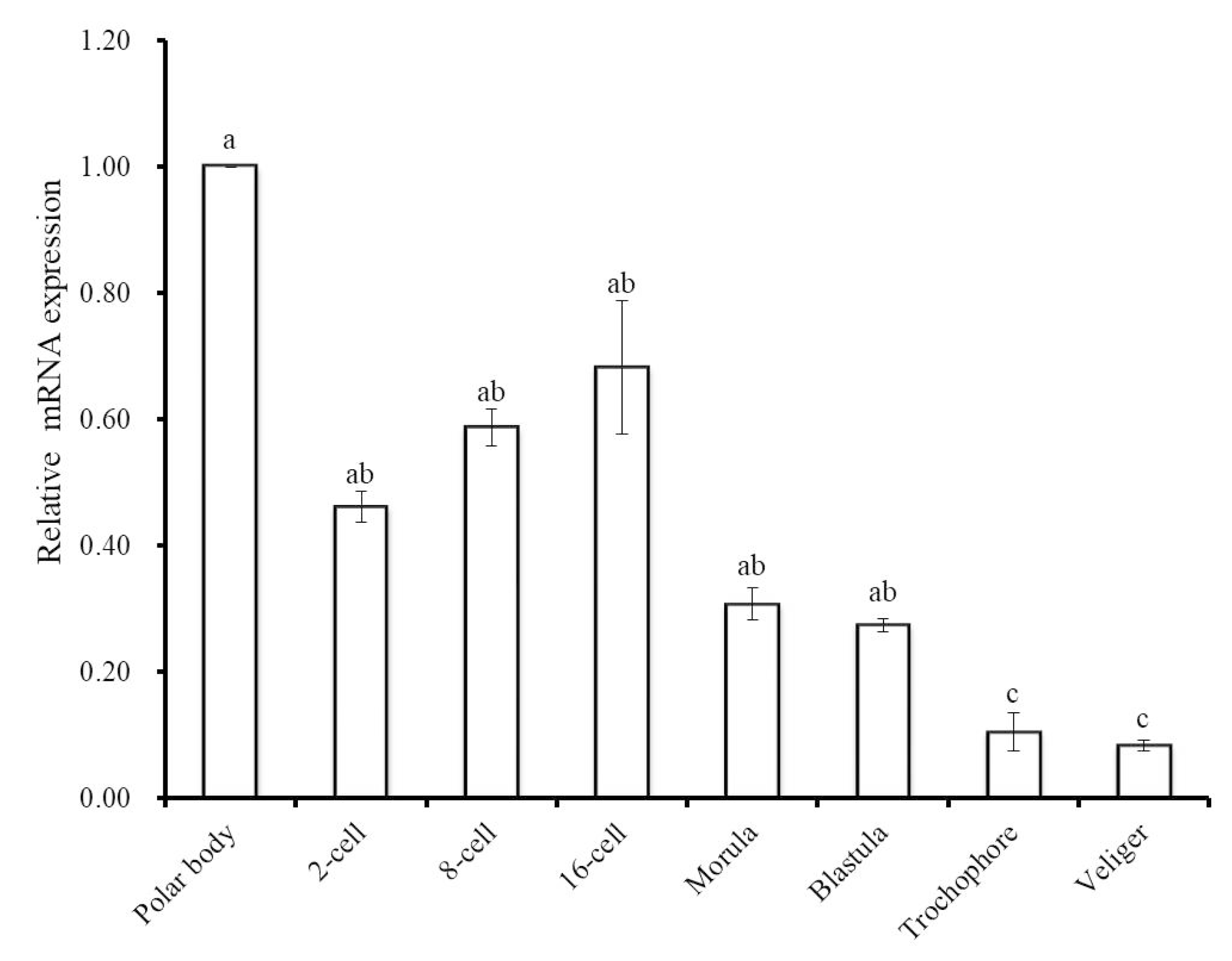
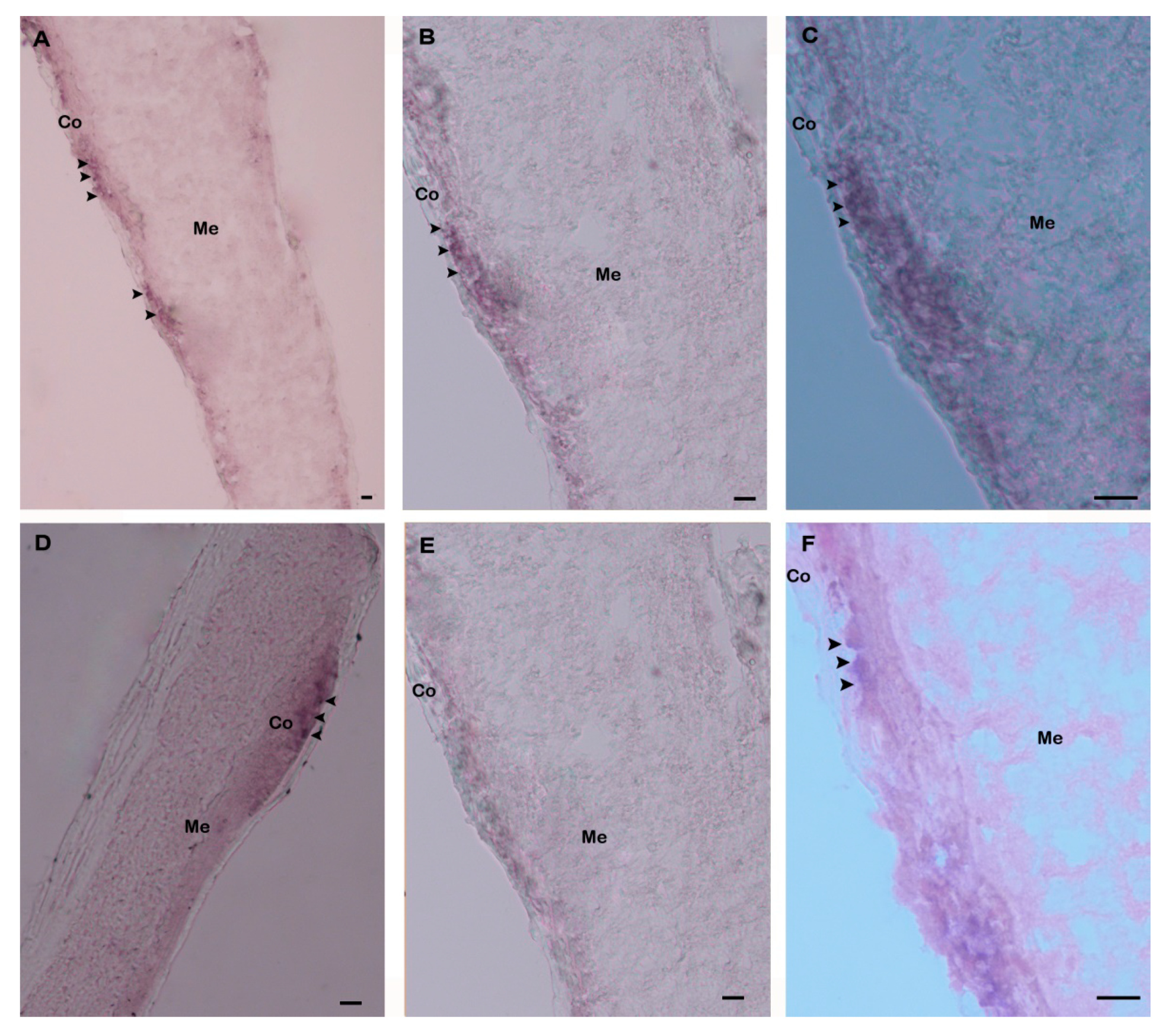
3.2. Expression Analysis of Hdh IGFBP-5
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rentería, M.; Gandhi, N.; Vinuesa, P.; Helmerhorst, E.; Mancera, R. A comparative structural bioinformatics analysis of the insulin receptor family ectodomain based on phylogenetic information. PLoS ONE 2008, 3, e3667. [Google Scholar] [CrossRef]
- Schlueter, P.J.; Peng, G.; Westerfield, M.; Duan, C. Insulin-like growth factor signaling regulates zebrafish embryonic growth and development by promoting cell survival and cell cycle progression. Cell Death Differ. 2007, 14, 1095–1105. [Google Scholar] [CrossRef]
- LeRoith, D. Editorial: Insulin-like growth factor I receptor signaling—Overlapping or redundant pathways? Endocrinology 2000, 141, 1287–1288. [Google Scholar] [CrossRef]
- Nakae, J.; Kido, Y.; Accili, D. Distinct and overlapping functions of insulin and IGF-I receptors. Endocr. Rev. 2001, 22, 818–835. [Google Scholar] [CrossRef] [PubMed]
- Wood, A.W.; Duan, C.; Bern, H.A. Insulin-like growth factor signaling in fish. Int. Rev. Cytol. 2005, 243, 215–285. [Google Scholar] [PubMed]
- Brogiolo, W.; Stocker, H.; Ikeya, T.; Rintelen, F.; Fernandez, R.; Hafen, E. An evolutionarily conserved function of the drosophila insulin receptor and insulin-like peptides in growth control. Curr. Biol. 2001, 11, 213–221. [Google Scholar] [CrossRef]
- Tatar, M.; Bartke, A.; Antebi, A. The endocrine regulation of aging by insulin-like signals. Science 2003, 299, 1346–1351. [Google Scholar] [CrossRef]
- Hwa, V.; Oh, Y.; Rosenfeld, R.G. The insulin-like growth factor-binding protein (IGFBP) Superfamily. Endocr. Rev. 1999, 20, 761–787. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Li, X.; Yu, Q.; Ning, X.; Dou, J.; Zou, J.; Zhang, L.; Wang, S.; Hu, X.; Bao, Z. A scallop IGF binding protein gene: Molecular characterization and association of variants with growth traits. PLoS ONE 2014, 9, e89039. [Google Scholar] [CrossRef]
- Duan, C.; Xu, Q. Roles of insulin-like growth factor (IGF) binding proteins in regulating IGF actions. Gen. Comp. Endocrinol. 2005, 142, 44–52. [Google Scholar] [CrossRef]
- Denley, A.; Cosgrove, L.J.; Booker, G.W.; Wallace, J.C.; Forbes, B.E. Molecular interactions of the IGF system. Cytokine Growth Factor Rev. 2005, 16, 421–439. [Google Scholar] [CrossRef] [PubMed]
- Clemmons, D.R. IGF binding proteins and their functions. Mol. Reprod. Dev. 1993, 35, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Duan, C. Specifying the cellular responses to IGF signals: Roles of IGF-binding proteins. J. Endocrinol. 2002, 175, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Allard, J.B.; Duan, C. IGF-binding proteins: Why do they exist and why are there so many? Front. Endocrinol. (Lausanne) 2018, 9, 117. [Google Scholar] [CrossRef]
- Wanscher, A.S.M.; Williamson, M.; Ebersole, T.W.; Streicher, W.; Wikström, M.; Cazzamali, G. Production of functional human insulin-like growth factor binding proteins (IGFBPs) using recombinant expression in HEK293 cells. Protein Expr. Purif. 2015, 108, 97–105. [Google Scholar] [CrossRef]
- Breves, J.P.; Fujimoto, C.K.; Phipps-Costin, S.K.; Einarsdottir, I.E.; Björnsson, B.T.; McCormick, S.D. Variation in branchial expression among insulin-like growth-factor binding proteins (igfbps) during Atlantic salmon smoltification and seawater exposure. BMC Physiol. 2017, 17, 2. [Google Scholar] [CrossRef]
- Firth, S.M.; Baxter, R.C. Characterisation of recombinant glycosylation variants of insulin-like growth factor binding protein-3. J. Endocrinol. 1999, 160, 379–387. [Google Scholar] [CrossRef]
- James, P.L.; Stewart, C.E.H.; Rotwein, P. Insulin-like growth factor binding protein-5 modulates muscle differentiation through an insulin-like growth factor-dependent mechanism. J. Cell Biol. 1996, 133, 683–693. [Google Scholar] [CrossRef]
- Cheng, H.L.; Shy, M.; Feldman, E.L. Regulation of insulin-like growth factor-binding protein-5 expression during schwann cell differentiation. Endocrinology 1999, 140, 4478–4485. [Google Scholar] [CrossRef]
- Pera, E.M.; Wessely, O.; Li, S.Y.; De Robertis, E.M. Neural and head induction by insulin-like growth factor signals. Dev. Cell 2001, 1, 655–665. [Google Scholar] [CrossRef]
- Salih, D.A.M.; Tripathi, G.; Holding, C.; Szestak, T.A.M.; Gonzalez, M.I.; Carter, E.J.; Cobb, L.J.; Eisemann, J.E.; Pell, J.M. Insulin-like growth factor-binding protein 5 (Igfbp5) compromises survival, growth, muscle development, and fertility in mice. Proc. Natl. Acad. Sci. USA 2004, 101, 4314–4319. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.; Allard, J.B. Insulin-like growth factor binding protein-5 in physiology and disease. Front. Endocrinol. (Lausanne) 2020, 11, 100. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Bai, Y.; Hebda, L.; Zhong, X.; Liu, J.; Kao, J.; Duan, C. Calcium deficiency-induced and TRP channel-regulated IGF1R-PI3K-Akt signaling regulates abnormal epithelial cell proliferation. Cell Death Differ. 2014, 21, 568–581. [Google Scholar] [CrossRef] [PubMed]
- Kusakabe, M.; Ishikawa, A.; Ravinet, M.; Yoshida, K.; Makino, T.; Toyoda, A.; Fujiyama, A.; Kitano, J. Genetic basis for variation in salinity tolerance between stickleback ecotypes. Mol. Ecol. 2017, 26, 304–319. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Wang, Z.; Yu, D.; Xu, Y. Molecular cloning and expression profiles of an insulin-like growth factor binding protein IGFBP5 in the pearl oyster, Pinctada fucata. J. Appl. Anim. Res. 2018, 46, 1395–1402. [Google Scholar] [CrossRef]
- Estes, J.A.; Lindberg, D.R.; Wray, C. Evolution of large body size in abalones (Haliotis): Patterns and implications. Paleobiology 2005, 31, 591–606. [Google Scholar] [CrossRef]
- Suleria, H.A.R.; Masci, P.P.; Gobe, G.C.; Osborne, S.A. Therapeutic potential of abalone and status of bioactive molecules: A comprehensive review. Crit. Rev. Food Sci. Nutr. 2017, 57, 1742–1748. [Google Scholar] [CrossRef]
- Ding, H.; Kharboutli, M.; Saxena, R.; Wu, T. Insulin-like growth factor binding protein-2 as a novel biomarker for disease activity and renal pathology changes in lupus nephritis. Clin. Exp. Immunol. 2016, 184, 11–18. [Google Scholar] [CrossRef]
- Moaeen-Ud-Din, M.; Bilal, G.; Reecy, J.M. Evolution of hypothalamus-pituitary growth axis among fish, amphibian, birds and mammals. Genetika 2015, 47, 665–677. [Google Scholar] [CrossRef]
- Kamangar, B.B.; Gabillard, J.C.; Bobe, J. Insulin-like growth factor-binding protein (IGFBP)-1, -2, -3, -4, -5, and -6 and IGFBP-related protein 1 during rainbow trout postvitellogenesis and oocyte maturation: Molecular characterization, expression profiles, and hormonal regulation. Endocrinology 2006, 147, 2399–2410. [Google Scholar] [CrossRef]
- Tripathi, G.; Salih, D.A.M.; Drozd, A.C.; Cosgrove, R.A.; Cobb, L.J.; Pell, J.M. IGF-independent effects of insulin-like growth factor binding protein-5 (Igfbp5) in vivo. FASEB J. 2009, 23, 2616–2626. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zhang, Z.; Zhang, L.; Wang, S.; Zou, Z.; Wang, G.; Wang, Y. Insulin-like growth factor binding protein 7, a member of insulin-like growth factor signal pathway, involved in immune response of small abalone Haliotis diversicolor. Fish Shellfish Immunol. 2012, 33, 229–242. [Google Scholar] [CrossRef] [PubMed]
- Sharker, M.R.; Kim, S.C.; Sumi, K.R.; Sukhan, Z.P.; Sohn, Y.C.; Lee, W.K.; Kho, K.H. Characterization and expression analysis of a GnRH-like peptide in the Pacific abalone, Haliotis discus hannai. Agri Gene 2020, 15, 100099. [Google Scholar] [CrossRef]
- Sharker, M.R.; Nou, I.S.; Kho, K.H. Molecular characterization and spatiotemporal expression of prohormone convertase 2 in the Pacific abalone, Haliotis discus hannai. PLoS ONE 2020, 15, e0231353. [Google Scholar] [CrossRef] [PubMed]
- Fariselli, P.; Riccobelli, P.; Casadio, R. Role of evolutionary information in predicting the disulfide-bonding state of cysteine in proteins. Proteins Struct. Funct. Genet. 1999, 36, 340–346. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER suite: Protein structure and function prediction. Nat. Methods 2014, 12, 7–8. [Google Scholar] [CrossRef]
- Wan, Q.; Whang, I.; Choi, C.Y.; Lee, J.S.; Lee, J. Validation of housekeeping genes as internal controls for studying biomarkers of endocrine-disrupting chemicals in disk abalone by real-time PCR. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2011, 153, 259–268. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Sharker, M.R.; Sukhan, Z.P.; Kim, S.C.; Lee, W.K.; Kho, K.H. Identification, characterization, and expression analysis of a serotonin receptor involved in the reproductive process of the Pacific abalone, Haliotis discus hannai. Mol. Biol. Rep. 2020, 47, 555–567. [Google Scholar] [CrossRef] [PubMed]
- Sharker, M.R.; Sukhan, Z.P.; Kim, S.C.; Lee, W.K.; Kho, K.H. Molecular identification, characterization, and expression analysis of a gonadotropin-releasing hormone receptor (GnRH-R) in Pacific abalone, Haliotis discus hannai. Molecules 2020, 25, 2733. [Google Scholar] [CrossRef] [PubMed]
- Honegger, B.; Galic, M.; Köhler, K.; Wittwer, F.; Brogiolo, W.; Hafen, E.; Stocker, H. Imp-L2, a putative homolog of vertebrate IGF-binding protein 7, counteracts insulin signaling in Drosophila and is essential for starvation resistance. J. Biol. 2008, 7, 10. [Google Scholar] [CrossRef] [PubMed]
- Rosen, O.; Weil, S.; Manor, R.; Roth, Z.; Khalaila, I.; Sagi, A. A crayfish insulin-like-binding protein: Another piece in the androgenic gland insulin-like hormone puzzle is revealed. J. Biol. Chem. 2013, 288, 22289–22298. [Google Scholar] [CrossRef]
- Chandler, J.C.; Aizen, J.; Elizur, A.; Hollander-Cohen, L.; Battaglene, S.C.; Ventura, T. Discovery of a novel insulin-like peptide and insulin binding proteins in the Eastern rock lobster Sagmariasus verreauxi. Gen. Comp. Endocrinol. 2015, 215, 76–87. [Google Scholar] [CrossRef]
- Li, F.; Bai, H.; Xiong, Y.; Fu, H.; Jiang, S.; Jiang, F.; Jin, S.; Sun, S.; Qiao, H.; Zhang, W. Molecular characterization of insulin-like androgenic gland hormone-binding protein gene from the oriental river prawn Macrobrachium nipponense and investigation of its transcriptional relationship with the insulin-like androgenic gland hormone gene. Gen. Comp. Endocrinol. 2015, 216, 152–160. [Google Scholar] [CrossRef]
- Bach, L.A.; Headey, S.J.; Norton, R.S. IGF-binding proteins—The pieces are falling into place. Trends Endocrinol. Metab. 2005, 16, 228–234. [Google Scholar] [CrossRef]
- Forbes, B.E.; McCarthy, P.; Norton, R.S. Insulin-like growth factor binding proteins: A structural perspective. Front. Endocrinol. (Lausanne) 2012, 3, 38. [Google Scholar] [CrossRef]
- Ali, E.S.; Hua, J.; Wilson, C.H.; Tallis, G.A.; Zhou, F.H.; Rychkov, G.Y.; Barritt, G.J. The glucagon-like peptide-1 analogue exendin-4 reverses impaired intracellular Ca2+ signalling in steatotic hepatocytes. Biochim. Biophys. Acta Mol. Cell Res. 2016, 1863, 2135–2146. [Google Scholar] [CrossRef]
- Swisshelm, K.; Ryan, K.; Tsuchiya, K.; Sager, R. Enhanced expression of an insulin growth factor-like binding protein (mac25) in senescent human mammary epithelial cells and induced expression with retinoic acid. Proc. Natl. Acad. Sci. USA 1995, 92, 4472–4476. [Google Scholar] [CrossRef]
- Castellanos, M.; Jiménez-Vega, F.; Vargas-Albores, F. Single IB domain (SIBD) protein from Litopenaeus vannamei, a novel member for the IGFBP family. Comp. Biochem. Physiol. Part D Genom. Proteom. 2008, 3, 270–274. [Google Scholar] [CrossRef]
- Neumann, G.M.; Bach, L.A. The N-terminal disulfide linkages of human insulin-like growth factor- binding protein-6 (hIGFBP-6) and hIGFBP-1 are different as determined by mass spectrometry. J. Biol. Chem. 1999, 274, 14587–14594. [Google Scholar] [CrossRef] [PubMed]
- Rosenzweig, S.A. What’s new in the IGF-binding proteins? Growth Horm. IGF Res. 2004, 14, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Nagalla, S.R.; Oh, Y.; Wilson, E.; Roberts, C.T.; Rosenfeld, R.G. Identification of a family of low-affinity insulin-like growth factor binding proteins (IGFBPs): Characterization of connective tissue growth factor as a member of the IGFBP superfamily. Proc. Natl. Acad. Sci. USA 1997, 94, 12981–12986. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Shi, Y.; He, M. Molecular identification of an insulin growth factor binding protein (IGFBP) and its potential role in an insulin-like peptide system of the pearl oyster, Pinctada fucata. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2017, 214, 27–35. [Google Scholar] [CrossRef]
- Grellier, P.; Berrebi, D.; Peuchmaur, M.; Babajko, S. The IGF system in neuroblastoma xenografts: Focus on IGF-binding protein-6. J. Endocrinol. 2002, 172, 467–476. [Google Scholar] [CrossRef]
- Landau, D.; Chin, E.; Bondy, C.; Domene, H.; Roberts, C.T.; Gronbaek, H.; Flyvbjerg, A.; Le Roith, D. Expression of insulin-like growth factor binding proteins in the rat kidney: Effects of long-term diabetes. Endocrinology 1995, 136, 1835–1842. [Google Scholar] [CrossRef]
- Zhou, J.; Li, W.; Kamei, H.; Duan, C. Duplication of the IGFBP-2 gene in teleost fish: Protein structure and functionality conservation and gene expression divergence. PLoS ONE 2008, 3, e3926. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharker, M.R.; Kim, S.C.; Hossen, S.; Kho, K.H. Characterization of Insulin-Like Growth Factor Binding Protein-5 (IGFBP-5) Gene and Its Potential Roles in Ontogenesis in the Pacific Abalone, Haliotis discus hannai. Biology 2020, 9, 216. https://doi.org/10.3390/biology9080216
Sharker MR, Kim SC, Hossen S, Kho KH. Characterization of Insulin-Like Growth Factor Binding Protein-5 (IGFBP-5) Gene and Its Potential Roles in Ontogenesis in the Pacific Abalone, Haliotis discus hannai. Biology. 2020; 9(8):216. https://doi.org/10.3390/biology9080216
Chicago/Turabian StyleSharker, Md. Rajib, Soo Cheol Kim, Shaharior Hossen, and Kang Hee Kho. 2020. "Characterization of Insulin-Like Growth Factor Binding Protein-5 (IGFBP-5) Gene and Its Potential Roles in Ontogenesis in the Pacific Abalone, Haliotis discus hannai" Biology 9, no. 8: 216. https://doi.org/10.3390/biology9080216
APA StyleSharker, M. R., Kim, S. C., Hossen, S., & Kho, K. H. (2020). Characterization of Insulin-Like Growth Factor Binding Protein-5 (IGFBP-5) Gene and Its Potential Roles in Ontogenesis in the Pacific Abalone, Haliotis discus hannai. Biology, 9(8), 216. https://doi.org/10.3390/biology9080216





